Abstract
One of the most fascinating questions in the field of neurobiology is to understand how neuronal connections are properly formed. During development, neurons extend axons that are guided along defined paths by attractive and repulsive cues to reach their brain target. Most of these guidance factors are regulated by heparan sulfate proteoglycans (HSPGs), a family of cell-surface and extracellular core proteins with attached heparan sulfate (HS) glycosaminoglycans. The unique diversity and structural complexity of HS sugar chains, as well as the variety of core proteins, have been proposed to generate a complex “sugar code” essential for brain wiring. While the functions of HSPGs have been well characterized in C. elegans or Drosophila, relatively little is known about their roles in nervous system development in vertebrates. In this chapter, we describe the advantages and the different methods available to study the roles of HSPGs in axon guidance directly in vivo in zebrafish. We provide protocols for visualizing axons in vivo, including precise dye labeling and time-lapse imaging, and for disturbing the functions of HS-modifying enzymes and core proteins, including morpholino, DNA, or RNA injections.
Keywords: Axon pathfinding, Sugar code, Syndecan, Glypican, Enzyme, Dye labeling, Injection, Mutant
1 Introduction
Brain connectivity and function depend on the proper development of long-range neuronal projections, which in turn relies on the guidance of individual axons as they elongate and grow. When they navigate to their targets, axons respond to diverse attractive and repulsive cues acting at a distance or locally by contact. Concomitantly or subsequently to this guidance process, refinement mechanisms involving pruning or degeneration correct axons that have deviated from the right path, thereby ensuring the formation of accurate neuronal circuits. While many growth factors and guidance cues regulate axon pathfinding, the combined information they provide does not seem sufficient to sculpt the entire neuronal network. Heparan sulfate proteoglycans (HSPGs) are cell-surface and extracellular core proteins with attached heparan sulfate (HS) glycans that are thought to play crucial roles in axon guidance. The diversity and structural complexity of their HS chains allow them to interact with many factors and orchestrate most if not all guidance pathways essential for neuronal wiring. In addition, accumulative observations indicate that core proteins also have functional specificities, and that cooperation between them and their HS chains appears essential for some HSPG functions in nervous system development.
1.1 Roles of HS Chains in Axon Guidance
Several biochemical experiments have shown that HS interacts with guidance molecules and is important for their functions. For instance, netrins were originally purified using heparin affinity columns [1], and their receptor DCC was shown to bind to HS chains in vitro [2]. Similarly, HS critically regulates the function of the guidance cue Slit and its receptor Robo by forming a ternary signaling complex at the surface of axons [3–8]. Additional morphogens such as Wnt, FGF, BMP, or Shh, whose role as guidance molecules has been later identified, also bind to HS with a high affinity [9]. The importance of HS in axon guidance has further been demonstrated in animal models by chemically or genetically modifying HS levels. In Xenopus, adding HS to the developing retinotectal pathway or removing HS with heparitinase prevents retinal axons from entering their brain target, the tectum [10, 11]. In mice, conditional depletion of HS in the nervous system induces severe guidance errors in major commissural tracts, revealing an essential role of HS in midline axon pathfinding [12]. Similarly in zebrafish, drastic reduction in HS induces many retinal axon guidance defects including projections into the forebrain, the hindbrain, and the opposite eye, as well as missorting of axons along the optic tract [13, 14]. Pathfinding of peripheral sensory neurons is also altered in mutants lacking HS [15]. Overall, these different interactions and functions designate HS as a “master-regulator” of axon guidance in vivo.
HS disaccharides are subject to a large number of modifications responsible for their high diversity. These modifications give HS its specific binding affinities and have thus been proposed to generate a “sugar code” for the recognition of guidance factors during development. At least 14 biochemical steps occur in HS chain synthesis. HS are synthesized in the Golgi, where HS polymerases generate a nonsulfated sugar backbone consisting of alternating N-acetylglucosamine and D-glucoronic acid repeats. Initiation and polymerization of this precursor disaccharide are catalyzed by glycosyltransferases of the exostosin family (Ext). Then several modifications occur nonuniformly along the HS chain, creating distinct and specific domains. The first step involves deacetylation and sulfation in N-acetylglucosamine catalyzed by the N-deacetylase- N-sulfotransferase (NDST) class of enzymes. Next, epimerases convert some glucuronic acid units to the isomeric iduronic acid. Then, sulfotransferases add sulfate to specific residues, creating in this way a unique HS fine structure. 2- O-sulfotransferases (Hs2st or 2-OST) attach sulfate to uronic acid residues, whereas 3- O - and 6- O -sulfotransferases (Hs3st and Hs6st or 3-OST and 6-OST) add it to glucosamine residues. Finally, 6- O -endosulfatases (sulf) “edit” HS chains at the plasma membrane by removing sulfate from defined domains. Consequently, these numerous modifications confer HS chains an exceptional diversity (up to 1036 types of HS isoforms) and thus, the potential to provide a large amount of information required for axon pathfinding.
The “sugar code” hypothesis has first emerged from the observation in C. elegans that mutants with different impaired HS modifying enzymes show distinct axon development phenotypes. C. elegans offers the great advantage of having only single orthologs of the various HS-modifying enzymes, making genetic manipulation and analysis of the resulting phenotype easier: deacetylation and epimerization are catalyzed by hst-1 and hse-5, respectively, while 2- O, 3- O, and 6- O sulfations are performed by hst-2, hst3.1 and hst3.2, and hst-6. Interestingly, distinct classes of neurons require the activity of hse-5, hst-2, and hst-6 in different combinations for their axon to be guided properly, while hst-3.1 and hst3.2 appear to regulate more refined steps of later differentiation, controlling branching in a context-dependent manner [16–19]. These results, together with biochemical studies dissecting the structural requirements for HS interaction with different factors, suggest that specific modifications of HS regulate the response of axons to different cues in an instructive manner [17, 20, 21]. It should be noted however that some enzymes can partially compensate each other, suggesting that the presumptive HS code may be degenerate [17, 22, 23]. While the contribution of specific HS motifs has been determined in C. elegans and Drosophila, it remains largely unknown in vertebrates. Many mouse mutants indeed have early embryonic patterning defects or die perinatally. Determining the roles of specific modifications is further complicated by the large number of isoforms for each class of enzymes. A few studies have nonetheless shown that retinal axons in mice lacking Hs2st or Hs6st1 make distinct errors at the chiasm, confirmed that different sulfations regulate specific aspects of axon pathfinding [24, 25].
1.2 Roles of HS Core Proteins
Syndecans (SDCs) and Glypicans (GPCs) are the two major families of cell surface core proteins highly expressed in the nervous system. Tetrapod genomes typically contain four SDC and six GPC genes, each expressed in a specific spatiotemporal expression pattern. In contrast, only one SDC and two GPCs have been identified in Drosophila and C. elegans. Functional studies in these two invertebrates revealed essential roles of SDC and GPCs in nervous system development. Mutation in the single sdc gene induces defects in midline axon guidance through impaired Slit/Robo signaling [5, 26–28]. Both SDC and the GPC dally-like are required for proper axon guidance and visual system function in Drosophila [29]. Finally, the GPC lon2 controls motor axon guidance in C. elegans [17]. To date, very little is known about similar roles in vertebrates. This lack of information is even more surprising considering that core proteins may influence HS levels or composition [30]. Mice lacking SDC3 show neural migration defects that may have made the detection of misguided axons difficult [31, 32]. Only one recent study in chick has demonstrated a role for GPC1 in mediating the repulsive response of postcrossing axons to Shh at the floorplate [33].
1.3 Studying HSPGs In Vivo in Zebrafish
Studying axon guidance in vivo in classical vertebrate models like the mouse presents several difficulties: the generation of knockout models is long and fastidious, and importantly, analysis of axonal trajectories is done a posteriori by fixing and labeling tissues. Observing axon turning, retracting, or degenerating during the course of their navigation thus proves to be very challenging. In contrast, the zebrafish offers several advantages [34, 35]. The optical transparency of zebrafish embryos allows a direct visualization of axons and is particularly suited for high-resolution imaging, especially time-lapse analysis. External fertilization and large clutches provide many embryos that can be observed at different stages. The recent characterization of the zebrafish genome is particularly suited for genetic analysis and allows the fast generation of mutants. Finally, chimeric individuals can easily be obtained by cell transplants. The zebrafish is thus a model of choice to study the roles of HSPGs in axon guidance in vivo, and test the “sugar code” hypothesis.
Several mutants of the HS synthetic pathways have already been identified in a screen for retinal axon guidance defects. Dackel (dak), Boxer (box), and Pinscher (pin) lack functional Ext2, Extl3, and Papst1 (a sulfate transporter), respectively, and can be compared to wild-type (WT) animals for testing the role of HS in axonal development [13, 14, 36–38]. The other HS-modifying enzymes, as well as SDC and GPC core proteins, have been cloned and can be tested for their functions in vivo during development [39–46]. In this chapter, we describe methods for (1) visualizing axon pathfinding directly in the embryo, and (2) down-regulating the expression of genes of interest to test HSPGs’ functions. Directly imaging transgenic embryos that express fluorescent proteins in the neurons of interest constitutes the easiest way to visualize axons as they develop (Fig. 1b). Some transgenes like Tg[elavl3:EGFP] [47] label most neurons and their projections, whereas others are more specific of a class of neurons. Available transgenic lines can be found in the Zfin database at http://zfin.org/action/fish/search. Alternatively, DNA encoding the transgene of interest can be injected at one cell stage in the embryo to either generate transgenic lines, or label transiently a subpopulation of neurons. This last approach is particularly useful to visualize a single axon, and is described in the method section. Finally, lipophilic carbocyanine dyes like DiI, DiA, DiD or DiO can be injected in fixed or live embryos and are particularly suited to label specific subpopulations of neurons (Fig. 1c–e). A detailed protocol for dye injection is provided in the method section. All these different approaches can be performed on the available mutants described above to test the role of HS in axon pathfinding. Heparinase can also be injected at specific places in the embryos to assess the effects of locally removing HS [15]. To further investigate the roles of specific enzymes or core proteins, one needs to experimentally manipulate the expression of corresponding genes. A first common approach is to inject stable antisense morpholino oligonucleotides (MOs) into one-cell stage embryos. MOs inhibit either protein translation when targeted near the start codon of mRNAs [48] or splicing of the pre-mRNAs when targeted to exon–intron or intron–exon boundaries [49]. Under good conditions, MOs can quickly reveal required functions for a targeted gene, though their use is subject to several caveats, such as loss of efficacy as they are diluted during development [50]. Another approach is to inject RNAs encoding sequence-specific synthetic nucleases called TALENs (Transcription Activator-Like Effector Nucleases) to generate targeted knockouts [51, 52]. TALENs combine a TAL effector DNA binding domain with a DNA cleavage domain to target specific sequences in the genome and induce mutations at any locus. Finally, DNA constructs encoding dominant negative forms of the protein of interest can be transiently or stably expressed. Spatial or temporal control can be provided by cell-specific promoters or the hsp70l heat shock promoter, respectively. Gain-of-function experiments can also be performed by overexpressing genes of interest at specific times or locations. At last, cell-autonomy of HSPGs’ function can be tested by expressing a WT gene in a specific tissue in the corresponding mutant embryo, and test whether the mutant phenotype is rescued. All these different approaches rely on the injection of MOs, RNA, or DNA in the embryo at one-cell stage [53], for which we provide a detailed protocol below.
Fig. 1.
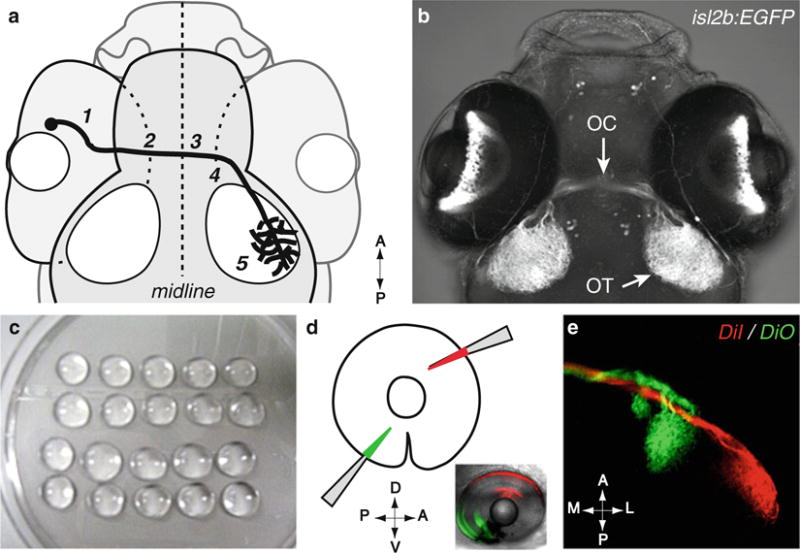
Methods for visualizing retinal axons. (a) Diagram of the retinal axon pathway. Retinal axons navigate to the optic nerve head (1), pass through the optic nerve and exit the eye (2), cross the midline at the chiasm (3), and grow dorsally along the optic tract (4) to reach the tectum (5). (b) Dorsal view of a Tg[isl2b:EGFP]zc7 transgenic embryo, in which EGFP is specifically expressed in all RGCs, allowing a direct visualization of retinal projections. Courtesy of A. Pittman. OC optic chiasm, OT optic tectum. (a, b) dorsal views, anterior up. Maximum intensity projection, confocal microscopy. (c–e) Focal injection of dyes in the retina allows visualization of retinal axons making topographic connections in the tectum. (c) Embryos are mounted laterally in low-melt agarose drops placed on a Petri dish lid. (d) DiI- (red) and DiO-(green) coated glass micropipettes are briefly inserted in a peripheral direction into the retina to label dorsonasal (DN in red) and ventrotemporal (VT in green) retinal neurons (method described in detail in Subheading 3.2). A anterior, P posterior, D dorsal, V ventral. (e) Dorsal view of the corresponding retinal axon projections in the brain target, the tectum. A anterior, P posterior, M medial, L lateral. Maximum intensity projection, confocal microscopy
2 Materials
2.1 Zebrafish Embryos
Wild-type (WT) and mutant embryos are obtained from natural matings, raised at 28.5 °C in E3 medium in the presence of 150 mM of 1-phenyl-2-thiourea (PTU) to prevent pigment formation, and staged by age and morphology [54]. They are dechorionated before dye injection experiments.
-
–
E3 medium: 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4
2.2 Material Needed for Dye Injection
-
–
DiI or DiO crystals (Molecular Probes)
-
–
Glass capillary (World Precision Instruments, Inc.) with an outer diameter of 1.0 mm and an inner diameter of 0.58 mm to prepare the microneedle for injections. Pull the capillary to make a microneedle with a final taper length of 9.0 mm and a tip size of 2 μm.
-
–
4 % PFA: 4 % paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, if the experiment is performed on fixed embryos (see Note 1).
-
–
Tricaine stock: 0.4 % tricaine, 10 mM HEPES, pH 7.4, if the experiment is performed on live embryos (see Note 2).
-
–
1 % low-melt agarose in water (for fixed embryos) or PBS (phosphate-buffered saline) for live embryos.
-
–
50 % and 80 % glycerol in water.
-
–
1 % low-melt agarose in E3/GN/tricaine if the experiment is performed on live embryos.
-
–
E3/GN/tricaine: 10 μg/ml gentamicin in E3 medium, 0.02 % tricaine.
-
–
A three-axis micromanipulator, a needle holder, and a microscope.
2.3 Material Needed for Injections at One-Cell-Stage
-
–
Glass capillary (World Precision Instruments, Inc.) with an outer diameter of 1.0 mm; the glass capillary is pulled into two needles that are stored on playdough lines in a 150 mm Petri dish.
-
–
Injection plate (Fig. 2a): pour approximately 40 mL of 2 % agarose in E3 medium in a 150 mm Petri dish on a level surface. Set the plastic mold for making slots (Adaptive Science Tools) teeth down into the agarose, and tap gently to eliminate bubbles. After the agarose sets, add a small amount of E3 medium, remove the mold, wrap the Petri dish in parafilm, and store at 4 °C.
-
–
A microinjector, a three-axis micromanipulator, a needle holder, and a microscope.
-
–
MOs: MOs are designed and made by Gene Tools, LLC. They are lyophilized when delivered and are then resuspended in Danieau’s solution. MOs in solution can be stored at −20 °C.
-
–
Danieau’s solution: 1.5 mM HEPES, 0.18 mM Ca(NO3)2, 0.12 mM MgSO4, 0.21 mM KCl, 17.4 mM NaCl.
-
–
DNA and mRNA are diluted in water. Plasmid DNA is purified with plasmid miniprep purification kits. Pure mRNA is made by in vitro transcription and purified on micro Bio-Spin chromatography columns (Bio-Rad).
-
–
Phenol red is used at a final concentration of 0.5 % as a marker dye for all solutions to be injected.
-
–
E3/GN: 10 μg/ml gentamicin in E3 medium.
Fig. 2.
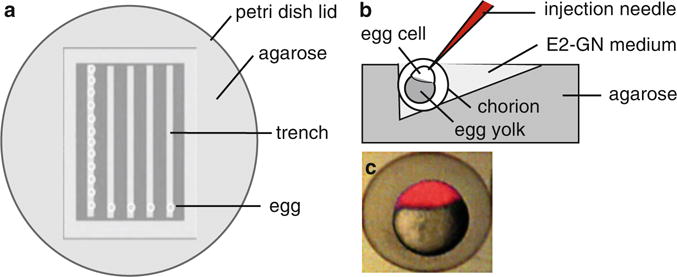
Injecting one-cell stage embryos. (a) Diagram of the injection plate. Agarose is poured in a Petri dish, and trenches are made with a plastic mold set into the agarose. Eggs collected at one-cell stage are aligned in the trenches, and covered with E3-GN medium. (b) Egg positioning for injection. The egg in its chorion is positioned so that its cell is oriented up. Using the micromanipulator (not shown), the needle is brought next to the egg. It is then moved in a smooth movement to pierce the surface of the chorion and enter the cell. (c) Picture showing an egg successfully injected in the cell with a solution labeled with phenol red
3 Methods
3.1 Precise Labeling with Injection of Lipophilic Dyes
Originally developed by Torsten Trowe [55], this method uses glass microneedles coated with lipophilic carbocyanine dyes to focally deposit dye into a region of interest. It can be adapted to label different population of neurons such as retinal neurons, as shown in Fig. 1 [14, 35, 56]. We provide a detailed protocol for labeling fixed embryos, but a similar approach can be used in live embryos (see Notes 3 and 4).
Fix zebrafish embryos at required stage in 4 % PFA at room temperature for the first 2 h, then at 4 °C for 10 h. Embryos can be kept at 4 °C as long as wanted for future experiments.
To coat the microneedle with dye, place a few dye crystals on a cover glass and melt them at 100 °C on a hot plate. Dip the tip of the microneedle horizontally into the dye paste and roll it to cover the tip equally on all sides. Wipe off as much dye from the tip as possible onto the cover glass.
Prepare 30 ml of 1 % low-melt agarose in water and keep on heating block at 37 °C to prevent from solidifying. Use a Petri dish lid to embed embryos for dye injection. Rinse embryos in water, transfer them onto the lid, and cover them with a drop of 1 % low-melt agarose. Orient them in an appropriate position, so that the region to be injected is easily accessible to the micropipette.
Use a standard pipette holder and three-axis micromanipulator to hold the dye-coated microneedle. Insert the microneedle into the region of interest by advancing in a peripheral direction at a roughly 45° angle (this angle usually allows good visualization and penetration of the tissue). The time during which the needle needs to be left in the tissue depends on how big the region of interest is or how many neurons need to be labeled. Leaving the needle for 5–20 s ensures a small injection site and labeling of few axons. The coated microneedle can be reused for several injections before it has to be coated with fresh dye again.
After finishing the injections, cover embedded embryos with water to avoid drying. This step also washes off excessive dye. Store the embryos for a few hours at room temperature for fast diffusion of the dye, or keep them at 4 °C overnight if slower diffusion is desired. Long incubation times can result in nonspecific diffusion of the dye, which can prevent clear imaging results later on.
Recover embryos from agarose drops using forceps, and rinse them with water. Transfer embryos to 50 % glycerol/H2O and incubate them under agitation for 3 h at 4 °C. Change the medium to 80 % glycerol/H2O, and store embryos at 4 °C overnight. Now that they are cleared, embryos can be mounted for confocal imaging in 80 % glycerol between two coverslips.
3.2 Injection in Embryos at One-Cell Stage
As mentioned earlier, this approach is used to inject MOs, RNA or/and DNA. Optimal concentrations for these different compounds are discussed in Subheading 4 (see Note 5).
The night before the experiment, set up fish in breeding tanks with dividers in place. The next morning, remove the dividers, wait about 20 min, and collect eggs using a strainer.
Align the eggs in the trenches of the injection plate using a transfer pipette (Fig. 2a). Cover the eggs with E3/GN, and position them such as the cell is oriented up (Fig. 2b) (see Note 6).
Load the needle with 3 μL of the solution to be injected (the media used to dilute MOs, RNA, and DNA are described in Subheading 2) (see Note 7). Insert the needle into the pipette holder and micromanipulator connected to the microinjector. Check that the micromanipulator is in a proper position to allow movement and adjustment of the needle. The needle should be positioned such as it is directed towards the egg and especially, its cell part (Fig. 2b). Bring the needle tip into the plane of view of the microscope and focus on the thinnest part of its tip.
Cut the needle at its tip with a pair of forceps, so that it is narrow enough to pierce the chorion but still capable of delivering a consistent volume. Press the foot pedal of the injector and monitor the size of the drop released in the medium. Volume can be adjusted by trimming the needle, adjusting the injection pressure, or the duration of injection. Injection volumes of 500 pL or 1 nL are typically used.
Ensure that the embryos are still at one-cell stage before pursuing the injections.
Using the micromanipulator, bring the needle next to the egg, pierce the surface of the chorion and enter the cell in one smooth movement. Inject the solution in the cell while being careful not to move to avoid tearing the membrane. After injecting, remove the needle, and slowly move the injection plate with your hand to proceed with the next egg. Always keep some eggs uninjected as a control.
After finishing, use E3 medium to move the injected eggs into a clean Petri dish. At the end of the day, remove dead or damaged embryos, and count the number of embryos you have injected (see Note 8).
Acknowledgments
F.E. Poulain is supported by a grant from the NINDS (K99-1K99NS083714-01).
Footnotes
Do not inject lipophilic dyes if embryos have been permealized with triton or tween for other experiments such as immunolabeling. Permeabilization would affect the diffusion of lipophilic dyes along plasma membranes, resulting in a blurry, nonspecific staining.
Injection of lipophilic dyes can also be performed on live embryos to visualize axons as they develop. In this case, anesthetize embryos in 0.015 % tricaine, and mount them in 1 % agarose, 0.015 % tricaine in E3/GN on a Petri dish lid (first pipet the embryo in the agarose tube, and then pipet the agarose containing the embryo on the lid, so that manipulation of embryos is kept at a minimum).
Using dyes diluted at a concentration of 3 or 4 % in DMSO can also be used and is often more effective for labeling live embryos. Keep the tube containing the diluted dye at 37 °C, so that the dye remains liquid, and fill an injecting needle connected to a microinjector with the dye solution. Embryos are mounted in the same way as described previously, but they are submerged with water only. It is important to limit the use of saline solutions like PBS, as salts are known to facilitate dye precipitation. Before inserting the microneedle into the region of interest and injecting the dye, use an agarose drop as a test to determine the parameters of injection corresponding to the volume that needs to be injected.
Injecting dyes in live embryos allows a direct visualization of elongating axons in vivo by time-lapse microscopy. To perform time-lapse imaging, mount embryos in 1 % low-melt agarose in E3 medium with 150 mM PTU and 0.015 % tricaine in a glass-bottomed Petri dish. Chamber temperature should be maintained at 28.7 °C using a heated stage. Using a confocal microscope, z-series can be acquired a regular intervals. It is important to keep the power of the lasers at a minimum, so that embryos survive the procedure. Maximal intensity projections for each time point can be compiled and aligned using ImageJ software and StackReg plugin [57].
Concentration of DNA, mRNA, and MO should be determined by the user. Usually, between 20 and 60 pg of DNA can be injected. Higher concentrations are often toxic, leading to embryos’ death. MOs are usually injected at a final concentration of 0.5–1 mM (which corresponds to around 4–8 ng, depending on the MO sequence). Finally, mRNA appears less toxic than DNA or MO and can be injected at higher concentrations ranging from 25 pg to 1 ng.
For good efficiency, DNA needs to be injected as early as possible at one-cell stage. When performing DNA injections, it is recommended to load the injection needle and prepare the injection set up before collecting eggs, so that DNA can be injected right away as soon as eggs are transferred to the injection plate. In contrast to DNA, MOs, and mRNAs can be injected in the egg yolk right below the cell for a similar efficiency.
The Tol2kit system is a powerful tool widely used to generate transgenic lines. It uses site-specific recombination-based cloning (multisite Gateway technology) to allow modular and quick assembly of constructs in a Tol2 transposon backbone. Plasmid DNA generated with this system is coinjected with transposase mRNA at one-cell stage [58].
As MOs can have off-target effects or can be diluted during development [59], performing several controls is required before drawing any conclusion about a gene’s function. At least two different MOs targeting distinct regions of the gene of interest should induce the phenotype. Alternatively or in complement, the MO phenotype should be rescued by coinjecting a synthetic mRNA encoding the protein from the targeted locus but not the MO target sequence (MO and mRNA are injected separately with different needles). Embryos injected with the MO alone should be compared to embryos injected with the mRNA alone or with both the mRNA and the MO.
References
- 1.Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 2.Bennett KL, Bradshaw J, Youngman T, Rodgers J, Greenfield B, Aruffo A, Linsley PS. Deleted in colorectal carcinoma (DCC) binds heparin via its fifth fibronectin type III domain. J Biol Chem. 1997;272:26940–26946. doi: 10.1074/jbc.272.43.26940. [DOI] [PubMed] [Google Scholar]
- 3.Hu H. Cell-surface heparan sulfate is involved in the repulsive guidance activities of Slit2 protein. Nat Neurosci. 2001;7:695–701. doi: 10.1038/89482. [DOI] [PubMed] [Google Scholar]
- 4.Ronca F, Andersen JS, Paech V, Margolis RU. Characterization of Slit protein interactions with glypican-1. J Biol Chem. 2001;276:29141–29147. doi: 10.1074/jbc.M100240200. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KG, Ghose A, Epstein E, Lincecum J, O’Connor MB, Van Vactor D. Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr Biol. 2004;14:499–504. doi: 10.1016/j.cub.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Ronca F, Linhardt RJ, Margolis RU. Structural determinants of heparan sulfate interactions with slit proteins. Biochem Biophys Res Commun. 2004;317:352–357. doi: 10.1016/j.bbrc.2004.03.059. [DOI] [PubMed] [Google Scholar]
- 7.Hussain SA, Piper M, Fukuhara N, Strochlic L, Cho G, Howitt JA, Ahmed Y, Powell AK, Turnbull JE, Holt CE, Hohenester E. A molecular mechanism for the heparan sulfate dependence of slit-robo signaling. J Biol Chem. 2006;281:39693–39698. doi: 10.1074/jbc.M609384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuhara N, Howitt JA, Hussain SA, Hohenester E. Structural and functional analysis of slit and heparin binding to immunoglobulin-like domains 1 and 2 of Drosophila Robo. J Biol Chem. 2008;283:16226–16234. doi: 10.1074/jbc.M800688200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strigini M. Mechanisms of morphogen movement. J Neurobiol. 2005;64:324–333. doi: 10.1002/neu.20164. [DOI] [PubMed] [Google Scholar]
- 10.Walz A, McFarlane S, Brickman YG, Nurcombe V, Bartlett PF, Holt CE. Essential role of heparan sulfates in axon navigation and targeting in the developing visual system. Development. 1997;124:2421–2430. doi: 10.1242/dev.124.12.2421. [DOI] [PubMed] [Google Scholar]
- 11.Irie A, Yates EA, Turnbull JE, Holt CE. Specific heparan sulfate structures involved in retinal axon targeting. Development. 2002;129:61–70. doi: 10.1242/dev.129.1.61. [DOI] [PubMed] [Google Scholar]
- 12.Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science. 2003;302:1044–1046. doi: 10.1126/science.1090497. [DOI] [PubMed] [Google Scholar]
- 13.Lee JS, von der Hardt S, Rusch MA, Stringer SE, Stickney HL, Talbot WS, Geisler R, Nusslein-Volhard C, Selleck SB, Chien CB, Roehl H. Axon sorting in the optic tract requires HSPG synthesis by ext2 (dackel) and extl3 (boxer) Neuron. 2004;44:947–960. doi: 10.1016/j.neuron.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 14.Poulain FE, Chien CB. Proteoglycan-mediated axon degeneration corrects pretarget topographic sorting errors. Neuron. 2013;78:49–56. doi: 10.1016/j.neuron.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Wolfson SN, Gharib A, Sagasti A. LAR receptor tyrosine phosphatases and HSPGs guide peripheral sensory axons to the skin. Curr Biol. 2012;22:373–382. doi: 10.1016/j.cub.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bülow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 17.Bülow HE, Tjoe N, Townley RA, Didiano D, van Kuppevelt TH, Hobert O. Extracellular sugar modifications provide instructive and cell-specific information for axon-guidance choices. Curr Biol. 2008;18:1978–1985. doi: 10.1016/j.cub.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tecle E, Diaz-Balzac CA, Bülow HE. Distinct 3-O-sulfated heparan sulfate modification patterns are required for kal-1-dependent neurite branching in a context-dependent manner in Caenorhabditis elegans. G3 (Bethesda) 2013;3:541–552. doi: 10.1534/g3.112.005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gysi S, Rhiner C, Flibotte S, Moerman DG, Hengartner MO. A network of HSPG core proteins and HS modifying enzymes regulates netrin-dependent guidance of D-type motor neurons in Caenorhabditis elegans. PLoS One. 2013;8:e74908. doi: 10.1371/journal.pone.0074908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shipp EL, Hsieh-Wilson LC. Profiling the sulfation specificities of glycosaminoglycan interactions with growth factors and chemotactic proteins using microarrays. Chem Biol. 2007;14:195–208. doi: 10.1016/j.chembiol.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Moniz HA, Walcott B, Moremen KW, Linhardt RJ, Wang L. Characterization of the interaction between Robo1 and heparin and other glycosaminoglycans. Biochimie. 2013 doi: 10.1016/j.biochi.2013.08.018. pii:S0300-9084(13)00290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamimura K, Koyama T, Habuchi H, Ueda R, Masu M, Kimata K, Nakato H. Specific and flexible roles of heparan sulfate modifications in Drosophila FGF signaling. J Cell Biol. 2006;174:773–778. doi: 10.1083/jcb.200603129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dejima K, Takemura M, Nakato E, Peterson J, Hayashi Y, Kinoshita-Toyoda A, Toyoda H, Nakato H. Analysis of Drosophila glucuronyl C-5 epimerase: implications for developmental roles of heparan sulfate sulfation compensation and 2-O sulfated glucuronic acid. J Biol Chem. 2013;288:34384–34393. doi: 10.1074/jbc.M113.499269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratt T, Conway CD, Tian NM, Price DJ, Mason JO. Heparan sulphation patterns generated by specific heparan sulfotransferase enzymes direct distinct aspects of retinal axon guidance at the optic chiasm. J Neurosci. 2006;26:6911–6923. doi: 10.1523/JNEUROSCI.0505-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway CD, Howe KM, Nettleton NK, Price DJ, Mason JO, Pratt T. Heparan sulfate sugar modifications mediate the functions of slits and other factors needed for mouse forebrain commissure development. J Neurosci. 2011;31:1955–1970. doi: 10.1523/JNEUROSCI.2579-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steigemann P, Molitor A, Fellert S, Jackle H, Vorbruggen G. Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by slit/robo signaling. Curr Biol. 2004;14:225–230. doi: 10.1016/j.cub.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Rhiner C, Gysi S, Frohli E, Hengartner MO, Hajnal A. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 2005;132:4621–4633. doi: 10.1242/dev.02042. [DOI] [PubMed] [Google Scholar]
- 28.Smart AD, Course MM, Rawson J, Selleck S, Van Vactor D, Johnson KG. Heparan sulfate proteoglycan specificity during axon pathway formation in the Drosophila embryo. Dev Neurobiol. 2011;71:608–618. doi: 10.1002/dneu.20854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawson JM, Dimitroff B, Johnson KG, Rawson JM, Ge X, Van Vactor D, Selleck SB. The heparan sulfate proteoglycans Dally-like and Syndecan have distinct functions in axon guidance and visual-system assembly in Drosophila. Curr Biol. 2005;15:833–838. doi: 10.1016/j.cub.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 30.Chen RL, Lander AD. Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J Biol Chem. 2001;276:7507–7517. doi: 10.1074/jbc.M008283200. [DOI] [PubMed] [Google Scholar]
- 31.Hienola A, Tumova S, Kulesskiy E, Rauvala H. N-Syndecan deficiency impairs neural migration in brain. J Cell Biol. 2006;174:569–580. doi: 10.1083/jcb.200602043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bespalov MM, Sidorova YA, Tumova S, Ahonen-Bishopp A, Magalhães AC, Kulesskiy E, Paveliev M, Rivera C, Rauvala H, Saarma M. Heparan sulfate proteoglycan syndecan- 3 is a novel receptor for GDNF, neurturin, and artemin. J Cell Biol. 2011;192:153–169. doi: 10.1083/jcb.201009136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson NH, Stoeckli ET. Sonic hedgehog regulates its own receptor on postcrossing commissural axons in a glypican1-dependent manner. Neuron. 2013;79:478–491. doi: 10.1016/j.neuron.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 34.Hutson LD, Campbell DS, Chien CB. Analyzing axon guidance in the zebrafish retinotectal system. Methods Cell Biol. 2004;76:13–35. doi: 10.1016/s0091-679x(04)76002-1. [DOI] [PubMed] [Google Scholar]
- 35.Poulain FE, Gaynes JA, Hörndli C, Law MY, Chien CB. Analyzing retinal axon guidance in zebrafish. Methods Cell Biol. 2010;100:3–26. doi: 10.1016/B978-0-12-384892-5.00001-3. [DOI] [PubMed] [Google Scholar]
- 36.Karlstrom RO, Trowe T, Klostermann S, Baier H, Brand M, Crawford AD, Grunewald B, Haffter P, Hoffmann H, Meyer SU, Muller BK, Richter S, van Eeden FJ, Nusslein-Volhard C, Bonhoeffer F. Zebrafish mutations affecting retinotectal axon pathfinding. Development. 1996;123:427–438. doi: 10.1242/dev.123.1.427. [DOI] [PubMed] [Google Scholar]
- 37.Trowe T, Klostermann S, Baier H, Granato M, Crawford AD, Grunewald B, Hoffmann H, Karlstrom RO, Meyer SU, Muller B, Richter S, Nusslein-Volhard C, Bonhoeffer F. Mutations disrupting the ordering and topographic mapping of axons in the retinotectal projection of the zebrafish, Danio rerio. Development. 1996;123:439–450. doi: 10.1242/dev.123.1.439. [DOI] [PubMed] [Google Scholar]
- 38.Clément A, Wiweger M, von der Hardt S, Rusch MA, Selleck SB, Chien CB, Roehl HH. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genet. 2008;4(7):e1000136. doi: 10.1371/journal.pgen.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: II. The 6-O-sulfotransferase family. Dev Dyn. 2006;235:3432–3437. doi: 10.1002/dvdy.20990. [DOI] [PubMed] [Google Scholar]
- 40.Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-O-sulfotransferase family. Dev Dyn. 2006;235:3423–3431. doi: 10.1002/dvdy.20991. [DOI] [PubMed] [Google Scholar]
- 41.Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: III. 2-O-sulfotransferase and C5-epimerases. Dev Dyn. 2007;236:581–586. doi: 10.1002/dvdy.21051. [DOI] [PubMed] [Google Scholar]
- 42.Kramer KL, Barnette JE, Yost HJ. PKCgamma regulates syndecan-2 inside-out signaling during Xenopus left-right development. Cell. 2002;111:981–990. doi: 10.1016/s0092-8674(02)01200-x. [DOI] [PubMed] [Google Scholar]
- 43.Arrington CB, Yost HJ. Extra- embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136:3143–3152. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofmeister W, Devine CA, Key B. Distinct expression patterns of syndecans in the embryonic zebrafish brain. Gene Expr Patterns. 2013;13:126–133. doi: 10.1016/j.gep.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Topczewski J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, Hammerschmidt M, Postlethwait J, Solnica-Krezel L. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev Cell. 2001;1:251–264. doi: 10.1016/s1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 46.Gorsi B, Whelan S, Stringer SE. Dynamic expression patterns of 6-O endosulfatases during zebrafish development suggest a subfunctionalisation event for sulf2. Dev Dyn. 2010;239:3312–3323. doi: 10.1002/dvdy.22456. [DOI] [PubMed] [Google Scholar]
- 47.Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, Miki N, Chitnis AB, Huh TL. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- 48.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 49.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 50.Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 51.Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, Ii, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen JN, Sweeney MF, Mably JD. Microinjection of zebrafish embryos to analyze gene function. J Vis Exp. 2009 doi: 10.3791/1115. pii:1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 55.Trowe T. Ph.D. thesis. Eberhard-Karls-Universität Tϋbingen; 2000. Analyse von Mutationen mit Einfluss aud die topographische Ordnung von Axonen im retinotektalen System des Zebrabärblings, Danio rerio. [Google Scholar]
- 56.Stacher Hörndli C, Chien CB. Sonic hedgehog is indirectly required for intraretinal axon pathfinding by regulating chemokine expression in the optic stalk. Development. 2012;139:2604–2613. doi: 10.1242/dev.077594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 58.Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 59.Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6:69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]


