Disatnik et al. identify mitochondrial DNA levels, 8-OHdG, and inflammation factors as potential peripheral biomarkers to follow progression and treatment response of Huntington’s disease.
Abstract
Huntington’s disease (HD) is a rare genetic disease caused by expanded polyglutamine repeats in the huntingtin protein resulting in selective neuronal loss. Although genetic testing readily identifies those who will be affected, current pharmacological treatments do not prevent or slow down disease progression. A major challenge is the slow clinical progression and the inability to biopsy the affected tissue, the brain, making it difficult to design short and effective proof of concept clinical trials to assess treatment benefit. In this study, we focus on identifying peripheral biomarkers that correlate with the progression of the disease and treatment benefit. We recently developed an inhibitor of pathological mitochondrial fragmentation, P110, to inhibit neurotoxicity in HD. Changes in levels of mitochondrial DNA (mtDNA) and inflammation markers in plasma, a product of DNA oxidation in urine, mutant huntingtin aggregates, and 4-hydroxynonenal adducts in muscle and skin tissues were all noted in HD R6/2 mice relative to wild-type mice. Importantly, P110 treatment effectively reduced the levels of these biomarkers. Finally, abnormal levels of mtDNA were also found in plasma of HD patients relative to control subjects. Therefore, we identified several potential peripheral biomarkers as candidates to assess HD progression and the benefit of intervention for future clinical trials.
Introduction
Huntington’s disease (HD) is a fatal autosomal-dominant neurodegenerative disease caused by an expanded trinucleotide CAG repeat in the gene encoding the huntingtin protein (MacDonald et al., 1993). HD is a progressive disease that affects middle-age carriers, and the severity of the disease correlates with the length of the CAG repeat (Lee et al., 2015). Patients affected by HD display a loss of neurons predominantly in the striatum and cortex that is progressively accompanied by a loss of voluntary and involuntary movements as well as psychiatric and cognitive disturbances. Patients usually die 10–15 yr after the onset of the disease because of immobility-induced complications. Currently, there is no cure for the disease, and no treatment effectively slows down the disease progression.
Since the discovery of the genetic basis for the disease, mutant huntingtin (mtHtt), in 1993 (MacDonald et al., 1993), there has been considerable effort toward developing therapeutic strategies for HD, and several compounds have shown beneficial effects in various HD cell and transgenic mouse models (Li et al., 2005; Ray and Shoulson, 2011; Guo et al., 2013). However, human trials in HD are time consuming because of the slow progression of the disease, its insidious onset, and patient-to-patient variability (MacDonald et al., 1993; Weir et al., 2011). There is also a need to include a large cohort of patients because many of the clinical assessments are quite subjective (e.g., psychiatric tests) and an inability to biopsy the affected tissue, neurons in the brain. Therefore, identification of peripheral biomarkers that correlate with HD progression and treatment intervention would greatly help assess the efficacy of experimental therapies in human clinical trials.
The neurological symptoms of HD are caused by the aggregation of mtHtt protein in neurons that causes, among other pathologies, mitochondrial dysfunction (Shao and Diamond, 2007; Costa and Scorrano, 2012). This, in turn, leads to loss of ATP and increase in oxidative stress (Costa and Scorrano, 2012; Guo et al., 2013; Hwang et al., 2015). Evidence from studies in human HD subjects and experimental HD mouse models suggests that mitochondrial dysfunction precedes neuropathology and clinical symptoms (Feigin et al., 2001; Ciarmiello et al., 2006; Song et al., 2011), indicating that mitochondrial impairment is an early event in the cascade of events leading to HD pathology. Notably, targeting impaired mitochondrial dysfunction has been shown in some cases to be a beneficial strategy to delay HD onset and to slow disease progression after its onset (Duan et al., 2014).
We recently reported that inhibition of mitochondrial dynamics impairment by a novel Drp1/Fis1 peptide inhibitor, P110 (Qi et al., 2013), rescued mtHtt-induced mitochondrial injury, corrected defects in mitochondrial function, and reduced neuronal cell death both in HD patient–derived neuronal cultures and in HD transgenic mouse brains (Guo et al., 2013). These findings provided further evidence for a causal role for mitochondrial damage in the pathogenesis of HD and demonstrated that blocking mitochondrial injury can reduce neuronal degeneration in HD models. Here, we used samples from R6/2 mice, an HD model, to identify biomarkers that correlate with HD disease progression and treatment benefit with P110 and included a pilot human study for one of these biomarkers, using plasma and spinal fluid samples from healthy subjects and HD patients.
Results
Alteration of mitochondrial DNA (mtDNA) in the brain and plasma of HD mice
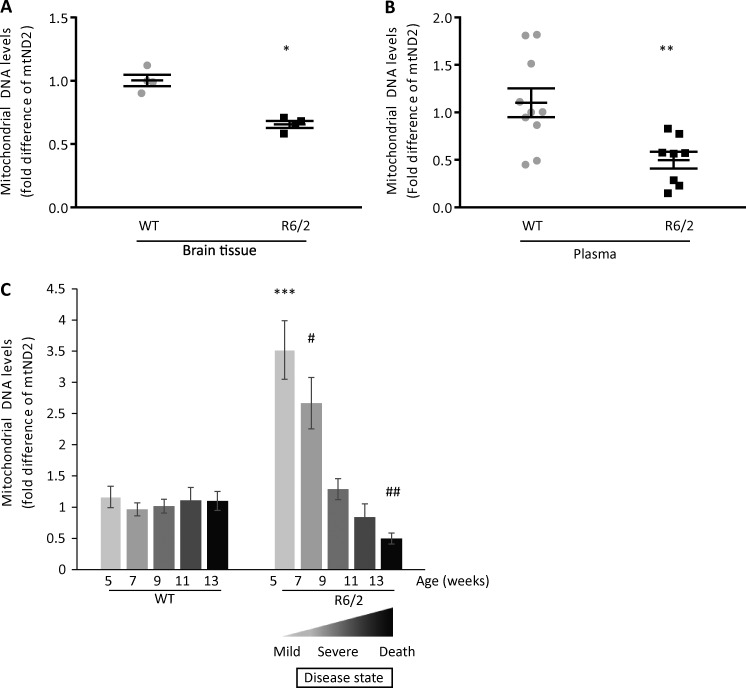
Because HD is associated with impaired mitochondrial integrity and excessive mitochondrial fission, we first evaluated the extent of mitochondrial loss in the brains of 13-wk-old R6/2 mice, an age that we previously found to exhibit severe HD-related symptoms (Guo et al., 2013). It was previously found that mitochondrial number in the brain decline by >50% in severe HD patients (Kim et al., 2010). As a surrogate measure for mitochondrial number in the brain, we measured the levels of the transcript of the mitochondrial gene, mtND2 (mitochondria-encoded NADH dehydrogenase 2; a subunit of complex 1 located at the inner mitochondrial membrane), using DNA reverse transcribed from RNA isolated from brain tissue of WT and R6/2 mice. Using real-time PCR, we found that the brains of 13-wk-old R6/2 mice had almost half the amount of mtND2 as compared with the brains of WT mice (Fig. 1 A).
Figure 1.
Analysis of mtDNA content in brain and plasma. (A) DNA synthetized from mouse brain tissue was used in real-time PCR with mtND2 primers targeting mouse mtDNA (n = 4 mice per group). (B) mtND2 DNA content in plasma samples from mouse blood collected at the age of 13 wk were assayed by real-time PCR (n = 10 WT and n = 8 R6/2 Tg mice). Housekeeping nuclear gene, GAPDH, was used for normalization. (C) Change in mtND2 levels over time in mice. Mouse plasma samples from WT and R6/2 mice were collected every 2 wk, and mtND2 levels were determined using real-time PCR. mtND2 levels decreased over time in R6/2 mice as compared with WT mice (n = 10 WT; n = 10 R6/2 [5–11-wk-old mice]; n = 9 [13-wk-old mice]). The data are presented as mean ± SEM of 2−ΔΔCT. *, P = 0.0006; **, P = 0.0055; ***, P = 0.00017 versus WT 5 wk; #, P = 0.00085 versus WT 7 wk; ##, P = 0.004 versus WT 13 wk (unpaired Student’s t test and one-way ANOVA).
Because mtDNA is quite resistant to degradation, we expected the content of mtDNA in the plasma to increase as dead neurons released mtDNA into the circulation. Therefore, we determined the levels of mtDNA in the plasma of 13-wk-old mice (the end stage of the disease; Mangiarini et al., 1996) using the mtND2 transcript in Fig. 1 A. GAPDH, a nuclear gene, was used as a control (Xia et al., 2009a,b; Budnik et al., 2013). Surprisingly, mtND2 levels in R6/2 plasma were reduced by 54% as compared with plasma of WT mice (Fig. 1 B).
A candidate biomarker should show a measurable response to the progression and severity of the disease. Thus, to evaluate the levels of mtND2 in mouse plasma during the course of 13 wk, we collected plasma every 2 wk and analyzed by real-time PCR mtND2 levels to determine whether they correlated with the progression of the disease. The results are shown as the mean of mtND2 levels (2−ΔΔCT) obtained from 10 mice per group at each age (Fig. 1 C). We observed that mtND2 levels in plasma of WT mice remained constant over time from 5 to 13 wk of age. However, mtND2 levels in R6/2 mice were 3.5-fold higher at 5 wk (before or early after symptoms onset; Mangiarini et al., 1996) and 2.5-fold higher at 7 wk as compared with WT mice, and these levels decreased over time to half of the WT levels by 13 wk, the end stage of the disease in this model (Mangiarini et al., 1996), as shown also in a separate cohort of mice in Fig. 1 B.
To determine whether the high levels of mtND2 in the presymptomatic mice occur in another HD mouse model, we analyzed mtND2 levels in a less severe mouse model of HD, YAC128 (Fig. S2 A). This mouse model exhibits a much slower disease progression as compared with R6/2 mice, with end stage and death occurring at about >1.5 yr (Slow et al., 2003). To match the study in 5-wk-old R6/2 mice, we used 6-mo-old YAC128 mice, as at that age, the YAC128 mice also exhibit no behavioral impairment. As expected from our data in R6/2 mice (Fig. 1 C), the levels of mtDNA in plasma of 6-mo-old YAC128 mice were 2.5-fold higher than WT mice of the same age (nine mice/group).
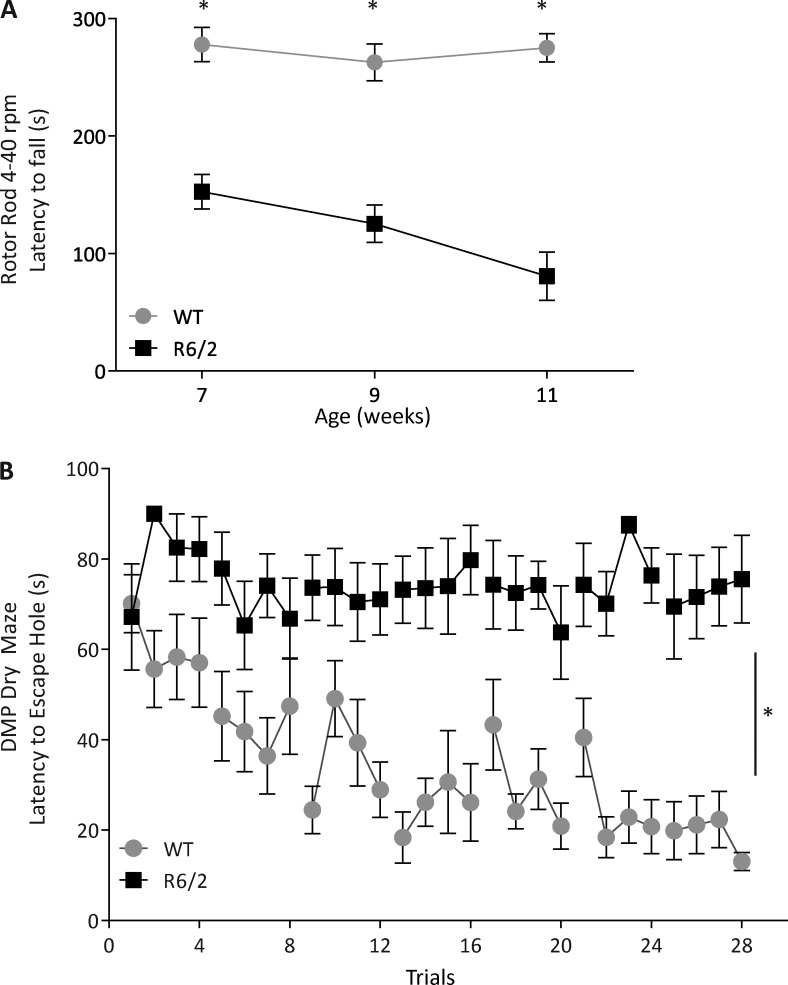
We previously reported that 13-wk-old R6/2 HD mice exhibited a severe behavioral deficit accompanied by mitochondrial loss (Fig. 1, A and B; Guo et al., 2013). Behavioral studies measured by several tests of 7-wk-old mice demonstrated an overall behavioral deficits of R6/2 mice as compared with WT mice (Fig. 2, A and B; and Fig. S1, A–D). The behavioral deficits, shown by a decrease in mobility (Fig. 2 A), became more severe with age (Note that behavioral studies in younger mice that have not been acclimated to an animal facility are less reliable. However, on arrival, at the age of 5 wk, the HD mice appear relatively unimpaired; Guo et al., 2013). 11-wk-old R6/2 mice demonstrated a large impairment in memory and learning skills, as shown by delay match-to-place dry maze (DMP-DM; Fig. 2 B). No behavioral deficits were noted in 5-wk-old R6/2 mice (Guo et al., 2013). Yet, 5-wk-old R6/2 mice had high levels of mtDNA in the plasma relative to WT controls (Fig. 1 C). These results suggest that evidence for maximal neuronal loss (as measured by decrease of mtDNA in the brain and increased mtDNA in the plasma) occurred earlier than motor and behavioral impairments. Collectively, these results show that the highest increase in mtDNA in the plasma preceded maximal behavioral deficits of R6/2 mice, consistent with the evidence that mitochondrial damage occurred at the early stage of HD (Kim et al., 2010). Therefore, assessing mtDNA in the plasma may be a useful marker to indicate early HD-associated pathology.
Figure 2.
Behavioral phenotype of R6/2 mice compared with WT. (A) R6/2 mice have significantly decreased latency to fall during the accelerating rotor rod test (4–40 rpm) starting at 7 wk of age and progressively exhibit a shorter latency as they age. WT mice maintained their latency to falling as they aged. n = 10 WT and n = 9 R6/2. *, P = 0.0001 WT versus R6/2 at respective week; two-way ANOVA, repeated measure with Bonferroni posthoc analysis. (B) 11-wk-old WT mice were able to find the location of the new escape box during the DMP-DM test, and their latency to finding the escape box decreased as more training days were conducted. The latency to find the escape box for 11-wk-old R6/2 mice remained stagnant during the four trials of training per day and across multiple days of testing. n = 10 WT and n = 9 R6/2. *, P = 0.0001 WT versus R6/2 at each trial; two-way ANOVA, repeated measure. Error bars represent mean ± SEM.
P110 treatment normalizes the amount of mtDNA in the plasma of HD mice
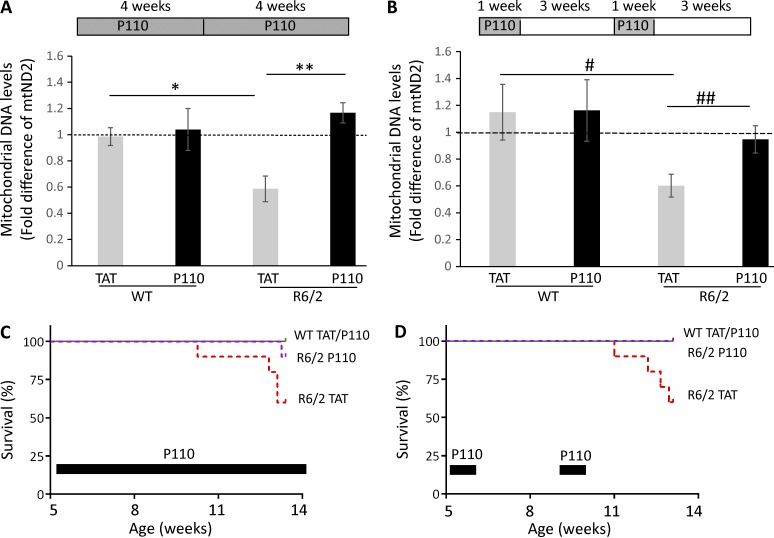
We previously described the beneficial effect of P110 treatment on HD mice (Guo et al., 2013). P110 is a heptapeptide conjugated to TAT47–57 (TAT, for peptide delivery) that inhibits the interaction between Drp1 and one of its adaptor proteins on the mitochondria, Fis1 (Guo et al., 2013). We showed that P110 inhibits excessive mitochondrial fission in several models of neurodegenerative disease as well as in a rat heart model of ischemia/reperfusion injury, without affecting basal (physiological) fission (Disatnik et al., 2013; Guo et al., 2013; Qi et al., 2013). To determine whether mtDNA levels in the plasma correlated with the benefit induced by P110, R6/2 mice were treated with P110 inhibitor peptide or TAT (vehicle control; each at 3 mg/Kg/d), delivered by a subcutaneous osmotic pump for 8 wk, as we described previously (Guo et al., 2013). As in separate cohorts (Fig. 1, B and C), 13-wk-old R6/2 mice exhibited a decrease of 50% in mtND2 in the plasma, and P110 treatment increased the levels of mtND2 levels by twofold, back to those of WT levels (Fig. 3 A).
Figure 3.
Beneficial effect of P110 treatment on R6/2 mice as measured by normalizing mtND2 levels in plasma. (A) WT and R6/2 mice were treated with TAT or P110 for 8 wk at 3 mg/kg/d. mtND2 levels were analyzed by real-time PCR. n = 9/group WT TAT or P110; n = 8 R6/2 TAT; n = 7 R6/2 P110. *, P = 0.004 versus WT TAT; **, P = 0.0005 versus R6/2 TAT. (B) WT and R6/2 mice were treated intermittently with TAT or P110 for 1 wk followed by 3 wk no treatment before another week of treatment was administered. Mice plasma samples were collected 3 wk after the end of the treatment, at the age of 13 wk. n = 9/group WT TAT and P110; n = 11 R6/2 TAT; n = 15 R6/2 P110. #, P = 0.017 versus WT TAT; ##, P = 0.021 versus R6/2 TAT (unpaired Student’s t test and one-way ANOVA). (A and B) Data are presented as mean ± SEM of 2−ΔΔCT. (C) Survival curve of WT and R6/2 mice treated with P110. The mice were treated with TAT and P110 for 8 wk. 4 mice out of 10 R6/2 TAT-treated mice and 2 mice out of 10 R6/2 P110-treated mice died at the age of 13 wk. The results are shown as a log-rank (Mantel-Cox) test; χ2 = 10.9; P = 0.0123. (D) The effect of intermittent treatment of P110 on WT and R6/2 mice over 8 wk. 4 out of 10 R6/2 mice treated with TAT died whereas 0 out of 10 R6/2 mice treated with P110 died at the age of 13 wk; no death occurred in the 10 TAT- and 10 P110-treated WT mice. The results are shown as a log-rank (Mantel-Cox) test; χ2 = 9.461; P = 0.0238.
We next determined the effect of intermittent P110 treatment consisting of 1 wk sustained treatment with P110 (3 mg/Kg/d) followed by no treatment for 3 wk, repeated twice for a total duration of 8 wk, as shown in Fig. 3 B. We found that even an intermittent P110 treatment was sufficient to increase the levels of mtND2 by twofold, close to WT levels (Fig. 3 B), similar to the results shown in Fig. 3 A using a separate cohort of mice. As previously reported (Guo et al., 2013), we found that P110 administered for 8 wk was beneficial and increased the survival of the R6/2 mice (Fig. 3 C). We also found that survival of R6/2 mice subjected to intermittent P110 treatment was also significantly increased (P = 0.0238; Fig. 3 D), indicating that intermittent treatment might be sufficient to correct mitochondrial function, thus protecting from neuronal cell loss. Finally, we observed that a 1-wk P110 treatment in 8-wk-old R6/2 mice was also sufficient to increase the levels of mtND2 in the plasma by twofold relative to TAT-treated R6/2 mice (Fig. S2 B), suggesting that plasma mtDNA correlates with treatment.
P110 treatment reduces the levels of oxidative DNA damage indicator in HD mice
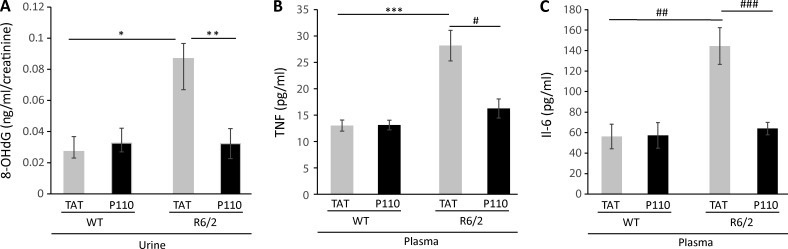
There are conflicting studies regarding the use of oxidative stress markers in plasma and urine, such as 8-hydroxy-deoxy-guanosine (8-OHdG). 8-OHdG is a product of guanine oxidation by oxidative stress (Shigenaga et al., 1989) that is found in the urine as a product of DNA excision repair. Urine and plasma from R6/2 mice have high levels of 8-OHdG (Bogdanov et al., 2001). We therefore evaluated the use of 8-OHdG as a biomarker for treatment benefit in urine of WT and R6/2 mice after 8 wk of P110 or TAT vehicle treatments (Note that because the mice were fragile, continual collection of urine as the disease progressed was not possible). The DNA damage product, 8-OHdG, measured by ELISA assay, was normalized to the levels of creatinine in each mouse urine sample to adjust for differences in water intake and urine volume. We found that 8-OHdG levels were threefold higher in 13-wk-old R6/2 mice relative to WT mice of the same age and that an 8-wk sustained P110 treatment of the R6/2 mice decreased the levels of 8-OHdG to WT levels (Fig. 4 A).
Figure 4.
DNA damage measurement in urine and inflammation markers in plasma of WT and R6/2 mice. (A) Analysis of 8-OHdG levels in urine samples of 13-wk-old WT and R6/2 mice that were treated with TAT or P110 for 8 wk. The levels of 8-OHdG were measured by ELISA. Creatinine levels in the same urine samples were determined for normalization. Increased levels of 8-OHdG in the urine of R6/2 mice were normalized to WT levels after P110 treatment. Data are presented as mean ± SEM (n = 9/group WT TAT and P110; n = 6 R6/2 TAT; n = 7 R6/2 P110). (B and C) TNF (B) and IL-6 (C) levels measured by ELISA in mice plasma of respective mice treated with TAT and P110 for 8 wk. Plasma samples were collected at 13 wk of age. TNFα: n = 7/group WT TAT and P110; n = 9 R6/2 TAT; n = 10 R6/2 P110. IL-6: n = 4/group WT TAT and P110; n = 9 R6/2 TAT; n = 10 R6/2 P110. Data are presented as mean ± SEM. *, P = 0.03 R6/2 TAT versus WT TAT; **, P = 0.043 R6/2 P110 versus R6/2 TAT (unpaired Student’s t test); ***, P = 0.0006 R6/2 TAT versus WT TAT; #, P = 0.0023 R6/2 P110 versus R6/2 TAT; ##, P = 0.01 R6/2 TAT versus WT TAT; ###, P = 0.0003 R6/2 P110 versus R6/2 TAT (unpaired Student’s t test and one-way ANOVA).
P110 treatment decreases the levels of inflammatory markers in plasma of HD mice
Activated monocytes are observed in presymptomatic HD patients (Crotti and Glass, 2015), and inflammation triggered by the presence of mtHtt was reported in mouse models of HD and in HD patients (Hsiao et al., 2013; Crotti and Glass, 2015; Politis et al., 2015; Träger et al., 2015). Inflammation is due, in part, to activation of microglia and recruitment of astrocytes associated with mtHtt, which leads to enhanced secretion of cytokines and chemokines by microglia (Crotti et al., 2014). Therefore, using ELISA, we measured the levels of two inflammatory cytokines: TNF and IL-6. The levels of both these cytokines were elevated in the plasma of 13-wk-old R6/2 mice by more than twofold relative to WT mice, and P110 treatment of R6/2 mice for 8 wk reduced their levels back to the levels of WT mice (Fig. 4, B and C).
P110 treatment reduces the levels of mtHtt aggregation and 4-HNE adducts in peripheral tissues of HD mice
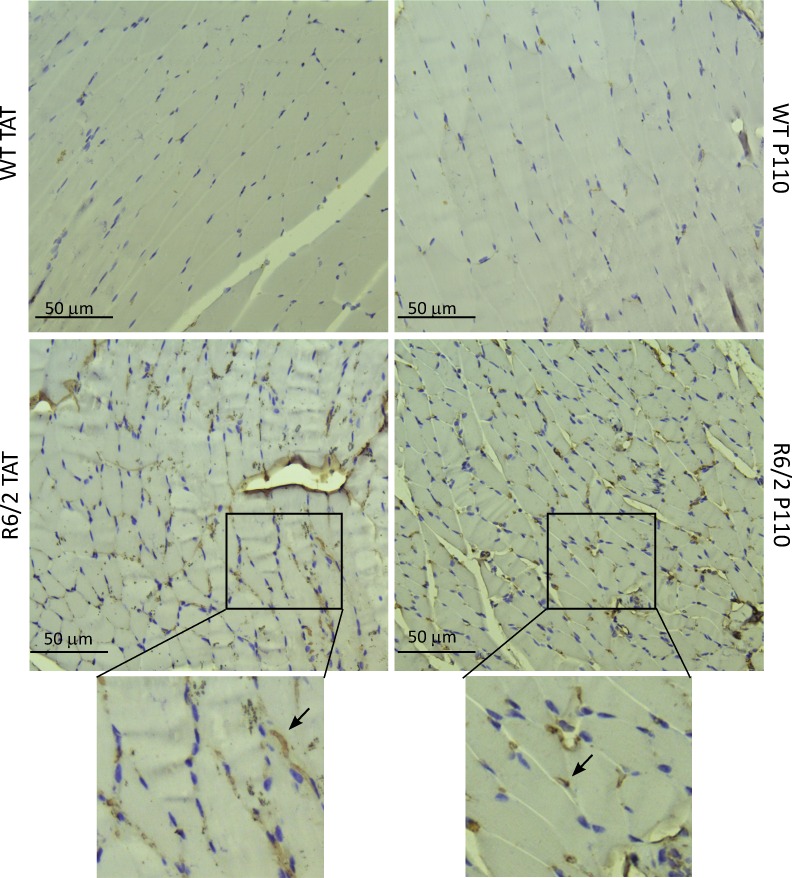
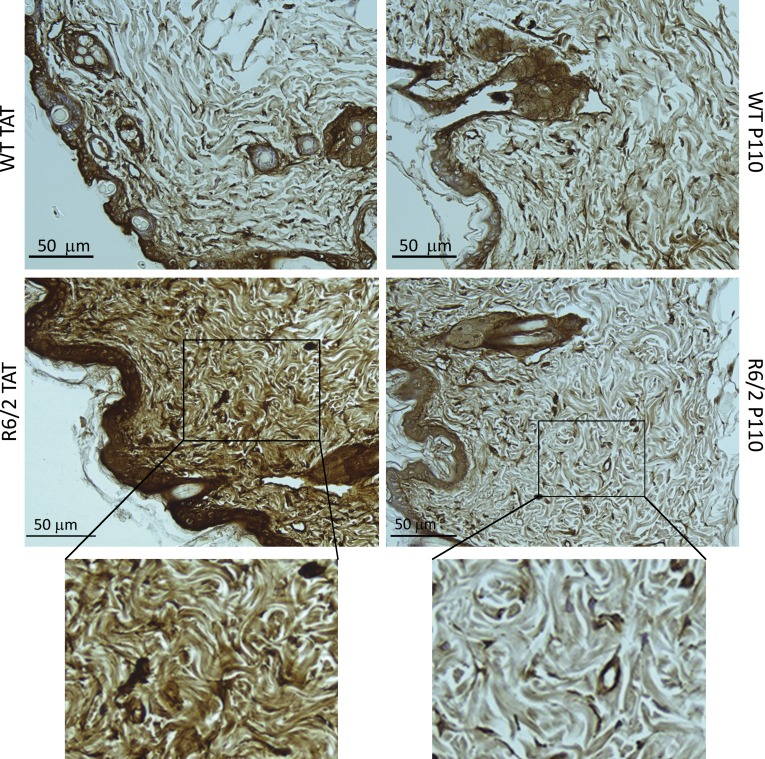
Aggregates of mtHtt were previously reported in the brains of human HD patients (DiFiglia et al., 1997) and R6/2 mice when measured at the age of 13 wk (Guo et al., 2013). However, non–central nervous system (non-CNS) tissues of the HD mice model also have mtHtt aggregates as well as evidence of oxidative stress (Sathasivam et al., 1999). Therefore, we determined the presence of mtHtt aggregates and 4-HNE adducts on proteins (an aldehydic product of lipid oxidation; Hardas et al., 2013) in skeletal muscle and skin of 13-wk-old R6/2 mice. We found an increase in mtHtt aggregates at the periphery of the muscle fibers (Fig. 5) and skin sections (Fig. 6) in TAT-treated R6/2 mice; an 8-wk P110 treatment of R6/2 mice correlated with decreased levels of these aggregates by 40% in the skin and 60% in the muscle tissue (Table 1). We also found skeletal muscle from R6/2 compared with WT mice to have a high number of nonmuscle nuclei (Fig. 5) that might reflect infiltration of inflammatory cells into this tissue. Data by Sathasivam et al. (1999) also show an HD muscle section with a high number of nuclei; however, the authors did not comment on that (Sathasivam et al., 1999). We also observed twofold-higher levels of 4-HNE immunoreactivity in R6/2 leg muscle as well as 36% increase in skin sections stained with 4-HNE, as compared with WT levels (Fig. S3, A and B; and Table 1). Those results indicate that non-CNS peripheral tissue can be used to follow the progression of HD.
Figure 5.
P110 reduces mtHtt aggregation in R6/2 skeletal muscle. Skeletal muscle sections were stained with anti-mtHtt (EM-48) antibody and hematoxylin (blue nuclei). More aggregates of mtHtt were found (arrows) in TAT-treated than P110-treated R6/2 mice. (Bottom) Magnification of boxed areas.
Figure 6.
P110 reduces mtHtt aggregation in R6/2 skin. Skin sections were analyzed for the presence of mtHtt in WT and R6/2 mice. P110 reduced the level of mtHtt aggregates. (Bottom) Magnification of boxed areas.
Table 1. Biomarkers in peripheral tissues.
| mtHtt staining | 4-HNE staining | |
|---|---|---|
| Muscle | ||
| WT TAT | 80.8 ± 2.0 | 45.1 ± 2.8 |
| WT P110 | 94.8 ± 0.5 | |
| R6/2 TAT | 143.3 ± 0.4a | 91.8 ± 2.5b |
| R6/2 P110 | 57.1 ± 0.6c | |
| Skin | ||
| WT TAT | 96.4 ± 4.2 | 95.3 ± 2.8 |
| WT P110 | 89.4 ± 3.3 | |
| R6/2 TAT | 141 ± 5.0d | 148.6 ± 7.0e |
| R6/2 P110 | 85.7 ± 3.5f |
Quantification of images of mtHtt and 4-HNE staining in muscle and skin. Images of the respective staining were obtained from three mice/group, and 16 areas of each section were analyzed. 58 areas of each section were analyzed for mtHtt staining in muscle. Data are presented as mean ± SEM.
P = 0.038 (WT TAT vs. R6/2 TAT)
P = 0.0026 (WT TAT vs. R6/2 TAT)
P = 0.027 (R6/2 TAT vs. R6/2 P110)
P = 0.019 (WT TAT vs. R6/2 TAT)
P = 0.018 (WT TAT vs. R6/2 TAT)
P = 0.015 (R6/2 TAT vs. R6/2 P110)
Alteration of mtDNA content in biofluids of HD patients
To begin determining whether the biomarkers identified in R6/2 mice can be applied in human studies, we obtained three each of human cerebrospinal fluid (CSF) samples collected from control or HD patients, ages 53–69 yr old; both males and females were included (VA Greater Los Angeles Healthcare Center). Two of the control subjects were reported to have a chronic obstructive pulmonary disease. These CSF samples, the only samples that were available to us, were used in a pilot study; as in mouse plasma, using real-time PCR, we measured the level of mtND2 levels in these samples. The differences in mtND2 levels were not significantly different between control and HD patients, probably because of the low number of samples, the wide range of age of the subjects, and variations in disease onset. However, there was a shift in the correlation line to the right for the three HD patient samples (Fig. S4 A). There was also a wider range of mtDNA levels in HD CSF samples as compared with non-HD patients (Fig. S4 B), but the significance of these differences could be determined only in a study that includes a much larger number of samples.
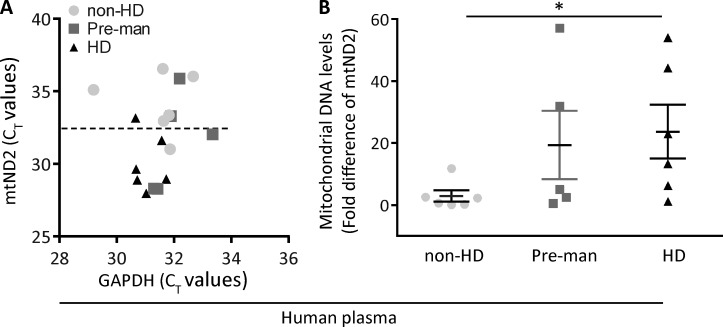
We then determined the levels of mtDNA in the plasma of HD patients and control subjects. However, even in this small sample group, we found a correlation between the severity of the disease and the levels of mtDNA (mtND2) measured in plasma of HD patients (Fig. 7, A and B); the increase shown in mtND2 in presymptomatic and symptomatic HD patients compared with control subjects was significant (P = 0.0415) and might correlate with the results obtained in R6/2 mice at the ages of 5–7 wk.
Figure 7.
mtDNA in HD patients and control subjects. (A) Distribution of mtND2 levels in plasma among the groups of human plasma were analyzed by real-time PCR. The scatter plot illustrates the CT values of GAPDH (x axis) against CT values of mtND2 (y axis) in human plasma of non-HD, premanifest (Pre-man), and HD subjects. CT values of mtND2 of HD plasma were lower and clustered below the 50% line compared with premanifest HD and non-HD groups. (B) mtND2 levels were significantly higher in HD plasma versus non-HD plasma (n = 6/non-HD and HD; n = 5/premanifest). Data are presented as mean ± SEM of 2−ΔΔCT. *, P = 0.0415 HD versus non-HD (one-way ANOVA).
Discussion
The challenge in conducting clinical trials using experimental therapeutics for HD patients is not the diagnosis of these patients; HD patients can unequivocally be identified via genetic testing for this dominant trait. The challenge is how a response to experimental treatment can be assessed early in the trial, considering that the main affected tissue responsible for the pathology is the brain. Furthermore, changes in affected individuals must occur from the time of conception, yet neurodegeneration symptoms are not apparent for >40 or 50 yr. Therefore, although ideally, therapeutic interventions should begin in presymptomatic subjects, it is prohibitively expensive to await several decades to assess the benefit of that intervention.
Here, we began exploring the possibility that peripheral biomarkers to assist in clinical trials in HD patients can be identified. We focused on biomarkers related to (a) mitochondrial and cell integrity (measuring mtDNA in the plasma; Chiu et al., 2003; Xia et al., 2009b; Nakahira et al., 2013), (b) mtHtt aggregation in the peripheral tissue (Sathasivam et al., 1999), and (c) evidence of increased oxidative stress, as measured by increased aldehydic load in human and mice brain tissues (levels of 4-HNE adducts; Lee et al., 2011) and DNA damage (as measured by the presence of a product of DNA repair in the urine, 8-OHdG; Browne et al., 1997). We found that the levels of all these parameters differed between WT mice and R6/2 HD mice. Importantly, the levels of these parameters in R6/2 mice were normalized by treatment with P110, a therapeutic intervention that reduces the symptoms and pathology in these animals (Guo et al., 2013); levels of these parameters in treated HD mice were brought close to WT levels. Therefore, all of these biomarkers appear to correlate with the improvement seen by therapeutic intervention in this animal model.
When focusing on the presence of mtDNA in plasma, we also noted that both sustained and intermittent treatments (1 wk on and 3 wk off, twice) were beneficial (Fig. 3, A and B), and a small study of 1-wk treatment at the onset of the disease (Fig. S2 B) also suggests that mtDNA in the plasma of these HD mice may correlate even with a short therapeutic intervention. If confirmed in human samples, mtDNA may be a useful biomarker to assist in determining the efficacy of a treatment in humans.
Mitochondrial dysfunction in HD is well documented (Song et al., 2011; Costa and Scorrano, 2012; Guo et al., 2013) as a main contributor to neurodegeneration and is associated with the accumulation of mtHtt protein at the mitochondria and in the nucleus (Oliveira, 2010; Costa and Scorrano, 2012). Progressive loss of striatal and cortical neurons mediates the cognitive and motor impairments in HD patients and in R6/2 mice (Ross and Tabrizi, 2011; Guo et al., 2013). A study in R6/2 mice showed a decrease of brain weight at 4 wk, thus preceding body weight loss and motor deficits (Sathasivam et al., 1999). Studies in both HD patients and HD transgenic mice also revealed that deficits in energy metabolism attributable to toxin-induced mitochondrial dysfunction play a key role in HD pathogenesis (Chen, 2011). Moreover, clinical evidence shows that metabolic impairment precedes neuropathology and clinical symptoms in HD patients (Koroshetz et al., 1997; Feigin et al., 2001; Ciarmiello et al., 2006; Powers et al., 2007), indicating that mitochondrial dysfunction and metabolic deficit are early events in HD. Together, our findings demonstrated that mitochondrial dysfunction and damage are associated with HD pathology, and they precede motor dysfunction in animal models and in patients.
Quantification of nuclear and mtDNA raised great interest as a noninvasive diagnostic for patients after trauma (Lam et al., 2004; Nakahira et al., 2013) and for diseases such as cancer and neurodegenerative diseases (Kohler et al., 2009; Xia et al., 2009a; Podlesniy et al., 2013). Our pilot study in plasma and CSF of HD patients, although greatly limited in its size, is encouraging, showing statistically significant higher mtDNA plasma levels in plasma of HD patients as compared with control subjects (Fig. 7, A and B). Because clinical information on these patients is not available to us, the correlation with changes in this biomarker and the severity of the disease (as we found in R6/2 mice; Fig. 1) cannot be made. Before evaluation of the use of any of this or any other potential biomarkers identified here as a surrogate for disease progression can be made, it is imperative that studies of more patients and, importantly, following the same patient over the progression of the disease and correlation with the severity of the symptoms be performed. Furthermore, according to a study by Chiu et al. (2003), collection of plasma and extraction of DNA protocols can affect the quantification of mtDNA. Thus, a detailed protocol for sample collection and processing needs to be established before testing mtDNA in patient’s plasma as a potential HD biomarker for therapeutic benefit.
Why do the plasma levels of mtDNA increase in HD mice and of patients with HD? The presence of mtDNA in the plasma may reflect lysis of neuronal cells (or other cells) and the release of their content into the plasma. mtDNA may also reflect active exosome-mediated release of damaged mitochondria (Shutt and McBride, 2013). The initial rise in levels of mtDNA and the subsequent decrease over the course of the disease (Fig. 1 C) probably reflect initially cell damage and subsequently decline in cell and/or mitochondrial number (Chen and Chan, 2009; Podlesniy et al., 2013). Future studies may directly address this question. Nevertheless, the finding that HD mice show a dynamic change in this biomarker that responds to P110 treatment and correlates with therapeutic effect and the finding that the same biomarker is abnormal in humans with HD is encouraging.
As discussed in this paper, the damage to mitochondria shown in HD patients and HD mice models causes oxidative damage to the DNA and can be measured by the presence of 8-OHdG, a principle marker of hydroxyl radical damage to DNA, in urine and plasma (Shigenaga et al., 1989). However, there are conflicting studies on the use of 8-OHdG as an oxidative stress marker: urine and plasma from R6/2 mice were found to have high levels of 8-OHdG using high-performance liquid chromatography with an electrochemical detection analytic technique in a study (Bogdanov et al., 1999, 2001). In a careful study on cohorts of presymptomatic and symptomatic HD patients (32 each), Borowsky et al. (2013) reported that plasma 8-OHdG was not found to be a biomarker of disease progression when using either liquid chromatography–mass spectrometry assay or liquid chromatography–electrochemical array assay. In contrast, our data using the ELISA method on mouse urine samples are consistent with the studies by Bogdanov et al. (2001) and Stack et al. (2007) using urine of R6/2 and Q82 mice, respectively, showing a 2.5-fold increase in 8-OHdG in the urine of 13-wk-old R6/2 HD mice relative to age-matched WT mice; we also show that 8-OHdG levels were lowered to WT mouse levels in R6/2 mice treated with P110 for 8 wk (Fig. 4 A). Our data are also consistent with five of six other patient sample analyses (Browne et al., 1997; Polidori et al., 1999; Hersch et al., 2006; Chen et al., 2007; Long et al., 2012; Ciancarelli et al., 2014). Therefore, the presence of 8-OHdG in the urine should also be evaluated as a potential biomarker for a trial of therapeutic intervention, especially when following changes in the same patient during the progression of the disease and following treatment.
DNA oxidation and mitochondria released into circulating fluids provoke inflammation in HD. In addition, excessive mitochondrial fragmentation in microglial cells leads to a proinflammatory state in the brain vasculature (Ferger et al., 2010), and activated microglial cells (Park et al., 2013) and several HD mouse models as well as HD patients have increased plasma cytokine levels (Björkqvist et al., 2008). Our findings that the inflammation markers TNF and IL-6 increased in R6/2 mice and that these were normal in R6/2 mice treated with P110 (Fig. 4, B and C) suggest that these could also be tested as potential biomarkers for a trial of therapeutic intervention in HD patients.
Although much of the HD research focuses on pathologies associated with the CNS, it is clear that peripheral tissues are also affected in the disease. Andre et al. (2014) evaluated in detail the advantages and disadvantages of several biomarkers used to follow HD patients. The authors listed biomarkers that may be considered today in HD. However, those biomarkers are either expensive (e.g., neuroimaging) or insensitive to premanifest HD patients (e.g., motor tests). The authors discuss the merit in measuring biomarkers in biofluids (e.g., CSF). Microarray analysis of global gene expression changes in blood of HD patients identified 773 changed genes with a wide range of functions (Borovecki et al., 2005).
Importantly, a recent large genome-wide association study analysis of ∼4,000 HD patients provides support for the potential biomarkers that we have identified in our study here. The genome-wide association study analysis was set out to identify genetic variations to explain the age of neurological symptoms onset in HD patients that differ from the predicted age of onset based on the length of the CAG repeat. 14 significant pathways clustered by gene membership into three groups of genes: the largest group includes genes related to DNA repair, the second relates to genes that affect mitochondrial organization, release of cytochrome c (indicative of mitochondrial damage), and mitochondrial fission, and the third relates to oxi-reductase activity (Lee et al., 2015). The authors suggested that these HD-modifying genes in humans identified validated therapeutic targets in humans. We would like to suggest that their study may have also identified validated biomarkers for HD, as shown here: association of the onset of the disease with genes regulating DNA repair can be reflected by the changes that we found in the product of DNA excision repair, 8-OHdG; association with genes regulating mitochondrial organization, damage, and fission can be reflected by the observed increase in mtDNA in the plasma and the benefit obtained by inhibiting excessive mitochondrial fission with P110; and association with genes regulating oxi-reductase activity can correlate with increased aldehydic load, as measured by protein adduction of 4-HNE, a product of oxidative stress.
Multiple models of HD have been developed, and R6/2 mice exhibit a fast progression of the disease, whereas YAC128 mice and other models show slower disease progression (Mochly-Rosen et al., 2014). However, because R6/2 mice benefited from treatment with P110, as did cells and neurons derived from HD patients (Guo et al., 2013), and because at least one of the biomarkers (mtND2) is also altered in a more slowly progressing disease (YAC128 mice) and in HD patients, our data appear promising. We have identified four potential biomarkers that correlate with disease progression and/or treatment benefits, all easily accessible. These are mtDNA in plasma, 8-OHdG in urine, levels of mtHtt aggregates in skin, and skeletal muscle and carbonylated (4-HNE) modified proteins in skin and muscle. These potential biomarkers should not be used for the diagnostics of HD patients, as they may correlate with other diseases. However, they are likely to be useful as surrogate markers for treatment benefit. Our mouse study also suggests that changes in these biomarkers should correlate with disease progression in patients. Thus, HD patients should be tested over time, and mtDNA levels should be compared with the values before or at the onset of the disease.
A very recent commentary by Townsend and Arron (2016) discusses the importance of identification of surrogate biomarkers to reduce the risk of failure in human clinical trials, even if they are distal from the targeted pathway. They point out that to achieve an early proof of concept in a slow progressing disease, it is necessary to have biomarkers that change upon treatment and conclude that a biomarker-guided clinical trial will reduce the risk of failure of drug candidates to reach patients. The goal of our study was to identify such potential peripheral biomarkers for HD that correlate well with disease progression and with treatment benefits. We strongly believe that this goal was achieved here and hope that our study will trigger a prospective clinical study of a large cohort of patients that will follow changes in these biomarkers as the disease progresses. We suggest that our work provides the basis for a larger study of patient samples that ultimately may identify the biomarkers that could be used as surrogate markers to determine the benefit of therapeutic intervention in diagnosed but asymptomatic HD patients to prevent or delay the onset of the disease.
Materials and methods
Peptide treatment in mouse model
All the experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Stanford University and were performed based on the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Hemizygous R6/2 HD mice and their WT littermates were purchased from The Jackson Laboratory and shipped to us at 5 wk of age. The animals used in the P110 treatment study were implanted with a 28-d osmotic pump (Alzet) containing TAT47–57 carrier control peptide or P110-TAT47–57 (P110 peptide; Qi et al., 2013), which delivered to the mice at a rate of 3 mg/Kg/d, as described previously (Guo et al., 2013). The first pump was implanted at 5 wk of age and replaced once, after 4 wk. For the intermittent treatment study, TAT or P110 were delivered in 5-wk-old mice using a 1-wk pump. After 3 wk with no treatment, a new pump was implanted for another week of treatment, and mice were sacrificed 3 wk later at the age of 13 wk.
Animal survival and behavior study
The overall survival during the study period was recorded, and the remaining mice were sacrificed when they reached 13 wk. All the behavior and survival tests and analyses were conducted by an experimenter who was blind to genotypes and drug groups. Detailed behavioral methods used in this study are listed below.
Blood collection and measurement of mtDNA levels from plasma
Mouse blood was collected by retroorbital bleeding. For the time-course experiment, 200 µl of blood samples was collected from alternate eyes every 2 wk from the age of 5 to 13 wk. In the P110 treatment study, 500 µl of blood was collected at 13 wk, just before euthanasia. Plasma was obtained by a single centrifugation step at 1,600 g for 10 min, as previously reported (Chiu et al., 2003). 100 µl of plasma samples was used to extract DNA, eluted in 60 µl of elution buffer using a viral DNA kit (QIAGEN). A 1:10 DNA dilution was used in the real-time PCR reaction.
Human plasma samples were obtained from B.R. Leavitt; six plasma samples were from control subjects, six from presymptomatic HD subjects (mtHtt gene carrier), and six from affected HD patients. Each group included three male and three female subjects. DNA was extracted using a viral DNA kit (QIAGEN), and a 1:2 dilution was used for the real-time PCR.
RNA extraction from brain tissue
100 mg of brain tissue was used for RNA isolation using an RNAquaeous kit (Ambion) as per the manufacturer’s protocol. 1 µg of total RNA was used for the synthesis of first-strand cDNA using a PrimeScript first-strand cDNA synthesis kit (Takara Bio Inc.), and 15 ng of cDNA was used as a template for the real-time PCR reaction.
Quantitative analysis of DNA in plasma by real-time PCR
5 µl of DNA from mouse plasma, human plasma, and mouse brain tissue (15 ng) were used as templates for real-time PCR analysis. For assessment of nuclear DNA present in the samples and normalization of the measurements, we used GAPDH as a housekeeping gene (18S ribosomal DNA was used as well, yielding similar results). For the mouse GAPDH gene (GenBank accession no. AC166162.6), we used forward 5′-GGACCTCATGGCCTACATGG-3′ and reverse 5′-TAGGGCCTCTCTTGCTCA-3′ primers. For the human GAPDH gene (GenBank accession no. AF275320.1), we used forward 5′-GTCGGAGTCAACGGATTTG-3′ and reverse 5′-CCATGTAGTTGAGGTCAATGAA-3′. To detect circulating mtDNA, we used mouse mtND2 gene (NCBI RefSeq accession no. NC_005089) with forward 5′-AACCCACGATCAACTGAAGC-3′ and reverse 5′-TTGAGGCTGTTGCTTGTGTG-3′. For human mtND2 (NCBI RefSeq accession no. NC_012920), we used forward 5′-CTATCTCGCACCTGAAAC-3′ and reverse 5′-GAGGGTGGATGGAATTAAG-3′. PCR was performed using an ABI/Life Technologies StepOnePlus real-time PCR instrument (Applied Biosystems) in a total volume of 20 µl, containing 5 µl of plasma DNA or 5 µl of 15 ng brain DNA, 10 µl of Fast Sybr green master mix (Applied Biosystems), and 1 µl of primers (forward + reverse) at 2 µM using cycles as follows: 95°C for 20 s, 40 cycles of 95°C for 3 s, and 57°C for 30 s followed by a melt curve at 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s We optimized the reaction using several primers for each of the target genes according to the melting curve of respective primers in the assay. Relative changes in gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), where ΔΔCT = (CT,mtND2 − CT,GAPDH)treatment − (CT,mtND2 − CT,GAPDH)control. For treated samples, evaluation of 2−ΔΔCT indicates the fold-changes in gene expression relative to untreated control.
Measurement of DNA damage (8-OHdG) by ELISA
Urine was collected from WT and R6/2 mice at 13 wk of age after 8 wk of treatment with TAT control or P110. Urine, diluted 1:100 in water, was assayed as described in the manufacturer’s protocol (Cell Biolabs) with a competitive ELISA assay kit for quantitative measurement of 8-OHdG. Urine samples were analyzed in parallel for creatinine content for normalization of 8-OHdG results according to the manufacturer’s protocol (Cell Biolabs). The results are expressed as ng/ml 8-OHdG/µmol/L creatinine levels.
TNF and IL-6 measurements
Plasma TNF and IL-6 levels were determined by a mouse TNF and IL-6 ELISA kit according to the manufacturer’s protocol (eBioscience) using 20 µl of plasma collected and prepared as described in the Blood collection and measurement of mtDNA levels of plasma section.
Immunohistochemistry in tissue sections
13-wk-old mice were sacrificed, and skeletal muscles (quadriceps and hamstrings) and skin from the top dorsal area after hair removal were fixed in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. Tissues were processed for paraffin embedment, and sections were used for immunohistochemical staining of mtHtt (EM-48; 1:200; EMD Millipore) and 4-HNE staining (1:200; Abcam) using an immunohistochemistry Select HRP/DAB kit (EMD Millipore). The images were viewed using a microscope (Leica Biosystems) with a 20× objective. 16 areas for skin mtHtt, 4-HNE sections, and muscle 4-HNE sections and 58 areas for muscle mtHtt sections from each image from three animals/group were analyzed using MATLAB scripting language. Pixels from the respective staining were separated from background pixels producing a binarized image (Selinummi et al., 2005).
YAC128 animal model
YAC128 (FVB-Tg[YAC128]53Hay/J; stock no. 004938) breeders (FVB/N genetic background) were purchased from The Jackson Laboratory. The YAC128 mice contain a full-length human huntingtin gene modified with a 128 CAG repeat expansion in exon 1. The mice were mated, bred, and genotyped in the animal facility of Case Western Reserve University. Male mice at the age of 6 mo were used in the study. All of the mice were maintained with a 12-h light/dark cycle (on 6 am; off 6 pm).
Animal model and behavioral tests
Two cohorts of male B6CBA-Tg (HDexon1)62Gpb/3J (R6/2) mice and their WT littermates from The Jackson Laboratory were used for behavioral phenotyping (stock no. 006494). Cohort 1 mice, of n = 10 WT and n = 10 R6/2 mice, were housed under a 12-h light/dark cycle (7:00 am light on; 7:00 pm light off). Cohort 1 mice were tested in a rotating rod test, y-maze spontaneous alternation test, and DMP-DM test during the light-on cycle of the day. Cohort 2 mice, of n = 10 WT and n = 10 R6/2 mice, were housed under a 12-h light/dark cycle (8:30 am light off; 8:30 pm light on). Cohort 2 mice were tested in a social discrimination test using a PhenoLab passive avoidance test and fear-conditioning test during the light-off cycle of the day. The mice were group housed three to five per cage and handled by an experimenter for 5 d before the behavioral experiment. In addition to having ad libitum access to regular food and water, wet chow on disposable weight boats was provided at the bottom of the cage. Because R6/2 mice have high sensitivity to vibration and noise, cages were hand-carried by the experimenters, and mice were habituated on a cart outside or inside the testing rooms 1 h before the tests. The experimenters were not aware of the genotype of the mice during the experiments. All behavioral procedures were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Stanford University and were performed based on the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All actions were considered for reducing discomfort of the animals throughout the study.
Rotating rod test
Mice motor learning and coordination were accessed using a Five Station Rota-Rod Treadmill (ENV-575M; Med Associates Inc.) during 7, 9, and 11 wk of age. 2 d before the first testing of the 7-wk-old mice, each mouse received three training trials. Each training trial was 60 s long at a fixed speed of 32 rounds per minute (rpm) with 5–10 min intertrial intervals (ITIs). During the testing, each mouse received two to three trials of 4-40 rpm accelerated speed. The maximum duration of each trial was 300 s with 15–20 min of ITIs. Mice were tested for a minimum of two trials per day but removed from the Rota-Rod and tested for a third trial if the following exclusion criteria were met: a mouse held onto the rod instead of walking on it for two consecutive revolutions, for three cumulative revolutions during the trial jumped off the rod instead of dropping off the rod because of lack of balance, or fell off the rod in <5 s. The mean latency of the two trials to fall off the Rota-Rod or the latency when the mice met the exclusion criteria were used for data analysis. The Rota-Rod was cleaned with 10% alcohol between trials. A total of 19 mice (n = 10 WT and n = 9 R6/2) were used in the test.
Y-maze spontaneous alternation test
Spontaneous alternations in mice were measured in a custom-built y-maze when the mice were 8 wk old. The maze was made of opaque white plastic and had three equal arms of 40-cm length, 8-cm width, and 15-cm height. Each arm was labeled with the letter A, B, or C. Mice were placed in the maze facing arms B and C. The first entry was excluded from data analysis because of the fact that the animals were led to this initial arm. The total number of entries and sequence of entries into the arms were recorded for 8 min. Entries into the arms were defined as when all four paws entered into a new arm of the maze and not when the mice moved to the center and returned to the same arm. The percentage of spontaneous alternation was calculated (Drew et al., 1973; Hughes, 2004). In brief, the experimenter analyzed the sequence of the arm entries A, B, and C in a set of three entries or a triad. Every triad with all three letters was considered as alternation (e.g., ABC, BCA, CAB), and percent spontaneous alternation was calculated using the number of alternation divided by the total possible triads times 100. For example, “ABCACAB.” Data were then broken into triads of entries; a sequence with repeating letters such as ABA or CAC would be scored as a nonalternation, whereas a sequence with all three letters, e.g., ABC or CBA, would be scored as an alternation. For our sample above, the first triad was ABC, and the second was BCA; however the third triad CAC would not be considered an alternation. In these sample data, there were a total of seven entries, five possible triads, and three alternations. Percent spontaneous alternation would be 3/5 × 100 = 60%. The y-maze was cleaned with 10% alcohol between each mouse. A total of 19 mice (n = 10 WT and n = 9 R6/2) were used in this test.
DMP-DM
The DMP-DM test was conducted using a custom-built circular-shaped platform 122 cm in diameter with 40 holes elevated 50 cm from the floor. The test consisted of 7 d of testing when the mice were 10 wk old. Each hole was 5 cm in diameter, and an escape tube filled with bedding was attached to only one of the holes. The hole with an escape tube was defined as the target escape hole (TEH). The remaining 39 holes without the escape tube were covered with a piece of plastic so the mice would not accidentally drop into the holes. A short lip was placed around the edge of the maze to prevent the animals from falling off the platform. High overhead lighting with 900 lux was used to create an aversive stimulus that would encourage the animals to seek out the target hole to escape from the light. The maze was surrounded by privacy blinds, and distinct visual cues were placed on the privacy blinds. An individual mouse was given a series of four trials per day to find the escape hole with 10–12 min of ITIs. The maximum duration for each trial was 90 s. The bright lights in the testing room were kept dim before the start of a trial. The subject mouse was placed under an opaque box in the pseudorandomized positions around the edge of the maze. The experimenter turned on the bright light after 10 s, and the box was removed to allow the mouse to find the TEH. If the mouse found and entered into the TEH before 90 s, the experiment was stopped. Mice that could not find the TEH or enter the escape tube were led to it by the experimenter and allowed to enter. Mice were allowed to remain in the tube for 10 s after each trial and returned to the home cage. After each trial, the apparatus was cleaned with 10% alcohol to eliminate odor cues. At the start of days 2–7, the location of the TEH was moved to a new escape hole, and everything else remained the same. Mice were tracked with Ethovision XT software (Noldus Information Technology), and latency to find the TEH, distance moved, and velocity were recorded. A total of 19 mice (n = 10 WT and n = 9 R6/2) were used in the test.
Social discrimination test using PhenoLab
The PhenoLab cages were custom-built cages made of acrylic plastic with 30 cm length × 30 cm width × 60 cm height. Mice were individually housed and habituated in the cages for 4 d before the social discrimination test. Infrared cameras were mounted on top of the cages to monitor the mice inside the cages. The mice were 6 wk old when they were introduced into the cages. All n = 10 WT and n = 10 R6/2 mice were tested simultaneously in 20 individual PhenoLab cages. Each cage was equipped with a food tray, water bottle, running wheel (ENV-044; Med Associates Inc.), and shelter box (red transparent polycarbonate). Mice had ad libitum access to all enrichments and were not disrupted by the experimenter during the habituation. In the subsequent social discrimination test, the running wheel was removed, and two identical stainless steel pencil cups (11 cm height × 10 cm diameter solid bottom, with stainless steel bars spaced 1 cm apart) were inverted and placed in two corners of the cage adjacent from one another. A novel object (plastic cap) and a novel young juvenile mouse, stranger 1, were placed under each cup, and the subject mouse was allowed to explore for 2 h. After 2 h, stranger 1 and the cup were repositioned to the corner where the novel object was located. The novel object was removed from the cage, and a second novel young juvenile mouse, stranger 2, was placed under the cup. Subject mice were allowed to explore the stranger 1 and stranger 2 mice for 10 min after stranger 2 was introduced into the cage. Both juvenile mice were 5-wk-old C57BL/6J male mice (stock no. 000664), and they were housed in different cages. Subject mice were tracked with Ethovision XT software (Noldus Information Technology), and the center of the subject mice within 4-cm virtual zones around the cups were used as the interaction time. A total of 19 mice (n = 10 WT and n = 9 R6/2) were used in the test.
Passive avoidance test
The passive avoidance test was conducted using a GIMINI avoidance system (San Diego Instruments) when the mice were 8 wk old. This automated system contained two compartments, which were separated by a drop door (gate). Both compartments had grid floors that could deliver electric shock, but one compartment was lighted, whereas the other was dark. The experiment consisted of 1 d of habituation, 1 d of training, and 2 d of testing. On habituation day, the mouse was placed in the lighted compartment. After 30-s acclimation, the gate was opened and the mouse was allowed to explore both compartments freely. The gate was programmed to close when the mouse entered the dark compartment to prevent the mouse from returning to the lighted compartment. The mouse was removed from the system and returned to the home cage after it entered the dark compartment. On the following day, training day, the mouse was placed in the light compartment. After 30 s of acclimation, the gate was opened and the mouse was allowed to explore both compartments freely. The gate was closed after it entered the dark compartment. 3 s after the gate was closed, an electric shock (0.5 mA for 2 s) was delivered. The mouse remained in the dark compartment for an additional 30 s before being removed and returned to the home cage. On the following day, day 1 testing day, the mouse was placed in the lighted compartment. After 5-s acclimation, the gate was opened. When the mouse entered the dark compartment, the gate was closed, the trial ended, and the mouse was returned to the home cage. 7 d after training, day 7 testing day, the same procedures were repeated as day 1 testing day. The maximum duration of each trial was 300 s after the gate was opened. The time between the gate opening and the mouse passing through the gate was recorded as latency time. The compartments were cleaned with 1% Virkon between each animal. A total of 19 mice (n = 10 WT and n = 9 R6/2) were used in the test.
Fear-conditioning test
The fear-conditioning test was conducted using fear-conditioning chambers (Coulbourn Instruments) when the mice were 11–12 wk old. The test consisted of day 1 training, day 2 cued testing, and day 3 contextual testing. During day 1 training and day 3 contextual testing, mice were tested in distinct context A (metal grid floor, square-shape clear chamber, yellow dim light, mint extract as odor cue, and 10% simple green solution to clean the chamber between each mouse). During day 2 cued testing, mice were tested in context B (plastic floor, round-shape opaque chamber, blue dim light, vanilla extract, 70% alcohol to clean the chamber between each mouse, and different testing room). On day 1 training, the mice were acclimated in the chamber for 200 s followed by 5× pairing of tones and shocks. The tones were 20-s duration, 2-kHz frequency, and 70-dB loud. The shocks were 2-s duration at 0.5-mA shock intensity. The time between a tone and a shock pairing was 18 s, and the ITIs between the tones were 100 s. The mice were removed from the chamber and returned to the home cage 80 s after the last tone. On day 2 cued testing, the mice were acclimated in the chamber for 200 s followed by three tones without any shock. The tones were 20-s duration, 2-kHz frequency, and 70-dB loud. The ITI between each tone was 100 s. The mice were removed from the chamber and returned to the home cage 80 s after the last tone. On day 3 contextual testing, the mice were placed in the context A testing chamber for 5 min without any tone or shock. Mice were returned to the home cage after the trial. The mice freezing behavior was recorded with a camera above the chamber, and freezing was defined as the complete lack of motion for a minimum of 0.75 s, as assessed by FreezeFrame software (Actimetrics). A total of 15 mice (n = 9 WT and n = 6 R6/2) were used in the test.
Human CSF collection and measurement of mtDNA levels
CSFs from three control and three HD subjects were obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, Los Angeles, CA. CSF samples were collected before removing the brain from the skull of postmortem patients using a lumbar puncture needle with a stylet to prevent brain tissue from entering the needle. After the calvarium was open and before removing the brain, the lumbar puncture needle was inserted into the lateral (either the left or right side) ventricle. At least 20-ml samples of CSF were obtained from the lateral ventricles (without blood contamination). The CSF was mixed gently by inverting the tube five times and then transferred into 15-ml centrifuge tubes before spinning at 1.5 g for 15 min at 4°C. The supernatant was aliquoted and stored at −80°C. MtDNA from 200 µl CSF was extracted using a viral DNA kit (QIAGEN) to minimize contamination of molecules present in CSF that inhibit the detection of DNA by PCR. 5 µl of undiluted CSF DNA was used in the real-time PCR reaction as described in the Blood collection and measurement of mtDNA levels from plasma section. 2−ΔΔCT indicates the fold-changes in gene expression.
Statistical analysis
Statistical analyses for behavior studies were processed using Prism software (version 5; GraphPad Software). Data are presented as mean ± SEM, and statistically significant was defined as P < 0.05. Repeated measures two-way ANOVA with Bonferroni posthoc tests was used for evaluation of the parameters in rotating rod tests and DMP-DMs. Unpaired Student’s t tests were used for y-maze total entries, passive avoidance tests, and fear-conditioning tests. One-sample Student’s t tests were used for spontaneous alternation comparing the mean alternation of each genotype to the hypothetical value of 50%. Paired Student’s t tests were used to compare the time spent in stranger 1 versus novel object zones during the sociability sessions for both genotypes. Wilcoxon nonparametric paired Student’s t tests were used in social novelty sessions for the WT mice, and paired Student’s t tests were used for the R6/2 mice when comparing the time spent in stranger 1 and stranger 2 zones during the social discrimination test. D’Agostino and Pearson omnibus normality tests were used to determine the normal distribution of the dataset. Kolmogorov-Smirnov tests were used to determine the normal distribution of the dataset for fear conditioning because the number of R6/2 mice was too small for the D’Agostino and Pearson omnibus normality test.
The results (Figs. 1–7) are presented as mean ± SE. Statistical analysis was assessed by unpaired Student’s t tests and one-way ANOVA using Prism (GraphPad Software). The standard Mantel-Cox log-rank test was used to assess survival. Repeated measures two-way ANOVA with Bonferroni posthoc test was used for evaluation of the parameters in rotating rod tests and DMP-DMs. All tests were performed in a blinded way. P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows behavioral analysis of R6/2 mice at 11 wk of age. Fig. S2 shows mtDNA levels in plasma of YAC128 mice. Fig. S3 shows 4-HNE staining of skeletal muscle and skin sections of 13-wk-old mice. Fig. S4 shows mtND2 levels in human HD CSF.
Supplementary Material
Acknowledgments
The authors thank Mr. Juan Harrison (Takeda) for advice and support. We thank the Human Brain and Spinal Fluid Resource Center for providing the CSF samples. We also thank Alberto Lovell from the Pan Facility at Stanford for helpful discussion on real-time PCR, Opher S. Kornfeld for analyzing immunohistochemistry images, and Yoni S. Rubin for statistical analysis. The Human Brain and Spinal Fluid Resource Center is sponsored by the National Institute of Neurological Disorders and Stroke (NINDS)/National Institute of Mental Health, National Multiple Sclerosis Society, and Department of Veterans Affairs. The animal behavioral study was supported partly by a NINDS center core grant (2 P30 NS069375 06). Note that although Takeda sponsored the cost of this project, it was designed and executed entirely at Stanford University, and Takeda has no ownership on the findings.
This work was supported by a National Institutes of Health (NIH) grant (HL52141) and TAKEDA Pharmaceuticals U.S.A. Innovation funding to D. Mochly-Rosen and an NIH grant (R01 NS088192) to X. Qi.
In September 2016, a patent on the design and application of a mitochondrial fission peptide inhibitor was licensed to MitoConix, a company that was founded by D. Mochly-Rosen. X. Qi and D. Mochly-Rosen own shares or share options in the company. The authors declare no additional competing financial interests.
Author contributions: M.-H. Disatnik conducted all experiments and wrote and revised the manuscript. N.L. Saw and M. Shamloo performed and analyzed the animal behavior study. A.U. Joshi performed the inflammation marker study and the short P110 study and revised the manuscript. B.R. Leavitt provided human plasma samples. X. Qi performed the YAC128 mice cohort biomarker study and revised the manuscript. D. Mochly-Rosen supervised the design and revised the manuscript.
Footnotes
Abbreviations used:
- CNS
- central nervous system
- CSF
- cerebrospinal fluid
- DMP-DM
- delay match-to-place dry maze
- HD
- Huntington’s disease
- ITI
- intertrial interval
- mtDNA
- mitochondrial DNA
- mtHtt
- mutant huntingtin
- mtND2
- mitochondria-encoded NADH dehydrogenase 2
- TEH
- target escape hole
References
- Andre R., Scahill R.I., Haider S., and Tabrizi S.J.. 2014. Biomarker development for Huntington’s disease. Drug Discov. Today. 19:972–979. 10.1016/j.drudis.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Björkqvist M., Wild E.J., Thiele J., Silvestroni A., Andre R., Lahiri N., Raibon E., Lee R.V., Benn C.L., Soulet D., et al. 2008. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J. Exp. Med. 205:1869–1877. 10.1084/jem.20080178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M.B., Beal M.F., McCabe D.R., Griffin R.M., and Matson W.R.. 1999. A carbon column-based liquid chromatography electrochemical approach to routine 8-hydroxy-2′-deoxyguanosine measurements in urine and other biologic matrices: a one-year evaluation of methods. Free Radic. Biol. Med. 27:647–666. 10.1016/S0891-5849(99)00113-6 [DOI] [PubMed] [Google Scholar]
- Bogdanov M.B., Andreassen O.A., Dedeoglu A., Ferrante R.J., and Beal M.F.. 2001. Increased oxidative damage to DNA in a transgenic mouse model of Huntington’s disease. J. Neurochem. 79:1246–1249. 10.1046/j.1471-4159.2001.00689.x [DOI] [PubMed] [Google Scholar]
- Borovecki F., Lovrecic L., Zhou J., Jeong H., Then F., Rosas H.D., Hersch S.M., Hogarth P., Bouzou B., Jensen R.V., and Krainc D.. 2005. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc. Natl. Acad. Sci. USA. 102:11023–11028. 10.1073/pnas.0504921102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowsky B., Warner J., Leavitt B.R., Tabrizi S.J., Roos R.A., Durr A., Becker C., Sampaio C., Tobin A.J., and Schulman H.. 2013. 8OHdG is not a biomarker for Huntington disease state or progression. Neurology. 80:1934–1941. 10.1212/WNL.0b013e318293e1a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., and Beal M.F.. 1997. Oxidative damage and metabolic dysfunction in Huntington’s disease: selective vulnerability of the basal ganglia. Ann. Neurol. 41:646–653. 10.1002/ana.410410514 [DOI] [PubMed] [Google Scholar]
- Budnik L.T., Kloth S., Baur X., Preisser A.M., and Schwarzenbach H.. 2013. Circulating mitochondrial DNA as biomarker linking environmental chemical exposure to early preclinical lesions elevation of mtDNA in human serum after exposure to carcinogenic halo-alkane-based pesticides. PLoS One. 8:e64413 10.1371/journal.pone.0064413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M. 2011. Mitochondrial dysfunction, metabolic deficits, and increased oxidative stress in Huntington’s disease. Chang Gung Med. J. 34:135–152. [PubMed] [Google Scholar]
- Chen H., and Chan D.C.. 2009. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum. Mol. Genet. 18:R169–R176. 10.1093/hmg/ddp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.M., Wu Y.R., Cheng M.L., Liu J.L., Lee Y.M., Lee P.W., Soong B.W., and Chiu D.T.. 2007. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem. Biophys. Res. Commun. 359:335–340. 10.1016/j.bbrc.2007.05.093 [DOI] [PubMed] [Google Scholar]
- Chiu R.W., Chan L.Y., Lam N.Y., Tsui N.B., Ng E.K., Rainer T.H., and Lo Y.M.. 2003. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin. Chem. 49:719–726. 10.1373/49.5.719 [DOI] [PubMed] [Google Scholar]
- Ciancarelli I., De Amicis D., Di Massimo C., Di Scanno C., Pistarini C., D’Orazio N., and Tozzi Ciancarelli M.G.. 2014. Peripheral biomarkers of oxidative stress and their limited potential in evaluation of clinical features of Huntington’s patients. Biomarkers. 19:452–456. 10.3109/1354750X.2014.935955 [DOI] [PubMed] [Google Scholar]
- Ciarmiello A., Cannella M., Lastoria S., Simonelli M., Frati L., Rubinsztein D.C., and Squitieri F.. 2006. Brain white-matter volume loss and glucose hypometabolism precede the clinical symptoms of Huntington’s disease. J. Nucl. Med. 47:215–222. [PubMed] [Google Scholar]
- Costa V., and Scorrano L.. 2012. Shaping the role of mitochondria in the pathogenesis of Huntington’s disease. EMBO J. 31:1853–1864. 10.1038/emboj.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A., and Glass C.K.. 2015. The choreography of neuroinflammation in Huntington’s disease. Trends Immunol. 36:364–373. 10.1016/j.it.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A., Benner C., Kerman B.E., Gosselin D., Lagier-Tourenne C., Zuccato C., Cattaneo E., Gage F.H., Cleveland D.W., and Glass C.K.. 2014. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat. Neurosci. 17:513–521. 10.1038/nn.3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., and Aronin N.. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 277:1990–1993. 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- Disatnik M.H., Ferreira J.C., Campos J.C., Gomes K.S., Dourado P.M., Qi X., and Mochly-Rosen D.. 2013. Acute inhibition of excessive mitochondrial fission after myocardial infarction prevents long-term cardiac dysfunction. J. Am. Heart Assoc. 2:e000461 10.1161/JAHA.113.000461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew W.G., Miller L.L., and Baugh E.L.. 1973. Effects of δ9-THC, LSD-25 and scopolamine on continuous, spontaneous alternation in the Y-maze. Psychopharmacology (Berl.). 32:171–182. 10.1007/BF00428688 [DOI] [PubMed] [Google Scholar]
- Duan W., Jiang M., and Jin J.. 2014. Metabolism in HD: still a relevant mechanism? Mov. Disord. 29:1366–1374. 10.1002/mds.25992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin A., Leenders K.L., Moeller J.R., Missimer J., Kuenig G., Spetsieris P., Antonini A., and Eidelberg D.. 2001. Metabolic network abnormalities in early Huntington’s disease: an [18F]FDG PET study. J. Nucl. Med. 42:1591–1595. [PubMed] [Google Scholar]
- Ferger A.I., Campanelli L., Reimer V., Muth K.N., Merdian I., Ludolph A.C., and Witting A.. 2010. Effects of mitochondrial dysfunction on the immunological properties of microglia. J. Neuroinflammation. 7:45 10.1186/1742-2094-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Disatnik M.H., Monbureau M., Shamloo M., Mochly-Rosen D., and Qi X.. 2013. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J. Clin. Invest. 123:5371–5388. 10.1172/JCI70911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardas S.S., Sultana R., Clark A.M., Beckett T.L., Szweda L.I., Murphy M.P., and Butterfield D.A.. 2013. Oxidative modification of lipoic acid by HNE in Alzheimer disease brain. Redox Biol. 1:80–85. 10.1016/j.redox.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch S.M., Gevorkian S., Marder K., Moskowitz C., Feigin A., Cox M., Como P., Zimmerman C., Lin M., Zhang L., et al. 2006. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2'dG. Neurology. 66:250–252. 10.1212/01.wnl.0000194318.74946.b6 [DOI] [PubMed] [Google Scholar]
- Hsiao H.Y., Chen Y.C., Chen H.M., Tu P.H., and Chern Y.. 2013. A critical role of astrocyte-mediated nuclear factor-κB-dependent inflammation in Huntington’s disease. Hum. Mol. Genet. 22:1826–1842. 10.1093/hmg/ddt036 [DOI] [PubMed] [Google Scholar]
- Hughes R.N. 2004. The value of spontaneous alternation behavior (SAB) as a test of retention in pharmacological investigations of memory. Neurosci. Biobehav. Rev. 28:497–505. 10.1016/j.neubiorev.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Hwang S., Disatnik M.H., and Mochly-Rosen D.. 2015. Impaired GAPDH-induced mitophagy contributes to the pathology of Huntington’s disease. EMBO Mol. Med. 7:1307–1326. 10.15252/emmm.201505256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., and Ferrante R.J.. 2010. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum. Mol. Genet. 19:3919–3935. 10.1093/hmg/ddq306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C., Radpour R., Barekati Z., Asadollahi R., Bitzer J., Wight E., Bürki N., Diesch C., Holzgreve W., and Zhong X.Y.. 2009. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol. Cancer. 8:105 10.1186/1476-4598-8-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroshetz W.J., Jenkins B.G., Rosen B.R., and Beal M.F.. 1997. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Ann. Neurol. 41:160–165. 10.1002/ana.410410206 [DOI] [PubMed] [Google Scholar]
- Lam N.Y., Rainer T.H., Chiu R.W., Joynt G.M., and Lo Y.M.. 2004. Plasma mitochondrial DNA concentrations after trauma. Clin. Chem. 50:213–216. 10.1373/clinchem.2003.025783 [DOI] [PubMed] [Google Scholar]
- Lee J., Kosaras B., Del Signore S.J., Cormier K., McKee A., Ratan R.R., Kowall N.W., and Ryu H.. 2011. Modulation of lipid peroxidation and mitochondrial function improves neuropathology in Huntington’s disease mice. Acta Neuropathol. 121:487–498. 10.1007/s00401-010-0788-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-M., Wheeler V.C., Chao M.J., Vonsattel J.P.G., Pinto R.M., Lucente D., Abu-Elneel K., Ramos E.M., Mysore J.S., Gillis T., et al. Genetic Modifiers of Huntington’s Disease (GeM-HD) Consortium . 2015. Identification of genetic factors that modify clinical onset of Huntington’s disease. Cell. 162:516–526. 10.1016/j.cell.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., Popovic N., and Brundin P.. 2005. The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx. 2:447–464. 10.1602/neurorx.2.3.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Long J.D., Matson W.R., Juhl A.R., Leavitt B.R., and Paulsen J.S.. PREDICT-HD Investigators and Coordinators of the Huntington Study Group . 2012. 8OHdG as a marker for Huntington disease progression. Neurobiol. Dis. 46:625–634. 10.1016/j.nbd.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N., et al. The Huntington’s Disease Collaborative Research Group . 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 72:971–983. 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., and Bates G.P.. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87:493–506. 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D., Disatnik M.H., and Qi X.. 2014. The challenge in translating basic research discoveries to treatment of Huntington disease. Rare Dis. 2:e28637 10.4161/rdis.28637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K., Kyung S.Y., Rogers A.J., Gazourian L., Youn S., Massaro A.F., Quintana C., Osorio J.C., Wang Z., Zhao Y., et al. 2013. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 10:e1001577 10.1371/journal.pmed.1001577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J.M. 2010. Nature and cause of mitochondrial dysfunction in Huntington’s disease: focusing on huntingtin and the striatum. J. Neurochem. 114:1–12. [DOI] [PubMed] [Google Scholar]
- Park J., Choi H., Min J.S., Park S.J., Kim J.H., Park H.J., Kim B., Chae J.I., Yim M., and Lee D.S.. 2013. Mitochondrial dynamics modulate the expression of pro-inflammatory mediators in microglial cells. J. Neurochem. 127:221–232. 10.1111/jnc.12361 [DOI] [PubMed] [Google Scholar]
- Podlesniy P., Figueiro-Silva J., Llado A., Antonell A., Sanchez-Valle R., Alcolea D., Lleo A., Molinuevo J.L., Serra N., and Trullas R.. 2013. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical Alzheimer disease. Ann. Neurol. 74:655–668. 10.1002/ana.23955 [DOI] [PubMed] [Google Scholar]
- Polidori M.C., Mecocci P., Browne S.E., Senin U., and Beal M.F.. 1999. Oxidative damage to mitochondrial DNA in Huntington’s disease parietal cortex. Neurosci. Lett. 272:53–56. 10.1016/S0304-3940(99)00578-9 [DOI] [PubMed] [Google Scholar]
- Politis M., Lahiri N., Niccolini F., Su P., Wu K., Giannetti P., Scahill R.I., Turkheimer F.E., Tabrizi S.J., and Piccini P.. 2015. Increased central microglial activation associated with peripheral cytokine levels in premanifest Huntington’s disease gene carriers. Neurobiol. Dis. 83:115–121. 10.1016/j.nbd.2015.08.011 [DOI] [PubMed] [Google Scholar]
- Powers W.J., Videen T.O., Markham J., McGee-Minnich L., Antenor-Dorsey J.V., Hershey T., and Perlmutter J.S.. 2007. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc. Natl. Acad. Sci. USA. 104:2945–2949. 10.1073/pnas.0609833104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Qvit N., Su Y.C., and Mochly-Rosen D.. 2013. A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci. 126:789–802. 10.1242/jcs.114439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D.E., and Shoulson I.. 2011. Huntington’s disease: Clinical experimental therapeutics. In Frontiers in Neuroscience. Lo D.C., and Hughes R.E., editors. CRC Press/Taylor & Francis, Boca Raton, FL. Chapter 12. [Google Scholar]
- Ross C.A., and Tabrizi S.J.. 2011. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10:83–98. 10.1016/S1474-4422(10)70245-3 [DOI] [PubMed] [Google Scholar]
- Sathasivam K., Hobbs C., Turmaine M., Mangiarini L., Mahal A., Bertaux F., Wanker E.E., Doherty P., Davies S.W., and Bates G.P.. 1999. Formation of polyglutamine inclusions in non-CNS tissue. Hum. Mol. Genet. 8:813–822. 10.1093/hmg/8.5.813 [DOI] [PubMed] [Google Scholar]
- Selinummi J., Seppälä J., Yli-Harja O., and Puhakka J.A.. 2005. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques. 39:859–863. 10.2144/000112018 [DOI] [PubMed] [Google Scholar]
- Shao J., and Diamond M.I.. 2007. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 16:R115–R123. 10.1093/hmg/ddm213 [DOI] [PubMed] [Google Scholar]
- Shigenaga M.K., Gimeno C.J., and Ames B.N.. 1989. Urinary 8-hydroxy-2′-deoxyguanosine as a biological marker of in vivo oxidative DNA damage. Proc. Natl. Acad. Sci. USA. 86:9697–9701. 10.1073/pnas.86.24.9697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutt T.E., and McBride H.M.. 2013. Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim. Biophys. Acta. 1833:417–424. 10.1016/j.bbamcr.2012.05.024 [DOI] [PubMed] [Google Scholar]
- Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. 2003. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 12:1555–1567. 10.1093/hmg/ddg169 [DOI] [PubMed] [Google Scholar]
- Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R., et al. 2011. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med. 17:377–382. 10.1038/nm.2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack E.C., Del Signore S.J., Luthi-Carter R., Soh B.Y., Goldstein D.R., Matson S., Goodrich S., Markey A.L., Cormier K., Hagerty S.W., et al. 2007. Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Genet. 16:1164–1175. 10.1093/hmg/ddm064 [DOI] [PubMed] [Google Scholar]
- Townsend M.J., and Arron J.R.. 2016. Reducing the risk of failure: biomarker-guided trial design. Nat. Rev. Drug Discov. 15:517–518. 10.1038/nrd.2016.124 [DOI] [PubMed] [Google Scholar]
- Träger U., Andre R., Magnusson-Lind A., Miller J.R., Connolly C., Weiss A., Grueninger S., Silajdžić E., Smith D.L., Leavitt B.R., et al. 2015. Characterisation of immune cell function in fragment and full-length Huntington’s disease mouse models. Neurobiol. Dis. 73:388–398. 10.1016/j.nbd.2014.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir D.W., Sturrock A., and Leavitt B.R.. 2011. Development of biomarkers for Huntington’s disease. Lancet Neurol. 10:573–590. 10.1016/S1474-4422(11)70070-9 [DOI] [PubMed] [Google Scholar]
- Xia P., An H.X., Dang C.X., Radpour R., Kohler C., Fokas E., Engenhart-Cabillic R., Holzgreve W., and Zhong X.Y.. 2009a Decreased mitochondrial DNA content in blood samples of patients with stage I breast cancer. BMC Cancer. 9:454 10.1186/1471-2407-9-454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Radpour R., Zachariah R., Fan A.X., Kohler C., Hahn S., Holzgreve W., and Zhong X.Y.. 2009b Simultaneous quantitative assessment of circulating cell-free mitochondrial and nuclear DNA by multiplex real-time PCR. Genet. Mol. Biol. 32:20–24. 10.1590/S1415-47572009000100003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.