Hartmann et al. show that, in narcolepsy, T cells exhibit a proinflammatory signature characterized by increased production of TNF, IL-2, and B cell–supporting cytokines.
Abstract
Narcolepsy type 1 is a devastating neurological sleep disorder resulting from the destruction of orexin-producing neurons in the central nervous system (CNS). Despite its striking association with the HLA-DQB1*06:02 allele, the autoimmune etiology of narcolepsy has remained largely hypothetical. Here, we compared peripheral mononucleated cells from narcolepsy patients with HLA-DQB1*06:02-matched healthy controls using high-dimensional mass cytometry in combination with algorithm-guided data analysis. Narcolepsy patients displayed multifaceted immune activation in CD4+ and CD8+ T cells dominated by elevated levels of B cell–supporting cytokines. Additionally, T cells from narcolepsy patients showed increased production of the proinflammatory cytokines IL-2 and TNF. Although it remains to be established whether these changes are primary to an autoimmune process in narcolepsy or secondary to orexin deficiency, these findings are indicative of inflammatory processes in the pathogenesis of this enigmatic disease.
Introduction
Narcolepsy type 1 (hereafter called narcolepsy) is a rare, chronic, and disabling neurological disease characterized by excessive daytime sleepiness, sudden loss of muscle tone triggered by emotions, and fragmented nocturnal sleep (Dauvilliers et al., 2007; Scammell, 2015). Narcolepsy is caused by defective neurotransmission via orexin (also named hypocretin) neuropeptides, resulting from a selective and irreversible loss of orexinergic neurons in the lateral hypothalamus. The disease etiology is largely unclear but narcolepsy has a strong genetic association with the human leukocyte antigen (HLA)-DQB1*06:02 allele, which is present in >98% of patients versus 18% of control Europeans (Mignot et al., 1997; Tafti et al., 2014). Additionally, narcolepsy is associated with other genes relevant to immune responses, most notably HLA class I alleles and T cell receptor α (TCRA) polymorphisms (Han et al., 2013; Ollila et al., 2015; Tafti et al., 2016). These findings provide the main reasoning behind why narcolepsy is generally considered an immune-mediated disorder involving autoaggressive lymphocytes. In line with this hypothesis, autoantibodies of unknown pathogenic relevance have been identified in some individuals with narcolepsy (Cvetkovic-Lopes et al., 2010; Bergman et al., 2014; Ahmed et al., 2015; Saariaho et al., 2015). Further supporting an autoimmune etiology is the fact that vaccination against the 2009 pandemic H1N1 influenza virus with the European preparation Pandemrix was associated with induction of narcolepsy in individuals harboring the HLA-DQB1*06:02 allele (Nguyen et al., 2016). Although these points provide indirect evidence for an immune-mediated pathogenesis in narcolepsy, studies identifying the involved immune cell types and investigating their function and phenotype remain scarce (Liblau et al., 2015).
The recent advent of high-dimensional single-cell analysis techniques such as mass cytometry (also called cytometry by time-of-flight [CyTOF]) now permits in-depth examination of multiple immune cell subtypes simultaneously and, together with algorithm-guided analysis approaches, provides an ideal platform for such explorative studies (Bendall et al., 2012).
Here, we analyzed PBMCs of a large cohort of narcolepsy patients and healthy control individuals harboring the HLA-DQB1*06:02 allele via mass cytometry. Using new and powerful automated algorithms enabled an explorative, hypothesis-generating interrogation of the phenotypic and functional immune signature of narcolepsy. For this, we established a 45-parameter panel, including 22 cell surface and 13 intracellular heavy metal–conjugated antibodies, which were chosen to (i) identify all major immune populations in peripheral blood of humans, (ii) determine their activation status, and (iii) analyze their maturation, chemokine receptor (CCR), and most importantly, cytokine expression patterns. Using this approach revealed a proinflammatory signature of T cells in narcolepsy patients, dominated by elevated levels of B cell–supporting cytokines.
Results
Single-cell mass cytometry for the analysis of immune populations in narcolepsy
We collected PBMCs from narcolepsy patients (n = 39), tightly age-, sex-, and HLA-DQB1–matched healthy donors (HD; n = 25), as well as patients with other hypersomnias (n = 11; Table 1 and more detailed in Table S1). This collection included narcolepsy patients with a wide range of disease durations (8–552 mo), as well as patients in which the onset of narcolepsy occurred after Pandemrix H1N1 influenza virus vaccination (n = 11) or independently of it. Additionally, the inclusion of patients with other hypersomnias allowed us to control for and directly compare the influence of nonautoimmune sleep disorders.
Table 1. Summary characteristics of patients and control subjects in this study.
| Patients | N | Age | Sex (male/female) | HLA-DQB1*06:02 positive | Prior Pandemrix immunization | Disease duration | CSF orexin levels <110 pg/ml (N/N tested) | CSF orexin levels >200 pg/ml (N/N tested) |
|---|---|---|---|---|---|---|---|---|
| yr | mo | |||||||
| Healthy donors | 25 | 47 (13–64) | 16/9 | 25 | ||||
| Narcolepsy type 1 | 39 | 42 (13–64) | 23/16 | 39 | 11 | 59 (8–552) | 32/32 | |
| Other hypersomnias | 11 | 25 (6–54) | 3/8 | 10/10 |
For age and disease duration, numbers indicate the median with the range in parentheses.
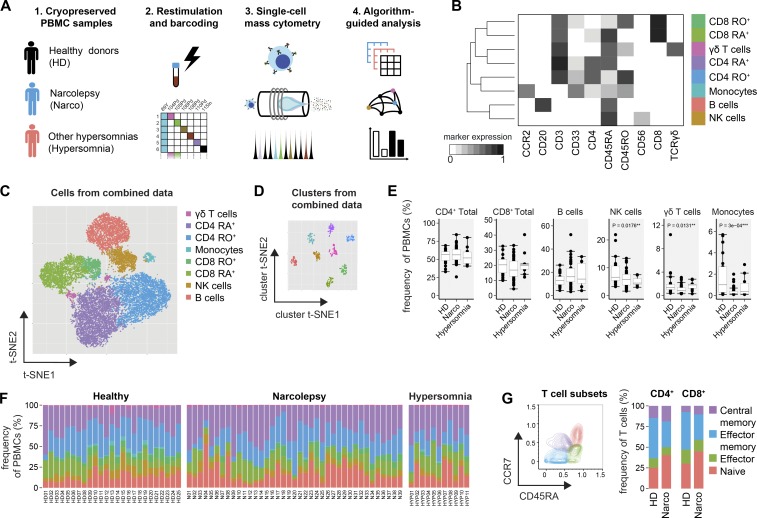
Given the uncertainty about potential autoantigens in narcolepsy, we brieftly restimulated the PBMC samples in an antigen-independent manner (PMA/ionomycin) and performed live-cell barcoding before the combined sample was stained with heavy metal–conjugated antibodies (Table S2) and acquired on a CyTOF2 mass cytometer (Fig. 1 A). After data preprocessing, the FlowSOM algorithm (Van Gassen et al., 2015) was used to identify the major immune cell populations in a data-driven and automated manner. FlowSOM assigned cells to clusters corresponding to the major immune populations (CD4+ and CD8+ T cells, γδT cells, B cells, NK cells, and monocytes, Fig. 1 B). Visualization of the high-dimensional data using the t-stochastic neighbor embedding (t-SNE) algorithm (Van Der Maaten and Hinton, 2008; Amir et al., 2013; Mair et al., 2016) corresponded well to the automatically defined populations, thus further verifying the FlowSOM clustering (Fig. 1 C). The defined populations from different samples were homogeneous in their expression levels of all analyzed markers and clearly distinct from other clusters (Fig. 1 D).
Figure 1.
High-dimensional mass cytometry for the analysis of immune populations in narcolepsy patients. (A) PBMCs of narcolepsy patients (Narco, n = 39), HD (n = 25) and patients with other hypersomnias (n = 11) were stained with heavy metal–labeled antibodies and acquired on a CyTOF2 mass cytometer. (B) Data from preprocessed, single, live cells was exported and used as input for the FlowSOM clustering algorithm. Only the 10 indicated surface markers were used in this initial clustering step. FlowSOM nodes were metaclustered (k = 8) and manually annotated based on the expression level of the lineage-associated markers. RO+, CD45RO+; RA+, CD45RA+. (C) The combined dataset was down-sampled to 20,000 cells and subjected to dimensionality reduction using the t-SNE algorithm. Again, only the 10 surface markers indicated in B were used as input dimensions. The manually annotated populations from C are overlaid as a color-dimension. (D) Instead of cells, clusters from the combined dataset and all measured parameters were used as an input for the t-SNE visualization. (E) Statistical comparison of the frequencies of immune populations and (F) sample-specific composition in narcolepsy patients and controls. Indicated p-values are comparing narcolepsy patients with HD. (G) T cells were automatically subdivided into naive, effector, effector memory, and central memory cells based on the expression of CD45RA and CCR7 using FlowSOM (k = 4, left). Composition of CD4+ and CD8+ T cell subsets in narcolepsy patients versus HD (right). Boxplots represent the interquartile range (IQR) with a black horizontal line indicating the median. Whiskers extend to the farthest data point within a maximum of 1.5× IQR. Values outside this range are plotted as points. All p-values were calculated using a nonparametric Mann-Whitney-Wilcoxon test. Controlling for multiple comparisons was performed with the Benjamini-Hochberg approach. **, significant with false discovery rate (FDR) < 5%; ***, significant with FDR < 1%.
First, we investigated whether the relative frequencies of the major immune cell populations are modulated in narcolepsy patients versus HD and hypersomnia patients (Fig. 1, E and F; and together with all measured frequencies in Table S3). We found the composition of the PBMCs to be largely comparable between patients and controls, with a slight reduction in the frequency of monocytes (1.0 ± 0.6% HD versus 0.2 ± 0.1% Narco, median ± SEM), as well as NK cells and γδT cells. To further define functional subsets of the previously identified immune lineages, we again used FlowSOM to automatically subdivide total CD4+ and CD8+ T cells into CD45RA+CCR7+ naive, CD45RA+CCR7− effector, CD45RA−CCR7− effector memory and CD45RA−CCR7+ central memory cells (Fig. 1 G). Narcolepsy patients were found to have higher proportions of naive CD4+ (37.6 ± 4.1% Narco vs. 22.7 ± 3.9% HD) and CD8+ T cells (46.7 ± 2.4% Narco vs. 23.7 ± 7.0% HD) co-occurring with fewer respective effector memory T cells.
Activation and increased production of TNF and B cell–supporting cytokines by Th cells
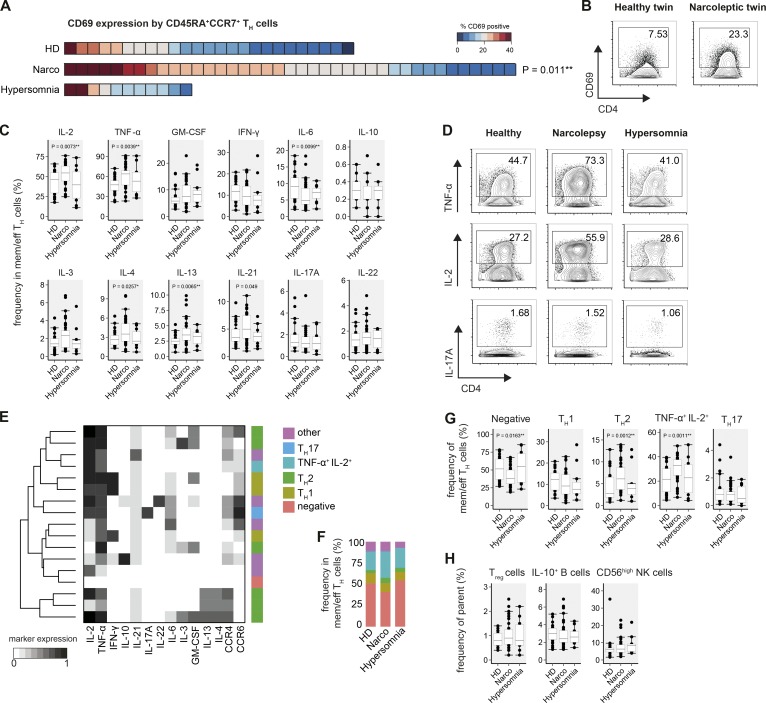
Having automatically defined different leukocyte populations, we next assessed whether their activation status and especially cytokine profiles are altered in narcolepsy. Starting with CD4+ Th cells, we found that CD69 expression was elevated on previously defined CD45RA+CCR7+ Th cells in narcolepsy patients (21.8 ± 1.3%) versus HD (11.3 ± 2.8%, Fig. 2 A). With the exception of two donors, this increase was not observed in patients with other sleep disorders (16.4 ± 2.7%). Interestingly, a higher frequency of CD69 expression was further confirmed in a monozygotic twin pair discordant for the disease (N09 and HD01), where the narcoleptic twin also expressed increased levels of CD69 on CD45RA+CCR7+ Th cells compared with the healthy sibling (Fig. 2 B).
Figure 2.
Activation and increased Th cell cytokine production in narcolepsy patients. FlowSOM-defined CD4+ Th cells of narcolepsy patients (Narco, n = 39), HD (n = 25), and patients with other hypersomnias (n = 11) were examined for CD69 expression and cytokine production. (A) Heat map depiction of the individual frequency of CD69+ cells in CD45RA+CCR7+ Th cells was sorted from highest to lowest frequency for all narcolepsy patients and controls within the cohort. Each square represents one individual. (B) CD69 expression by CD45RA+CCR7+ Th cells in a monozygotic twin pair discordant for narcolepsy. (C) Manually determined frequencies of cytokine+ antigen-experienced (effector, effector memory, and central memory) Th cells. (D) Production of TNF, IL-2, and IL-17A by Th cells as in C. (E) FlowSOM (k = 17) was used to cluster Th cells based on their cytokine-production profiles. The major Th cell subsets were manually annotated using these cytokine production profiles in combination with their CCR expression. (F) Group-specific composition and (G) statistical comparison of the frequencies of Th cell subsets in narcolepsy patients and controls. (H) Frequencies of the indicated regulatory immune cell populations were defined by manual gating. T reg cells were defined as CD25+CD127−IL-2− Th cells. Boxplots represent the IQR with a black horizontal line indicating the median. Whiskers extend to the farthest data point within a maximum of 1.5× IQR. Values outside this range are plotted as points. All p-values were calculated using a nonparametric Mann-Whitney-Wilcoxon test and are comparing narcolepsy patients with HD. Controlling for multiple comparisons was performed with the Benjamini-Hochberg approach. *, significant with FDR < 10%; **, significant with FDR < 5%.
Given the high-dimensional nature of mass cytometry, we were able to simultaneously analyze the production of up to 12 cytokines on antigen-experienced CD4+ T cells. Manually defining cytokine-positive cells, we found the frequency of TNF and IL-2–producing cells to be increased in antigen-experienced Th cells from narcolepsy patients (63.9 ± 4.2% TNF and 54.6 ± 2.7% IL-2) versus HD (47.5 ± 2.3% TNF and 46.2 ± 5.9% IL-2). Importantly, patients with other hypersomnias resembled HD (53.1 ± 8.7% TNF and 39.7 ± 12.0% IL-2; Fig. 2 C). Additionally, the frequency of several Th2-associated cytokines, such as IL-4 and IL-13, was increased in narcolepsy patients (3.5 ± 0.5% IL-4 and 3.5 ± 0.2% IL-13) versus HD (2.6 ± 0.3% IL-4 and 2.5 ± 0.2% IL-13). To a lesser extent, IL-3 did also show an increased frequency in narcolepsy patients. Again, patients with other hypersomnia did not show these augmented frequencies. The frequency of IL-6–producing Th cells was reduced in narcolepsy patients compared with controls, whereas IL-21–producing cells displayed a trend toward a higher frequency in narcolepsy patients (5.0 ± 0.7% Narco vs. 3.5 ± 0.4% HD; P = 0.049).
The frequencies of IFN-γ+ and IL-17A+ T cells, which are heavily implicated in other autoimmune diseases, including multiple sclerosis (Dendrou et al., 2015) and psoriasis (Becher and Pantelyushin, 2012; Perera et al., 2012), were not overtly altered in narcolepsy patients, as was the production of the immunosuppressive IL-10 by Th cells (Fig. 2, C–G). A more detailed analysis of individual subpopulations provided similar results for effector, effector memory, and central memory Th cells, demonstrating that the augmented cytokine production in narcolepsy patients is not driven by a specific subset (unpublished data).
To identify Th cell subsets based on the coexpression of the aforementioned cytokines in a more unbiased and data-driven approach, we again made use of the FlowSOM clustering algorithm. FlowSOM identified several subsets of Th1 and Th2 cells, as well as Th cells resembling Th17, Th22, and regulatory T (T reg) cell subsets (Fig. 2, E–H). Using this data-driven subset identification, we found the fraction of cytokine-negative cells to be markedly reduced in narcolepsy patients (37.8 ± 2.7%), compared to patients with other hypersomnias (54.9 ± 14.3%) or HD (51.5 ± 6.0%). Instead, narcolepsy patients had a strongly increased frequency of TNF+IL-2+, but IFN-γ−, Th cells (32.8 ± 1.8% Narco vs. 21.6 ± 4.0% HD). Confirming the results obtained using manual gating, this automated clustering also identified increased frequencies of Th2 cells in narcolepsy patients (6.1 ± 0.9%) vs. HD (2.7 ± 0.5%).
Augmented cytokine production might stem from changes in the frequency of regulatory immune subsets. However, we did not find any overt modulation of the frequency of T reg cells, IL-10–producing B cells, or CD56high NK cells in narcolepsy patients versus controls (Fig. 2 H).
Expansion of B cell–supporting CD8+ T cell populations
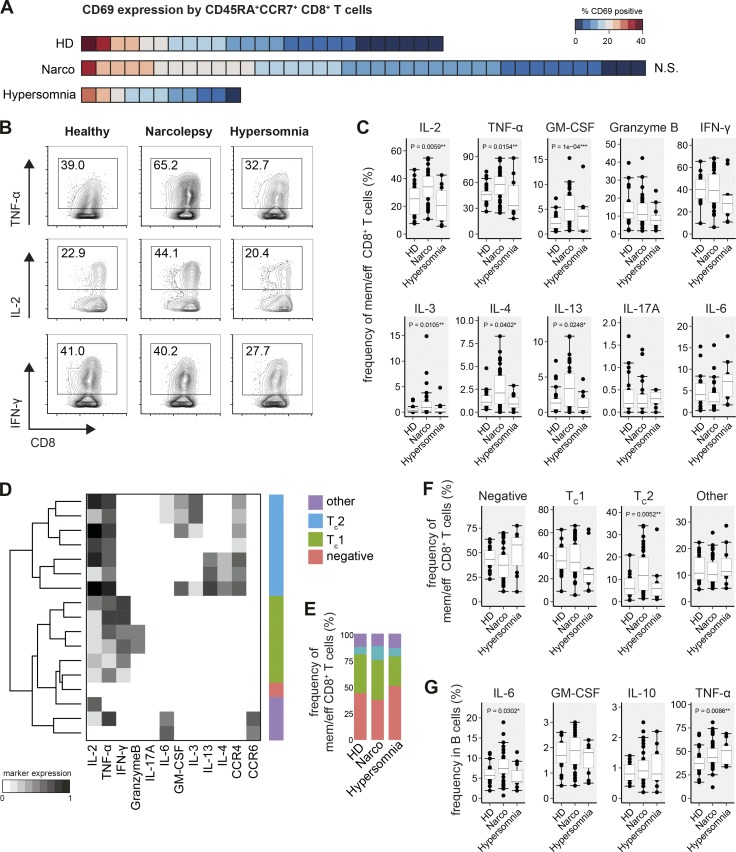
Besides CD4+ Th cells, CD8+ T cells are also thought to play a major role in CNS inflammation and autoimmunity (Waisman et al., 2015; Yshii et al., 2015), and in an experimental model of narcolepsy (Bernard-Valnet et al., 2016). Based on the activation and overt modulation of cytokine production by CD4+ T cells, we analyzed whether in narcolepsy patients, CD8+ T cell subsets are activated and show altered cytokine production profiles. Comparing CD69 expression on CD45RA+CCR7+ CD8+ T cells in narcolepsy patients versus HD, we did not find significant differences in the activation state of this population (Fig. 3 A).
Figure 3.
Expansion of B cell-supporting CD8+ T cells in narcolepsy. FlowSOM-defined CD8+ T cells of narcolepsy patients (Narco, n = 39), HD (n = 25), and patients with other hypersomnias (n = 11) were examined for CD69 expression and cytokine production. (A) Heat map depiction of the frequency of CD69+ cells in CD45RA+CCR7+ CD8+ T cells was sorted from highest to lowest frequency for all narcolepsy patients and controls. Each square represents one individual. (B) Manually determined frequencies of cytokine+ antigen-experienced (effector, effector memory, and central memory) CD8+ T cells and (C) statistical comparison of the frequencies of cytokine+ cells. (D) FlowSOM (k = 17) was used to cluster antigen-experienced CD8+ T cells based on their cytokine-production profiles. The major CD8+ T subsets were manually annotated using these cytokine production profiles in combination with their CCR expression. (E) Group-specific composition and (F) statistical comparison of the frequencies of CD8+ T cell subsets in narcolepsy patients and controls. (G) Manually gated frequencies of cytokine+ B cells. Boxplots represent the IQR with a black horizontal line indicating the median. Whiskers extend to the farthest data point within a maximum of 1.5× IQR. Values outside this range are plotted as points. All p-values were calculated using a nonparametric Mann-Whitney-Wilcoxon test and compare narcolepsy patients with HD. Controlling for multiple comparisons was performed with the Benjamini-Hochberg approach. *, significant with FDR < 10%; **, significant with FDR < 5%; ***, significant with FDR < 1%.
We found increased frequencies of TNF and IL-2 producing CD8+ T cells in narcolepsy patients (57.7 ± 3.7% TNF and 34.0 ± 2.3% IL-2) compared with HD (45.7 ± 2.7% and 25.4 ± 4.1%, respectively; Fig. 3, B and C). Additionally, GM-cerebrospinal fluid (CSF) and type 2 cytokines (IL-3, IL-4, and IL-13) were also augmented in CD8+ T cells of narcolepsy patients. Similar to CD4+ T cells, the levels of IFN-γ– and IL-17A–producing cells were not differentially regulated in narcolepsy patients versus control groups. Production of IL-6 or Granzyme B by CD8+ T cells was also comparable between narcolepsy patients (10.8 ± 1.9% Granzyme B) and HD (11.8 ± 2.1% Granzyme B; Fig. 3 C). Notably, the CD8+ T cell cytokine profile was similar between patients with other hypersomnias and HD and thus very distinct from that of narcolepsy patients (Fig. 3, B and C).
Next, again using an algorithm-guided approach, we clustered antigen-experienced CD8+ T cells based on their cytokine production profiles and annotated these clusters into TC1, TC2, cytokine-negative, and other subtypes (Fig. 3 D). Comparing the frequencies of these CD8+ T cell subsets, we found a striking increase in TC2-like cells in narcolepsy patients (11.7 ± 1.7%) compared with HD (5.8 ± 1.7%) and to patients with other hypersomnias (5.9 ± 2.0%; Fig. 3, E and F). This increase seemed to be at the expense of cytokine-negative cells, which were decreased in narcolepsy. The frequency of TC1-like cells was comparable in all three groups (Fig. 3, E and F).
Given the prominent production of B cell–supporting cytokines by CD4+ and CD8+ T cells, we examined cytokine expression profiles of B cells in narcolepsy patients (Fig. 3 G). The frequency of B cells producing IL-6 (7.4 ± 0.6% Narco vs. 5.7 ± 0.5% HD) and TNF (43.3 ± 2.0% Narco vs. 37.3 ± 1.9% HD) was augmented in narcolepsy patients; however, increased TNF production was also observed in patients with other hypersomnias (50.7 ± 7.0%).
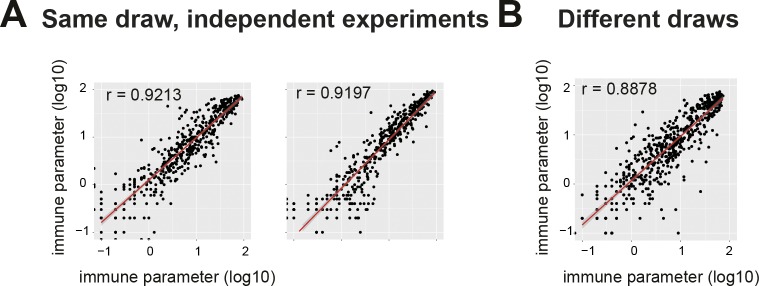
Technical stability of narcolepsy immune phenotyping
To confirm the narcolepsy-associated immune phenotypes, we performed repeated measurements of narcolepsy samples from the same and different blood draws (Fig. 4). Frequencies of immune phenotypes from independent measurements of separate aliquots taken from a single blood draw displayed a strong correlation, thus demonstrating the technical reproducibility of the method (Fig. 4 A). Further, samples drawn from patients on different occasions also displayed virtually identical immune phenotypes (Fig. 4 B), resembling the correlation of repeated measurements of the same samples.
Figure 4.
Stability of the narcolepsy-related immune phenotypes across independent experiments and different draws. Log-transformed frequencies of all previously defined immune phenotypes assessed by multiple measurements. (A) Different aliquots from the same draws were repeatedly measured in independent experiments (two independent repetitions; n = 10, left; n = 12, right). (B) Repeated measurements of narcolepsy samples from different blood draws (n = 12). Correlation coefficients were calculated using a linear regression model.
Narcolepsy-associated immune phenotypes in Pandemrix-vaccinated patients
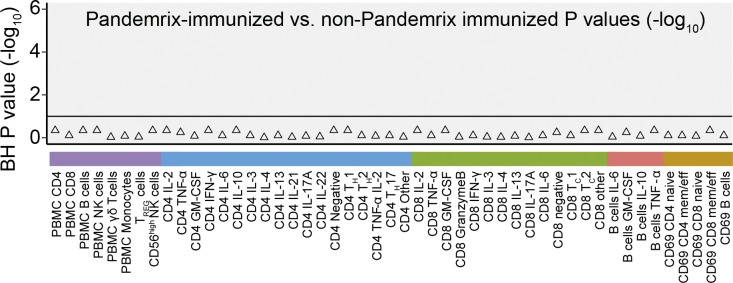
The cohort of narcolepsy patients comprised patients with a broad range of disease durations (8–552 mo). Among patients with shorter disease duration (median, 44 mo; range, 8–72 mo) were individuals in whom the onset of narcolepsy occurred after Pandemrix immunization (n = 11; Narco Pan) and patients who did not receive the Pandemrix vaccine (n = 12). Comparing these groups of patients matched for disease duration and age, we found that Pandemrix-vaccinated and non–Pandemrix-vaccinated patients displayed no statistically significant difference in the aforementioned immune phenotypes (Fig. 5).
Figure 5.
Immune phenotypes of narcolepsy patients stratified for Pandemrix vaccination. Benjamini-Hochberg corrected log-transformed p-values derived from Mann-Whitney-Wilcoxon tests comparing Pandemrix-vaccinated (n = 11) with non-Pandemrix vaccinated narcolepsy patients (n = 12). Only narcolepsy patients matched for disease duration and age were included in this comparison. The horizontal line indicates p-values that are considered significant with a FDR < 10%.
Narcolepsy-associated immune phenotypes are influenced by age and disease duration
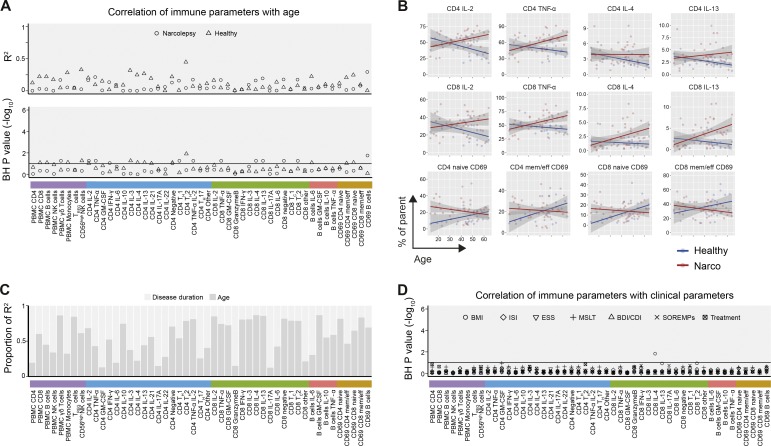
Next, we determined whether the immune alterations observed in narcolepsy patients versus controls are associated with any clinical characteristic (Table S1). First, using linear modeling, we assessed the influence of age and disease duration on immune phenotypes in narcolepsy patients and HD. We found a detectable influence of age to several immune frequencies in narcolepsy patients and HD, accounting in part for the variance in these phenotypes (median R2 = 0.1; Fig. 6 A).
Figure 6.
Narcolepsy-associated immune phenotypes are influenced by age and disease duration. Immune phenotypes of all narcolepsy patients (n = 39) and HD (n = 25) were included in this analysis. (A) R2-values (top) and –log10 of Benjamini-Hochberg–adjusted p-values (bottom) of linear regression models relating immunological phenotypes with age in narcolepsy patients and HD. The horizontal line indicates p-values that are considered significant with a FDR < 10%. (B) Changes of selected immune phenotypes for narcolepsy patients (red) and HD (blue) with age. The shaded area represents the 95% confidence interval of the linear model. (C) Proportion of R2 for narcolepsy patients (n = 39) explained by disease duration and age. (D) Benjamini-Hochberg-adjusted p-values of linear models relating immunological phenotypes in narcolepsy patients with clinical parameters.
Interestingly, narcolepsy-associated cytokine production by CD4+ and CD8+ T cells tended to increase with age in patients but not in HD (Fig. 6 B). In contrast, these trends were reversed for CD69 expression on T cells. As expected, the age of the narcolepsy patients correlated with disease duration. Therefore, we constructed a linear model using both variables as predictors to assess their relative importance. Disease duration and age both contributed almost equally to the explained variance in the immune phenotype of the narcolepsy patients (mean proportion of R2 0.44 and 0.56, respectively; Fig. 6 C).
Narcolepsy, like many other chronic diseases, has several clinical manifestations and comorbidities which may influence immune parameters. To control for potential confounding factors to the reported immunological phenotypes, we next investigated the contribution of patients’ clinical and neurophysiological characteristics, such as subjective (using the Epworth sleepiness scale [ESS]) and objective measures of daytime sleepiness (using the multiple sleep latency test [MSLT] and the number of sleep-onset rapid-eye-movement periods [SOREMPs]), disturbed sleep at night (using the insomnia severity index [ISI]), body-mass index (BMI), and self-reported depression (using the Beck depression inventory or children’s depression inventory [BDI/CDI]), as well as the patient’s treatment status (Fig. 6 D). We found that only IL-4 production by CD8+ T cells correlated with one of these parameters (BMI), whereas we did not observe any substantial influence of all other parameters on any of the analyzed immunological phenotypes (Fig. 6 D).
Discussion
In this study, we performed a comprehensive, explorative analysis of narcolepsy type 1 patients in which we studied numerous immunological phenotypes at the single-cell level. To do so, we made use of mass cytometry, which in combination with unbiased, data-driven analysis methods, has proven to be a valuable tool in the hypothesis-free identification of cellular phenotypes associated with cancer (Behbehani et al., 2015; Levine et al., 2015) or clinical recovery after surgery (Gaudillière et al., 2014). Using this approach in the context of a more elusive, putative autoimmune disease, we found complex patterns of immune activation in narcolepsy patients. Central to many of these narcolepsy-associated phenotypes was their association with B cell–mediated immune responses. This is in line with previous indications of autoantibodies in narcolepsy (Cvetkovic-Lopes et al., 2010; Bergman et al., 2014; Ahmed et al., 2015; Saariaho et al., 2015) and recent studies that further support an important role of B cells in the pathogenesis of T cell–driven CNS autoimmunity (Krumbholz et al., 2012; Flach et al., 2016).
Specifically, we found that CD4+ and CD8+ T cells of narcolepsy patients produced elevated levels of several cytokines involved in B cell differentiation, including IL-4, IL-13, and IL-21. In addition, we found strikingly elevated frequencies of TNF and IL-2–producing CD4+ and CD8+ T cells in narcolepsy patients. Similarly, B cells of narcolepsy patients also expressed TNF at a higher frequency than HD. These findings are in line with TNF being previously implicated in narcolepsy by genetic association (Hohjoh et al., 1999) and by increased plasma levels in narcolepsy patients (Himmerich et al., 2006; Tanaka et al., 2014). Interestingly, by performing linear modeling, we found this narcolepsy-specific polarization of cytokine production to become more pronounced in patients with longer disease duration and higher age. This analysis could not be performed in patients with other hypersomnias, as this group was underpowered to specifically study the impact of age on the measured immune parameters. However, the age-dependent progression of cytokine production in narcolepsy patients is striking and matches previous studies reporting increased levels of IL-4 in the CSF of narcolepsy patients with long disease duration (Dauvilliers et al., 2014), but not patients with short disease duration (Kornum et al., 2015).
This might reflect the presence of an underlying inflammatory milieu in narcolepsy patients, which is absent in nondiseased controls. Several factors could contribute to such an environment, among them disturbance of sleeping patterns (Vgontzas et al., 1997; Ackermann et al., 2012; Aho et al., 2013; Lasselin et al., 2015), circadian rhythm (Scheiermann et al., 2013; Spitzer et al., 2015), treatment with psychostimulants (Filiano et al., 2016), and associated metabolic changes (Wolowczuk et al., 2008). To control for these potential effects, we included patients with sleep disorders that exhibit similar manifestations and comorbidities and undergo overlapping treatment. Importantly, these hypersomnia patients were immunologically distinct from narcolepsy patients and did not present increased cytokine production found in narcolepsy patients. However, type 1 narcolepsy patients tend to have more severe sleepiness and more disturbed sleep compared with patients with other hypersomnia. Therefore, we directly modeled the potential influence of sleepiness, disturbed night-time sleep, depressive symptoms, BMI, and treatment status on all reported immune phenotypes. Besides IL-4 expression by CD8+ T cells, we did not find any correlation of these clinical parameters with the narcolepsy-associated immune phenotypes. Apart from these patient characteristics, it is possible that independent from narcolepsy, the genetic background contributes to this immune polarization over time. However, to control for this effect, we matched the HD with regard to the major susceptibility gene variant, presence of the HLA-DQB1*06:02 allele.
Contrasting with the increase of narcolepsy-associated cytokine patterns over time, elevated expression of CD69 on T cells from narcolepsy patients was most pronounced in younger patients with short disease duration, and decreased over time. Together with the modulation of cytokine production patterns, this suggests a progressive shift from early T cell activation to functional polarization. Longitudinal follow up of cohorts of patients with narcolepsy should help clarify this aspect.
Narcolepsy patients matched for age and short disease duration but grouped into Pandemrix-vaccinated and non–Pandemrix-vaccinated were found to be largely indistinguishable by single-cell cytometry (Carr et al., 2016). In agreement with studies on genetic (Nguyen et al., 2016) and clinical (Pizza et al., 2014) features of post-Pandemrix versus sporadic narcolepsy, our results support a shared pathophysiology regardless of vaccine association. Whether exposure to similar environmental factors (for instance vaccination vs. natural infection) is the reason for the lack of detectable differences between Pandemrix-vaccinated and non–Pandemrix-vaccinated patients remains to be determined. Lastly, although mass cytometry enabled us to simultaneously analyze 45 parameters and use this data to define numerous immunological phenotypes, this study mostly focused on lymphocyte activation, maturation, and cytokine production and did not include parameters necessary to study B cell maturation/activation in depth.
This analysis included a monozygotic twin pair where both siblings received Pandemrix-vaccination but only one developed narcolepsy. Although the narcolepsy-associated augmented CD69 expression was confirmed in this single pair, the cellular composition was found to be similar in both twins. Study of such monozygotic twin pairs in a clinical context can be extremely valuable. However, given the relative rarity of narcolepsy, only a single twin pair could be included here, which limits the possibility to draw firm conclusions regarding the pathogenesis of narcolepsy in genetically identical individuals.
Given technical considerations and the limitation in sample availability of this rare disorder, this study addressed changes in the recirculating immune system but not the tissue-infiltrating leukocyte compartment of the CNS directly. However, recent studies suggest a previously underappreciated functional link between the CNS and peripheral immunity (Kim et al., 2015; Louveau et al., 2015), and we thus believe that these results will be highly beneficial to guide future studies in generating more precise hypotheses which can be verified, e.g., in the CSF of patients.
Because of the unknown nature of a potential autoantigen in narcolepsy, we could not investigate the antigen-specificity of lymphocyte populations. This remains an aspect of great importance and it will be interesting to see whether the above-indicated expansion of certain T cell subsets would concur with a specific pattern of clonal expansion.
A surprisingly high frequency of T cells in patients with narcolepsy exhibited an inflammatory signature characterized by elevated expression of activation markers or cytokine production. This frequency by far exceeds the expected frequency of (auto)antigen-specific T cells as detected by MHC/peptide tetramer staining in other contexts (Pepper et al., 2010; Yu et al., 2015). It is unlikely, due to lack of sensitivity of current tools, to selectively analyze disease-causing autoreactive T cells, although innovative tools are now available to test this hypothesis provided autoantigens and HLA-binding peptide thereof, are identified (Huang et al., 2016). More likely, disease progression impacts on general immune cell activation and function. Precedent for broad T cell alterations in organ-specific autoimmune diseases has been reported, although the exact mechanisms involved still escape current understanding (McKinney et al., 2015).
Although sharing many clinical characteristics with type 1 narcolepsy, patients with other hypersomnias do not suffer from the lack of orexin-producing neurons and associated cataplexy episodes, both of which are used for the diagnosis of narcolepsy. For this study, we measured CSF orexin levels for the majority of narcolepsy patients (32 of 39) and patients with other causes of hypersomnia (10 of 11), whereas the remaining were classified based on clinical parameters and the documented presence or absence of cataplexy according to International Classification of Sleep Disorders, third edition guidelines (American Academy of Sleep Disorders, 2014). The impact of chronic depletion of the orexin neuropeptide on immune cell activation is unknown. It is tempting to speculate that orexin can affect immune cell biology and that its chronic deficiency in patients with narcolepsy somehow promotes general immune dysregulation.
Collectively, we believe that this high-dimensional, single-cell analysis approach represents a comprehensive account of the immunological signature found in narcolepsy patients. It revealed narcolepsy-associated immune activation and increased B cell–supporting cytokine production, the magnitude of which is dependent on disease duration and age. These findings thus offer insight into the nature of the underlying history and current peripheral immune-milieu in this elusive disease. In addition, it provides a rationale for the further investigation of lymphocytes and associated effector mechanisms as therapeutic targets in narcolepsy.
Materials and methods
Patient samples
Patients (Table 1 and Table S1) were characterized and their peripheral blood was collected at the National Reference Center for Orphan Sleep Disorders (Gui-de-Chauliac Hospital, Montpellier, France). Cryopreservation of PBMCs from patients and from healthy volunteers was performed at Institut National de la Santé et de la Recherche Médicale U1043 (Toulouse, France) within 24 h of blood collection. Narcolepsy type 1 was diagnosed according to International Classification of Sleep Disorders, third edition based on the presence of excessive daytime sleepiness, clear-cut cataplexy, ≥2 SOREMPs, and mean sleep latency <8 min on the MSLT, or CSF orexin levels <110 pg/ml. A video documentation of clear-cut cataplexy was available to confirm the diagnosis in all patients without CSF orexin measurement. All patients were HLA-DQB1*06:02 positive. Samples defined as other sleep disorders showed normal CSF orexin concentrations, i.e., >200 pg/ml. All patients gave their written informed consent to take part in the research program, which was approved by the local ethics committee (Comité de Protection des Personnes–Ile de France 06). All mass cytometry experiments and their initial analysis were performed blinded.
Genotyping
HLA typing of narcolepsy patients was performed by PCR sequence-specific oligonucleotides and four-digit allele assignments were performed by Luminex xMAP technology. To resolve ambiguous typing or degeneracy, direct PCR-sequencing with the following primers was used: forward, 5′-CCCGCAGAGGATTTCGTGTT-3′; reverse, 5′-AACTCCGCCCGGGTCCC-3′. Internal control primers: forward, 5′-TGCCAAGTGGAGCACCCAA-3′; reverse, 5′-GCATCTT-GCTCTGTGCAGAT-3′. Genomic DNA was amplified at 94°C for 15 min, followed by 35 cycles of 94°C for 1 min, 63°C for 1 min, 72°C for 1 min, and 72°C for 10 min. The amplification produced a 217-bp band in the presence of DQB1*06:02, and a 700–730-bp internal control band.
Ex vivo reactivation of PBMCs
Cryopreserved PBMCs were stored in liquid nitrogen until thawing in a 37°C water bath for 7 min. Cells were resuspended gently in 1 ml of prewarmed cell culture medium (CCM; RPMI-1640 [PAN biotech], 10% FCS [Biochrom]), 1× l-glutamine, and 1× penicillin/streptomycin [both Life Technologies]) supplemented with 1:10,000 benzonase (Sigma-Aldrich), transferred into tubes containing 10 ml of CCM, and centrifuged (300 rcf, 7 min, 24°C). Washing with CCM was repeated once before counting and adjusting to 106 cells/ml in CCM and resting overnight at 37°C. To determine cytokine production by mass cytometry, cells were stimulated with 50 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) and 500 ng/ml ionomycin (Sigma-Aldrich) in the presence of 1x Brefeldin A (BD) for 4 h at 37°C.
Antibodies
Monoclonal anti–human antibodies (Table S2) were purchased either preconjugated to heavy-metal isotopes from Fluidigm or from commercial suppliers in purified form and conjugated in house using the Maxpar X8 chelating polymer kit (Fluidigm) according to the manufacturer's instructions.
Live cell barcoding
To reduce inter-sample staining variability, minimize sample-handling time, and reduce antibody consumption, we made use of the live-cell barcoding approach (Mei et al., 2015). Therefore, isothiocyanobenzyl-EDTA (p-SCN-EDTA; Dojindo) was loaded with five different palladium isotopes (104Pd, 105Pd, 106Pd, 108Pd, and 110Pd) and one indium isotope (115In; all from Trace Sciences International) as previously described (Zunder et al., 2015) and subsequently used to label anti–human CD45 (BioLegend). In addition, preconjugated 89Y-CD45 (Fluidigm) was used as a seventh barcoding reagent. To obtain robust barcoding results with even staining intensity on all samples, either a 6-choose-2 or a 7-choose-3 scheme was used, resulting in 15 or 35 different combinations of two heavy metal–labeled CD45 antibodies, respectively. PBMCs were labeled with metal-tagged CD45 antibodies after ex vivo reactivation at 37°C for 25 min in cell staining medium (CSM; RPMI-1640, 4% FCS) on a rotating shaker (500 rpm). Samples were washed twice in CSM and combined into a single reaction vessel for further staining steps.
Surface staining
After live cell barcoding, the combined sample was centrifuged, supernatant was aspirated, and cells were resuspended in 300 µl of antibodies (Table S2) in CSM. Cells were stained for 20 min at 37°C on a rotating shaker (500 rpm) to optimize CCR staining (Berhanu et al., 2003). Dead cells were excluded by adding 1 ml of 2.5 µM cisplatin (Sigma-Aldrich) in PBS at the end of the staining period and incubating for 2 min on ice (Fienberg et al., 2012). Afterward, the sample was washed once in PBS supplemented with 0.5% BSA (PBS/BSA; Sigma-Aldrich).
Intracellular cytokine staining
Surface-stained cells were fixed by resuspending them in 1.6% paraformaldehyde (PFA; Electron Microscopy Sciences) for 30 min at 4°C on a rotary shaker (500 rpm). Afterward, the sample was washed twice with permeabilization buffer (PBS, 0.5% saponin, 2% BSA, and 0.01% sodium azide [all from Sigma-Aldrich]), the supernatant was aspirated and the sample was resuspended in 300 µl of antibody mixture (Table S2) in permeabilization buffer for 1 h at 4°C on a rotary shaker (500 rpm). The sample was washed with permeabilization buffer, supernatant was aspirated, and the cells were resuspended in 500 µl of iridium intercalator solution (Fluidigm) overnight. Next, the sample was washed twice with PBS/BSA, once with ddH2O, resuspended at 0.5–106 cells/ml and filtered through a cell strainer cap (Falcon) before acquisition.
Acquisition on CyTOF2 mass cytometer and data preprocessing
Samples were acquired on a CyTOF2 mass cytometer (DVS Sciences/Fluidigm). Quality control and tuning processes were performed on a daily basis before acquisition. Data from different days was normalized by adding five-element beads (Fluidigm) to the sample immediately before acquisition and using the Matlab-based normalization software as described previously (Finck et al., 2013). Only events with Event_length between 10 and 100 were exported. Before downstream analysis, live cells were exported by manual gating on Event_length, DNA (191Ir and 193Ir), and live cells (195Pt) using FlowJo software (Tree Star). Next, cells were assigned to their initial samples using a dedicated debarcoding software (Zunder et al., 2015). Bead-normalized files were imported into the R environment and transformed using an inverse hyperbolic sine (arcsinh) function with a cofactor of 5 (Bendall et al., 2011). To equalize the contribution of each marker in subsequent automated data analysis steps, we performed background subtraction and percentile normalization (Levine et al., 2015). Therefore, we normalized all data to the 99.5th percentile of the combined (barcoded) sample. This preserves inter-sample variability in maximum expression values, which might be biologically relevant. Markers expressed with a low frequency of PBMCs (mostly cytokines) were normalized to the 99.995th percentile.
Automated population identification in high-dimensional data analysis
After preprocessing, all samples from all cohorts were down sampled to a maximum of 100’000 cells per sample, combined into one file and subjected to the FlowSOM clustering algorithm to identify meaningful immunological populations. We choose FlowSOM over other clustering options for several reasons: (i) in our hands it identified clusters which efficiently represented immune populations that we expected to be present based on prior biological knowledge, (ii) its availability within the R environment which enabled us to perform large parts of the analysis in a streamlined way, (iii) it was able to perform clusterings within a few minutes on a standard desktop computer enabling us to run it multiple times and thus compare the influence of different input parameters, especially the k-value used in the meta-clustering step. FlowSOM was run on the combined sample to facilitate the identification of small populations that could be overlooked when clustering is performed on each single sample. After the initial clustering step, resulting nodes were subjected to meta-clustering. The respective k-value was chosen manually and in a way so that we were still able to identify all populations that were expected to be present, based on general biological knowledge. Heat maps display median expression levels for the indicated populations and were drawn using the ggplot2 R package. Dendrograms were calculated using hierarchical clustering. Correlation coefficients were calculated using the stats package.
Statistical analysis
Population frequencies or medians in narcolepsy and HD samples were compared using a nonparametric Mann-Whitney-Wilcoxon test. Samples with a low live cell count were excluded from the final analysis. In case we tested for the modulation of multiple markers or clusters at once, we controlled for multiple comparisons by using the Benjamini-Hochberg approach using R (Noble, 2009; McDonald, 2014). P-values were reported together with an estimate of the false discovery rate with which the p-values would be considered significant. Linear regression was performed with the R base function lm and the relative contributions were calculated with the relaimpo package (Grömping, 2006).
Online supplemental material
Table S1 shows clinical and demographic characteristics of all individuals evaluated in this study. Table S2 shows the heavy metal-labeled antibody panel for mass cytometry. Table S3 shows frequencies of the reported immune phenotypes for all individuals evaluated in this study. Tables S1–S3 are available as Excel files.
Supplementary Material
Acknowledgments
We acknowledge Raphaelle Romieu-Mourez and Pierre-Emmanuel Paulet of the Toulouse immunomonitoring platform for the PBMC biobank and we thank V. Tosevski (Mass Cytometry Facility, University of Zurich) for technical assistance and helpful discussions regarding all aspects of mass cytometry.
This study was supported by grants from the Swiss National Science Foundation 316030_150768 (to B. Becher) and 310030_146130 (to B. Becher), Toulouse University (Idex grant Emergence 2013 to R.S. Liblau), ANR (CE14-0016-03 to Y. Dauvilliers and R.S. Liblau), the Occitanie Region (to X.-H. Nguyen), and Toulouse (R.S. Liblau) and Montpellier (Y. Dauvilliers) centers of excellence in neurodegenerative diseases.
The authors declare no competing financial interests.
Author contributions: F.J. Hartmann, R.S. Liblau, and B. Becher designed the experiments and analyzed the data. F.J. Hartmann performed the experiments. D. Mrdjen, E. Galli, and C. Krieg assisted with the experiments. Y. Dauvilliers characterized all the patients with sleep disorders involved in this study and revised the manuscript. R. Bernard-Valnet, C. Quériault, and X.-H. Nguyen collected and characterized PBMCs from all the subjects studied. L.M. Weber and M.D. Robinson provided support for data analysis. F.J. Hartmann, R.S. Liblau, and B. Becher wrote the manuscript. R.S. Liblau and B. Becher supervised and funded the study.
Footnotes
Abbreviations used:
- BDI
- Beck’s depression inventory
- BMI
- body mass index
- CCM
- cell culture medium
- CCR
- chemokine receptor
- CDI
- children’s depression inventory
- CSF
- cerebrospinal fluid
- CSM
- cell staining medium
- CyTOF
- cytometry by time-of-flight
- ESS
- Epworth sleepiness scale
- FDR
- false discovery rate
- HD
- healthy donor
- IQR
- interquartile range
- ISI
- insomnia severity index
- MSLT
- multiple sleep latency test
- SOREMP
- sleep-onset rapid-eye-movement period
- T reg cell
- regulatory T cell
- t-SNE
- t-stochastic neighbor embedding
References
- Ackermann K., Revell V.L., Lao O., Rombouts E.J., Skene D.J., and Kayser M.. 2012. Diurnal rhythms in blood cell populations and the effect of acute sleep deprivation in healthy young men. Sleep. 35:933–940. 10.5665/sleep.1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.S., Volkmuth W., Duca J., Corti L., Pallaoro M., Pezzicoli A., Karle A., Rigat F., Rappuoli R., Narasimhan V., et al. 2015. Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2. Sci. Transl. Med. 7:294ra105 10.1126/scitranslmed.aab2354 [DOI] [PubMed] [Google Scholar]
- Aho V., Ollila H.M., Rantanen V., Kronholm E., Surakka I., van Leeuwen W.M.A., Lehto M., Matikainen S., Ripatti S., Härmä M., et al. 2013. Partial sleep restriction activates immune response-related gene expression pathways: experimental and epidemiological studies in humans. PLoS One. 8:e77184 10.1371/journal.pone.0077184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Academy of Sleep Disorders 2014. International Classification of Sleep Disorders. Third edition Sateia M., editor. American Academy of Sleep Medicine, Darien, IL. 383 pp. [Google Scholar]
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher B., and Pantelyushin S.. 2012. Hiding under the skin: Interleukin-17-producing γδ T cells go under the skin? Nat. Med. 18:1748–1750. 10.1038/nm.3016 [DOI] [PubMed] [Google Scholar]
- Behbehani G.K., Samusik N., Bjornson Z.B., Fantl W.J., Medeiros B.C., and Nolan G.P.. 2015. Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov. 5:988–1003. 10.1158/2159-8290.CD-15-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I., et al. 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332:687–696. 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall S.C., Nolan G.P., Roederer M., and Chattopadhyay P.K.. 2012. A deep profiler’s guide to cytometry. Trends Immunol. 33:323–332. 10.1016/j.it.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P., Adori C., Vas S., Kai-Larsen Y., Sarkanen T., Cederlund A., Agerberth B., Julkunen I., Horvath B., Kostyalik D., et al. 2014. Narcolepsy patients have antibodies that stain distinct cell populations in rat brain and influence sleep patterns. Proc. Natl. Acad. Sci. USA. 111:E3735–E3744. 10.1073/pnas.1412189111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhanu D., Mortari F., De Rosa S.C., and Roederer M.. 2003. Optimized lymphocyte isolation methods for analysis of chemokine receptor expression. J. Immunol. Methods. 279:199–207. 10.1016/S0022-1759(03)00186-8 [DOI] [PubMed] [Google Scholar]
- Bernard-Valnet R., Yshii L., Quériault C., Nguyen X.-H., Arthaud S., Rodrigues M., Canivet A., Morel A.-L., Matthys A., Bauer J., et al. 2016. CD8 T cell-mediated killing of orexinergic neurons induces a narcolepsy-like phenotype in mice. Proc. Natl. Acad. Sci. USA. 113:10956–10961. 10.1073/pnas.1603325113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A., and Liston A.. 2016. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 17:461–468. 10.1038/ni.3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovic-Lopes V., Bayer L., Dorsaz S., Maret S., Pradervand S., Dauvilliers Y., Lecendreux M., Lammers G.-J., Donjacour C.E.H.M., Du Pasquier R.A., et al. 2010. Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J. Clin. Invest. 120:713–719. 10.1172/JCI41366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvilliers Y., Arnulf I., and Mignot E.. 2007. Narcolepsy with cataplexy. Lancet. 369:499–511. 10.1016/S0140-6736(07)60237-2 [DOI] [PubMed] [Google Scholar]
- Dauvilliers Y., Jaussent I., Lecendreux M., Scholz S., Bayard S., Cristol J.P., Blain H., and Dupuy A.-M.. 2014. Cerebrospinal fluid and serum cytokine profiles in narcolepsy with cataplexy: a case-control study. Brain Behav. Immun. 37:260–266. 10.1016/j.bbi.2013.12.019 [DOI] [PubMed] [Google Scholar]
- Dendrou C.A., Fugger L., and Friese M.A.. 2015. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15:545–558. 10.1038/nri3871 [DOI] [PubMed] [Google Scholar]
- Fienberg H.G., Simonds E.F., Fantl W.J., Nolan G.P., and Bodenmiller B.. 2012. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A. 81:467–475. 10.1002/cyto.a.22067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano A.J., Xu Y., Tustison N.J., Marsh R.L., Baker W., Smirnov I., Overall C.C., Gadani S.P., Turner S.D., Weng Z., et al. 2016. Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature. 535:425–429. 10.1038/nature18626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R., Simonds E.F., Jager A., Krishnaswamy S., Sachs K., Fantl W., Pe’er D., Nolan G.P., and Bendall S.C.. 2013. Normalization of mass cytometry data with bead standards. Cytometry A. 83:483–494. 10.1002/cyto.a.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach A.-C., Litke T., Strauss J., Haberl M., Gómez C.C., Reindl M., Saiz A., Fehling H.-J., Wienands J., Odoardi F., et al. 2016. Autoantibody-boosted T-cell reactivation in the target organ triggers manifestation of autoimmune CNS disease. Proc. Natl. Acad. Sci. USA. 113:3323–3328. 10.1073/pnas.1519608113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudillière B., Fragiadakis G.K., Bruggner R.V., Nicolau M., Finck R., Tingle M., Silva J., Ganio E.A., Yeh C.G., Maloney W.J., et al. 2014. Clinical recovery from surgery correlates with single-cell immune signatures. Sci. Transl. Med. 6:255ra131 10.1126/scitranslmed.3009701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grömping U. 2006. Relative Importance for Linear Regression in R: The Package relaimpo. J. Stat. Softw. 17:1–27. 10.18637/jss.v017.i01 [DOI] [Google Scholar]
- Han F., Faraco J., Dong X.S., Ollila H.M., Lin L., Li J., An P., Wang S., Jiang K.W., Gao Z.C., et al. 2013. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 9:e1003880 10.1371/journal.pgen.1003880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmerich H., Beitinger P.A., Fulda S., Wehrle R., Linseisen J., Wolfram G., Himmerich S., Gedrich K., Wetter T.C., and Pollmächer T.. 2006. Plasma levels of tumor necrosis factor alpha and soluble tumor necrosis factor receptors in patients with narcolepsy. Arch. Intern. Med. 166:1739–1743. 10.1001/archinte.166.16.1739 [DOI] [PubMed] [Google Scholar]
- Hohjoh H., Nakayama T., Ohashi J., Miyagawa T., Tanaka H., Akaza T., Honda Y., Juji T., and Tokunaga K.. 1999. Significant association of a single nucleotide polymorphism in the tumor necrosis factor-alpha (TNF-alpha) gene promoter with human narcolepsy. Tissue Antigens. 54:138–145. 10.1034/j.1399-0039.1999.540204.x [DOI] [PubMed] [Google Scholar]
- Huang J., Zeng X., Sigal N., Lund P.J., Su L.F., Huang H., Chien Y.H., and Davis M.M.. 2016. Detection, phenotyping, and quantification of antigen-specific T cells using a peptide-MHC dodecamer. Proc. Natl. Acad. Sci. USA. 113:E1890–E1897. 10.1073/pnas.1602488113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Yan J., Wu W., Zhang G., Zhang Y., and Cai D.. 2015. Rapid linkage of innate immunological signals to adaptive immunity by the brain-fat axis. Nat. Immunol. 16:525–533. 10.1038/ni.3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornum B.R., Pizza F., Knudsen S., Plazzi G., Jennum P., and Mignot E.. 2015. Cerebrospinal fluid cytokine levels in type 1 narcolepsy patients very close to onset. Brain Behav. Immun. 49:54–58. 10.1016/j.bbi.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumbholz M., Derfuss T., Hohlfeld R., and Meinl E.. 2012. B cells and antibodies in multiple sclerosis pathogenesis and therapy. Nat. Rev. Neurol. 8:613–623. 10.1038/nrneurol.2012.203 [DOI] [PubMed] [Google Scholar]
- Lasselin J., Rehman J.U., Åkerstedt T., Lekander M., and Axelsson J.. 2015. Effect of long-term sleep restriction and subsequent recovery sleep on the diurnal rhythms of white blood cell subpopulations. Brain Behav. Immun. 47:93–99. 10.1016/j.bbi.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir A.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. 2015. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 162:184–197. 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liblau R.S., Vassalli A., Seifinejad A., and Tafti M.. 2015. Hypocretin (orexin) biology and the pathophysiology of narcolepsy with cataplexy. Lancet Neurol. 14:318–328. 10.1016/S1474-4422(14)70218-2 [DOI] [PubMed] [Google Scholar]
- Louveau A., Smirnov I., Keyes T.J., Eccles J.D., Rouhani S.J., Peske J.D., Derecki N.C., Castle D., Mandell J.W., Lee K.S., et al. 2015. Structural and functional features of central nervous system lymphatic vessels. Nature. 523:337–341. 10.1038/nature14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair F., Hartmann F.J., Mrdjen D., Tosevski V., Krieg C., and Becher B.. 2016. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur. J. Immunol. 46:34–43. 10.1002/eji.201545774 [DOI] [PubMed] [Google Scholar]
- McDonald J. 2014. Handbook of Biological Statistics. Third edition Sparky House Publishing, Baltimore, Maryland. [Google Scholar]
- McKinney E.F., Lee J.C., Jayne D.R.W., Lyons P.A., and Smith K.G.C.. 2015. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 523:612–616. 10.1038/nature14468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H.E., Leipold M.D., Schulz A.R., Chester C., and Maecker H.T.. 2015. Barcoding of live human peripheral blood mononuclear cells for multiplexed mass cytometry. J. Immunol. 194:2022–2031. 10.4049/jimmunol.1402661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignot E., Hayduk R., Black J., Grumet F.C., and Guilleminault C.. 1997. HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep. 20:1012–1020. [PubMed] [Google Scholar]
- Nguyen X.-H., Saoudi A., and Liblau R.S.. 2016. Vaccine-associated inflammatory diseases of the central nervous system: from signals to causation. Curr. Opin. Neurol. 29:362–371. 10.1097/WCO.0000000000000318 [DOI] [PubMed] [Google Scholar]
- Noble W.S. 2009. How does multiple testing correction work? Nat. Biotechnol. 27:1135–1137. 10.1038/nbt1209-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila H.M., Ravel J.-M., Han F., Faraco J., Lin L., Zheng X., Plazzi G., Dauvilliers Y., Pizza F., Hong S.-C., et al. 2015. HLA-DPB1 and HLA class I confer risk of and protection from narcolepsy. Am. J. Hum. Genet. 96:136–146. 10.1016/j.ajhg.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M., Linehan J.L., Pagán A.J., Zell T., Dileepan T., Cleary P.P., and Jenkins M.K.. 2010. Different routes of bacterial infection induce long-lived Th1 memory cells and short-lived TH17 cells. Nat. Immunol. 11:83–89. 10.1038/ni.1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera G.K., Di Meglio P., and Nestle F.O.. 2012. Psoriasis. Annu. Rev. Pathol. 7:385–422. 10.1146/annurev-pathol-011811-132448 [DOI] [PubMed] [Google Scholar]
- Pizza F., Peltola H., Sarkanen T., Moghadam K.K., Plazzi G., and Partinen M.. 2014. Childhood narcolepsy with cataplexy: comparison between post-H1N1 vaccination and sporadic cases. Sleep Med. 15:262–265. 10.1016/j.sleep.2013.09.021 [DOI] [PubMed] [Google Scholar]
- Saariaho A.-H., Vuorela A., Freitag T.L., Pizza F., Plazzi G., Partinen M., Vaarala O., and Meri S.. 2015. Autoantibodies against ganglioside GM3 are associated with narcolepsy-cataplexy developing after Pandemrix vaccination against 2009 pandemic H1N1 type influenza virus. J. Autoimmun. 63:68–75. 10.1016/j.jaut.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Scammell T.E. 2015. Narcolepsy. N. Engl. J. Med. 373:2654–2662. 10.1056/NEJMra1500587 [DOI] [PubMed] [Google Scholar]
- Scheiermann C., Kunisaki Y., and Frenette P.S.. 2013. Circadian control of the immune system. Nat. Rev. Immunol. 13:190–198. 10.1038/nri3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M.H., Gherardini P.F., Fragiadakis G.K., Bhattacharya N., Yuan R.T., Hotson A.N., Finck R., Carmi Y., Zunder E.R., Fantl W.J., et al. 2015. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science. 349:1259425 10.1126/science.1259425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafti M., Hor H., Dauvilliers Y., Lammers G.J., Overeem S., Mayer G., Javidi S., Iranzo A., Santamaria J., Peraita-Adrados R., et al. 2014. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 37:19–25. 10.5665/sleep.3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafti M., Lammers G.J., Dauvilliers Y., Overeem S., Mayer G., Nowak J., Pfister C., Dubois V., Eliaou J.-F., Eberhard H.-P., et al. 2016. Narcolepsy-Associated HLA Class I Alleles Implicate Cell-Mediated Cytotoxicity. Sleep. 39:581–587. 10.5665/sleep.5532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Honda M., Toyoda H., and Kodama T.. 2014. Increased plasma IL-6, IL-8, TNF-alpha, and G-CSF in Japanese narcolepsy. Hum. Immunol. 75:940–944. 10.1016/j.humimm.2014.06.023 [DOI] [PubMed] [Google Scholar]
- Van Der Maaten L., and Hinton G.. 2008. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9:2579–2605. 10.1007/s10479-011-0841-3 [DOI] [Google Scholar]
- Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., and Saeys Y.. 2015. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 87:636–645. 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Vgontzas A.N., Papanicolaou D.A., Bixler E.O., Kales A., Tyson K., and Chrousos G.P.. 1997. Elevation of plasma cytokines in disorders of excessive daytime sleepiness: role of sleep disturbance and obesity. J. Clin. Endocrinol. Metab. 82:1313–1316. 10.1210/jcem.82.5.3950 [DOI] [PubMed] [Google Scholar]
- Waisman A., Liblau R.S., and Becher B.. 2015. Innate and adaptive immune responses in the CNS. Lancet Neurol. 14:945–955. 10.1016/S1474-4422(15)00141-6 [DOI] [PubMed] [Google Scholar]
- Wolowczuk I., Verwaerde C., Viltart O., Delanoye A., Delacre M., Pot B., and Grangette C.. 2008. Feeding our immune system: impact on metabolism. Clin. Dev. Immunol. 2008:639803 10.1155/2008/639803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yshii L., Gebauer C., Bernard-Valnet R., and Liblau R.. 2015. Neurons and T cells: Understanding this interaction for inflammatory neurological diseases. Eur. J. Immunol. 45:2712–2720. 10.1002/eji.201545759 [DOI] [PubMed] [Google Scholar]
- Yu W., Jiang N., Ebert P.J.R., Kidd B.A., Müller S., Lund P.J., Juang J., Adachi K., Tse T., Birnbaum M.E., et al. 2015. Clonal deletion prunes but does not eliminate self-specific αβ CD8(+) T lymphocytes. Immunity. 42:929–941. 10.1016/j.immuni.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B., et al. 2015. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10:316–333. 10.1038/nprot.2015.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.