Abstract
ASAH1 gene encodes for acid ceramidase that is involved in the degradation of ceramide into sphingosine and free fatty acids within lysosomes. ASAH1 variants cause both the severe and early-onset Farber disease and rare cases of spinal muscular atrophy (SMA) with progressive myoclonic epilepsy (SMA-PME), phenotypically characterized by childhood onset of proximal muscle weakness and atrophy due to spinal motor neuron degeneration followed by occurrence of severe and intractable myoclonic seizures and death in the teenage years. We studied two subjects, a 30-year-old pregnant woman and her 17-year-old sister, affected with a very slowly progressive non-5q SMA since childhood. No history of seizures or myoclonus has been reported and EEG was unremarkable. The molecular study of ASAH1 gene showed the presence of the homozygote nucleotide variation c.124A>G (r.124a>g) that causes the amino acid substitution p.Thr42Ala. Biochemical evaluation of cultured fibroblasts showed both reduction in ceramidase activity and accumulation of ceramide compared with the normal control. This study describes for the first time the association between ASAH1 variants and an adult SMA phenotype with no myoclonic epilepsy nor death in early age, thus expanding the phenotypic spectrum of ASAH1-related SMA. ASAH1 molecular analysis should be considered in the diagnostic testing of non-5q adult SMA patients.
Introduction
Acid ceramidase (N-acylsphingosine amidohydrolase; EC 3.5.1.23) is a lysosomal enzyme consisting of a nonglycosylated alpha subunit and a glycosylated beta subunit which catalyzes the degradation of ceramide into sphingosine and fatty acid.1 It is codified by ASAH1 gene located on 8p22 chromosome (MIM #613468).2
At least 20 variants in the ASAH1 gene have been reported as a cause of the devastating early-onset Farber lipogranulomatosis (FL; MIM #228000; http://www.hgmd.org).3, 4
ASAH1 variants associated with FL severely reduce the activity of the enzyme, typically to less than one-tenth of normal.3 As a result, the enzyme cannot break down ceramides properly that accumulate in the lysosomes of various cells, including lungs, liver, muscles, brain, cartilage and bone.
Up to now, four variants in the ASAH1 gene have been associated with spinal muscular atrophy with progressive myoclonic epilepsy (SMA-PME; MIM #159950): c.850G>T, p.(Gly284Ter); c.456A>C, p.(Lys152Asn); c.125C>T, p.(Thr42Met); the large deletion c.(?_-197)_(*1118_?)del.5, 6
SMA-PME is an autosomal recessive condition characterized by muscle weakness and wasting and a combination of drug-resistant seizures and uncontrollable muscle jerks starting in childhood.5, 6, 7 Variants causing SMA-PME result in reduced acid ceramidase activity (one-third of normal) and ceramide accumulation damaging cells less extensively than FL.5, 6 However, the disorder is progressive and patients die in the teenage years.
Here, we report the first adult SMA phenotype with no myoclonic epilepsy owing to the novel homozygous c.124A>G (r.124a>g), p.(Thr42Ala) ASAH1 variant, thus expanding the phenotypical spectrum of ASAH1-related SMA.
Patients and methods
Clinical report
Patient 1
A 30-year-old, sixth week pregnant woman started to complain of very slowly progressive difficulty in walking and climbing stairs, carrying heavy objects and lifting her arms above the head since age of three. Thereafter, she experienced postural tremor, diffuse muscle pain, muscle cramps in legs and hands, and progressive scoliosis. No history of seizures or myoclonus was reported.
She was born in Pakistan from consanguineous parents (first cousins). One younger sister suffered from similar clinical picture while both parents and three other sisters had no neuromuscular complains (Figure 1a).
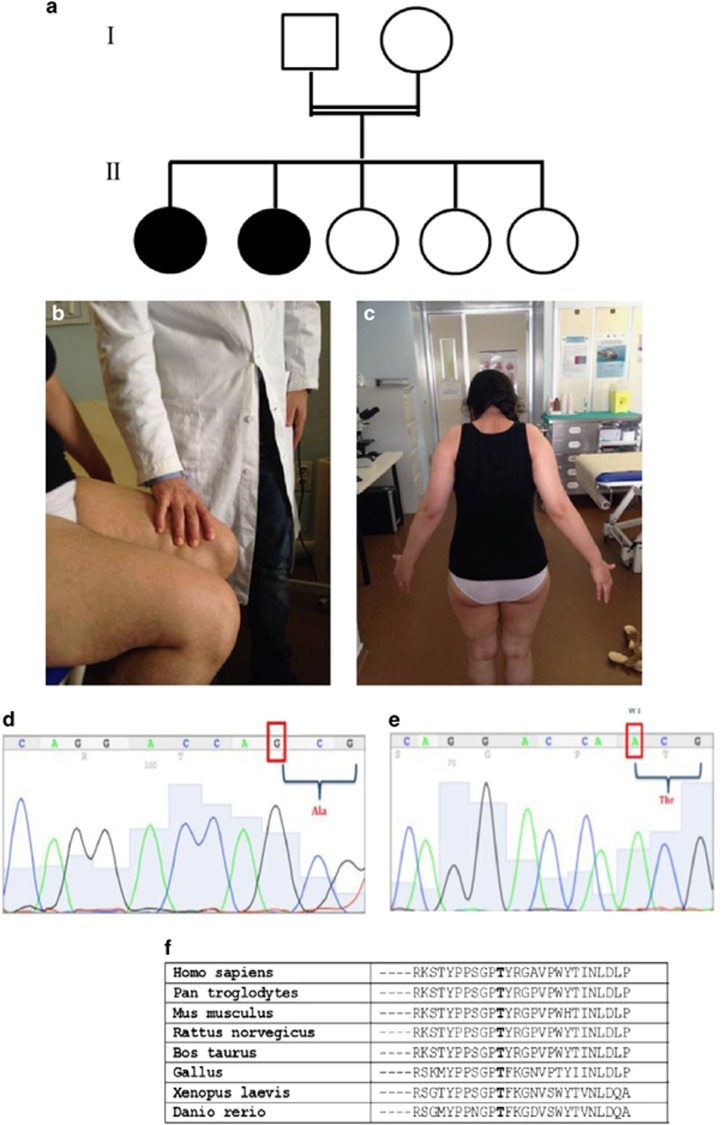
Figure 1.
(a) Family tree showing the two affected sisters (filled black circles), born from a first-degree consanguineous parents (double horizontal line). (b, c) Neurological examination in patient #1 showed weakness in lifting the thigh against slight resistance (b) and upper limb abduction possible only up to 30 degrees (c). (d, e) Sanger sequencing of ASAH1 exon 2 shows the homozygous nucleotide change c.124A>G in patient #1; the Sanger sequence trace from a normal control is shown in (e). (f) Evolutionary comparison of the protein sequences flanking the missense variant p.(Thr42Ala) identified in the human ASAH protein with their orthologous counterparts in seven vertebrates. The residue Thr42 (T) is bold typed.
Neurological examination showed waddling gait and proximal upper and lower limb weakness (2/5 MRC; Figure 1b) and atrophy. She got up from a sitting position with support, but she was unable to stand up after squatting; upper limb abduction was possible only up to 30 degrees (Figure 1c); postural distal tremor at upper limbs was present; deep tendon reflexes were retained; no contractures or foot deformities were present.
EEG, brain MRI, sensory and motor evoked potentials were normal. Respiratory function tests showed a mild restrictive ventilation impairment. Sensorineural deafness was not reported.
Routine blood tests including CK, lactate, antibodies to gliadin and transglutaminase, prolactin, ANA, ENA, ANCA, PTH and thyroid function were normal. Hexosaminidase A assay was also normal.
EMG and muscle biopsy analyses suggested a chronic neurogenic process as usually seen in SMA, but the search for SMN1 gene deletions and point variants was negative. She has given birth without complications in January 2014.
Patient 2
The younger sister of patient #1 (age 17) experienced at school age slowly worsening difficulty in running, climbing stairs and holding weights, and early fatigue in physical activities. Because she developed a significant scoliosis over time, she wore a removable back-brace.
At the neurological examination positive Gower maneuver, inability to rise from the squatting position, slight proximal weakness in the upper limbs (4-/5 MRC) and, more importantly, in the lower limbs (3/5 MRC), atrophy in the distal third of the legs and the lower third of the quadriceps bilaterally were present. Muscle tone was normal. Brisk reflexes to the four limbs were present with no Babinski sign.
No history of seizures or myoclonus was reported and EEG was unremarkable. Brain and cord MRI was normal with evidence of marked left-convex scoliosis to the dorsal-lumbar region with fulcrum in L1 (Coob angle of about 45°).
EMG showed chronic neurogenic changes as well as muscle biopsy. Motor and sensory evoked potentials were normal. Searching for SMN1 deletions or intragenic variants resulted negative.
Skin fibroblast cultures
Skin biopsy was obtained from patient #1 and a culture of fibroblasts was established according to standard procedures. The Farber fibroblast cell line derived from a female patient with the lysosomal deficiency of the acid ceramidase owing to the skipping of the ASAH1 gene exon 6 associated with a still unknown allele.
The fibroblast cell lines from a healthy donor and from the Farber patient were obtained from the ‘Telethon Cell Line and DNA Biobank from Patients affected by Genetic Diseases' at Giannina Gaslini Institute, Genoa, Italy.8
Ethical issues
Following ethical guidelines, the patient's and healthy donors' samples were obtained for analyses, research and storage with their written informed consent.
Molecular studies
Molecular analysis of ASAH1 gene was performed on both the patients. No other relatives for the purpose of this study were available.
Genomic DNA was extracted from venous peripheral blood lymphocytes, according to standard procedures in patients and controls. ASAH1 gene exons and exon–intron boundaries were PCR-amplified, using specific primers designed by reference to the genomic sequence (GenBank accession No. NG_008985.1).
Total RNA was extracted from patient fibroblasts using an RNeasy mini kit (QIAGEN, Courtaboeuf, France) and reverse-transcribed by means of The SuperScript VILO cDNA Synthesis Kit (Life Technologies, ThermoFisher Scientific, Waltham, MA, USA).
RT-PCR was performed using sets of primers designed by reference to the ASAH1 mRNA sequence (GenBank accession No. NM_177924.3).
PCR and RT-PCR products were purified and directly sequenced using an ABI Prism 3130XL DNA automated sequencer (Applied Biosystems, Waltham, MA, USA).
Real-time PCR was performed on 7300-Real-Time PCR System Instrument (Applied Biosystems), with Fast-Start Universal SYBR Green Master Mix (ROX, Roche, Basel, Switzerland) for exon 2 of ASAH1 gene, in order to rule out a potential allele drop-out.
Nucleotide numbering is derived from cDNA ASAH1 sequences (GenBank accession no. NM_177924.3). The variants are described according to current variant nomenclature guidelines (http://www.hgvs.org/mutnomen), ascribing the A of the first ATG translational initiation codon as nucleotide +1.
To predict the functional effect of the novel variant p.(Thr42Ala), we queried PolyPhen software (http://genetics.bwh.harvard.edu/pph2/).
The novel detected variation was studied by searching in EVS database (evs.gs.washington.edu/), 1000 Genomes (http://www.1000genomes.org/) and dbSNP server (http://www.ncbi.nlm.nih.gov/SNP) and by screening 280 alleles, including 100 of Pakistan origin, from healthy control subjects.
ASSP algorithm (wangcomputing.com/assp/) was used for alternative splice site prediction as the novel variant occurred close to the 5' splice site, and ESE Finder Software 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home) for evaluating disruption of putative exonic splicing enhancers (ESEs), which are binding sites for human SR proteins SF2/ASF, SC35, SRp40 and SRp55.9, 10
A next generation sequencing analysis of a neuromuscular gene panel was conducted in order to rule out other possible causative genetic factors associated with the patient's phenotype.
We used a Nextera Rapid Capture method with Studio Design software (Illumina, Inc., San Diego, CA, USA) for customizing a gene panel for the analysis of the following genes: AARS (100%), ACTA1 (100%), ALS2 (85%), ANG (100%), AP5Z1 (98%), APEX1 (100%), ARHGEF28 (83%), ATP2B3 (100%), BAG3 (99%), BSCL2 (100%), CLN8 (100%), CRYM (83%), CYP27A1 (95%), CYP7B1 (81%), DAO (85%), DCTN1 (100%), DHTKD1 (94%), DPP6 (80%), ELP3 (94%), EPHA4 (94%), ERBB4 (83%), EWSR1 (88%), FEZF2 (87%), FUS (98%), GLA (100%), GLE1 (100%), GRN (100%), HEXA (93%), HNRNPA1 (92%), HNRNPA2B1 (91%), HNRNPA3 (94%), HSPB3 (100%), IGHMBP2 (99%), KIF5A (98%), L1CAM (100%), MAPT (100%), MFN2 (100%), OPTN (87%), PDYN (100%), PFN1 (100%), PNPLA6 (98%), POLG (95%), PRNP (100%), PRPH (87%), REEP1 (87%), SETX (87%), SIGMAR1 (93%), SOD1 (100%), SPG11 (81%), SPG21 100%), SPG7 (89%), SQSTM1 (85%), SS18L1 (90%), TAF15 (83%), TARDBP (97%), TREM2 (100%), TRPV4 (100%), TUBA4A (97%), UBA1 (100%), UBQLN2 (97%), VAPB (85%), VCP (98%), ZFYVE26 (100%). Obtained sequences have been aligned to the reference genome (GRCh37/hg19) using MiSeq software. Filtering process modality is available upon request. Gene coverage at >20 × reading depth is indicated in brackets. Data analysis was obtained using the following softwares: Illumina MiSeq Reporter vs 2.4.60, Illumina Variant Studio vs 2.2, Qiagen CLC Genomics Workbench vs 7.0. Variants with MAF>1% reported in the dbSNP database, 1000 Genome and EVS were considered benign variants and excluded from the report.
Evaluation of enzymatic activity of acid ceramidase on cultured fibroblasts
The activity of total acid ceramidase was determined in pathological (SMA-PME and FL) and control fibroblasts as previously described using substrate composed of mixed micelles of palmityl-sphingosine (Ceramide C16) 97 μM, 90 nCi of palmitoyl- [3-3H] sphingosine (specific radioactivity 2 Ci/mmol), 0.05% Triton X-100 and 0.1% sodium cholate solubilized in 100 mM reaction buffer sodium acetate (pH 4.5).11
The radioactive sphingosine produced was detected by lipid extraction followed by the separation by high performance thin layer chromatography using the solvent system: chloroform, metanol, ammonia in the volume ratio 80:20:2. The radioactive lipids were visualized by digital autoradiography and identified by the use of authentic standards. Both radioactive sphingosine and ceramide were quantified using digital software M3vison.
Evaluation of the cell ceramide content on cultured fibroblasts
Cell ceramide content was evaluated as previously reported with some modifications.12 After two-phase partitioning of total lipid extracts, the organic phases were subjected to alkali treatment.13 Lipids were separated by high performance thin layer chromatography using the solvent systems hexane, chloroform, acetone, acetic acid 20:70:20:4. Lipids were visualized by spraying the high performance thin layer chromatography with anisaldehyde and quantified by densitometry. The estimation of the ceramide content was performed by comparison of the densitometry obtained from the ceramide in the sample and that associated with different amount 0.1–2 μg of C16 standard ceramide.
HPLC-MS/MS analyses
Mass spectrometry analysis was carried out using a ThermoQuest Finnigan LCQDeca ion-trap mass spectrometer (Finnigan MAT, San Jose, CA, USA) equipped with an electrospray ionization ion source, an Xcalibur data system and a TSP P4000 quaternary pump HPLC apparatus as previously reported.14
Results
Molecular studies
The molecular study of ASAH1 gene showed in exon 2 the presence of the novel homozygous nucleotide change c.124A>G predicted to cause the amino acid substitution p.(Thr42Ala) in both patients (Figure 1d and e). The ASAH1 exon 3 also carried the known benign variant c.214G>A, p.(Val72Met) (dbSNP, rs1071645), in homozygosis as well (data not shown). As the parents were not available for the analysis, a potential allele drop-out resulting in an apparent homozygosis, was excluded by real-time PCR quantification performed on the genomic region encompassing the c.124A>G variant.
The variant was not detected in a screen of 280 alleles, including 100 of Pakistani origin, from normal individuals, nor was it present in the EVS (evs.gs.washington.edu/) and 1000 Genomes (http://www.1000genomes.org/), while in dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/), it was present as rs779888892 (‘G' minor allele freq. =0.00002500). PolyPhen software gives a score of 0.529.
Additional support for the functional/pathological significance of this missense variant came from the high degree of evolutionary conservation, in various orthologous ASAH1 proteins, of the amino acid residue (Thr42) involved (Figure 1f).
We investigate the possibility of altered splicing for the variant c.124A>G using the ‘in silico' predictions showing a reduction of the constitutive donor site score and the creation of a newly putative cryptic acceptor (Supplementary Figure 1). In the same vein, the nucleotide substitution (c.124A>G) altered the score for recognition site of SRSF2 (also known as SC35) and caused the loss of recognition site of SRSF1 (also known as ASF1), both members of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, which constitute part of the spliceosome (Supplementary Figure 2).
On the basis of these in silico predictions, an alteration of the splicing process was expected for the c.124A>G variant. RT-PCR analysis of ASAH1 mRNA isolated from the fibroblasts of the patient #1 revealed instead the presence of normally sized 1185-bp transcripts harboring the homozygous causative r.124a>g, p.(Thr42Ala) and benign r.214g>a, p.(Val72Met) variants.
The variant c.124A>G and the associated phenotype were submitted to the Leiden Open Variant Database 3.0 (patient ID #00057786).
The complete next generation sequencing data analysis process revealed neither the presence of variants of possible pathological significance nor the presence of Variant of Unknown Significance, even though these results do not exclude the presence of additional variants in the genomic regions at lower coverage or the occurrence of rearrangements not detectable with next generation sequencing-based technique.
Evaluation of enzymatic activity of acid ceramidase, of cell ceramide content and HPLC-MS/MS analyses on cultured fibroblasts
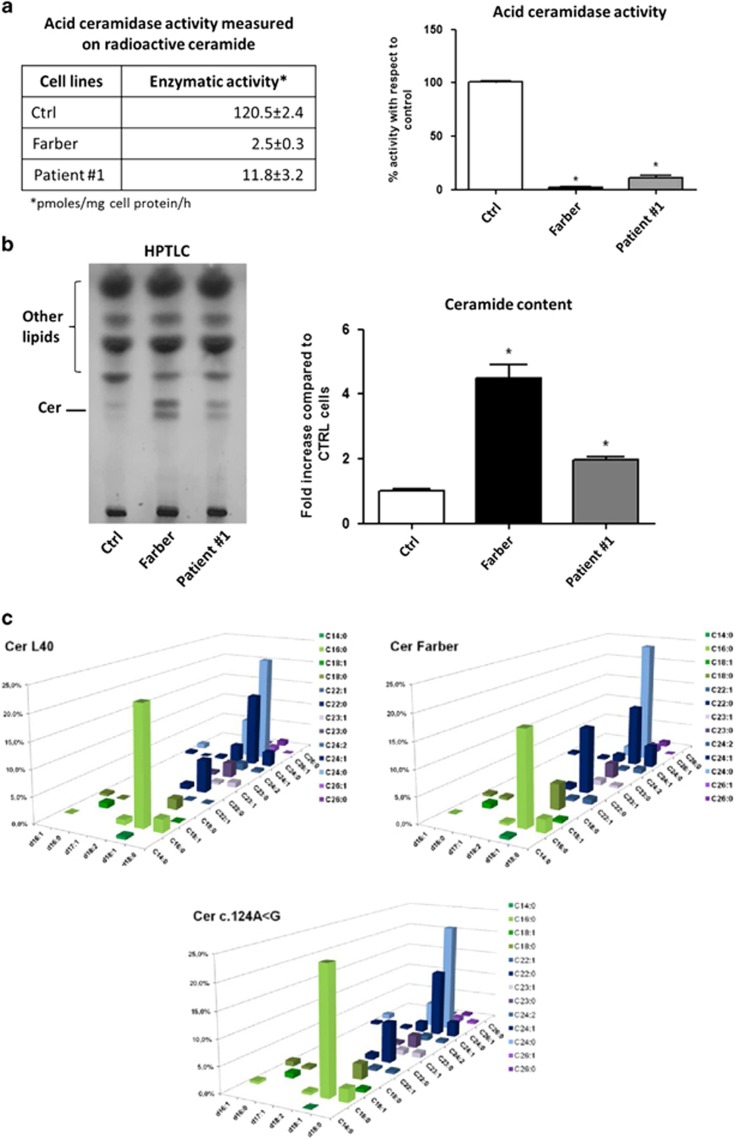
Figure 2a summarizes the results obtained in the cell lines derived from the patient #1, a normal control individual and a Farber patient. Compared with the normal control (120.5±2.4 pmoles/mg cell protein/h), the residual ceramidase activity was 2.5±0.3 pmoles/mg cell protein/h in the Farber fibroblasts (2% of control) and 11.8±3.2 pmoles/mg cell protein/h in the patient #1 carrying the c.124A>G (10% of control).
Figure 2.
(a) Acid ceramidase activity measured in normal and pathological fibroblasts. On the left, the table reports the results of the enzymatic activity in human fibroblasts derived from healthy subject (Ctrl), patient affected by Farber disease (Farber) and patient #1. Enzyme activity, measured using radioactive ceramide, is expressed as pmoles of sphingosine produced/mg of cell protein/h. On the right, the graph reports the percentages of the residual acid ceramidase activity in the pathological cells compared with the control cells. The experiments were performed in triplicate and analyzed by Bonferroni post hoc test. *P<0.01. (b) Ceramide content in normal and pathological fibroblasts. On the left, representative high performance thin layer chromatography (HPTLC) and on the right, quantitative analysis of the ceramide expressed as fold increase with respect to Ctrl. The analyses performed in triplicate show that the fibroblasts of the patient Farber and of the patient #1 display a fivefold and twofold increase in ceramide, respectively, compared with the control cell line. (c) Ceramide molecular species in normal and pathological fibroblasts determined by MS. The x axis reports the long-chain base content of sphinganine, sphingosine or sphingosine with a second double bond in an unknown position; z axis reports the length and saturation of the fatty acid and the y axis reports the percentage of each ceramide species. Data are the average of three different experiments, with SD never exceeding 15% of the average values.
Consistent with these data, Figure 2b shows that the fibroblasts of the patient Farber and of the patient #1 display a fivefold and twofold increase in ceramide, respectively, compared with the control cell line.
As shown in Figure 2c, 25 species of ceramide, containing 6 different long-chain bases and 12 fatty acids, were identified. The most abundant long-chain bases in the control cell line were d18:1 (almost 75%) and d18:2 (~14%). Also in the SMA-PME and FL fibroblasts the most representative was d18:1, but a slight reduction was found for d18:2 (~9% in content) in fibroblasts carrying the c.124A>G and more evident in Farber cell line (~5%).
According to the literature,14 the most representative fatty acids are C24:0 (~ 30%), C16:0 (~25%) and C24:1 (~20%). Interestingly in Farber cell line, the content of ceramide with C22:0 fatty acid was twofold increased if compared with the control and the c.124A>G fibroblasts.
Discussion
Variants in ASAH1 are responsible for rare cases of SMA-PME, an autosomal recessive disorder clinically characterized by childhood onset of proximal muscle weakness and atrophy due to spinal motor neuron degeneration and occurrence of myoclonic seizures.5, 6, 7, 15 Phenotype is considered clinically homogenous: patients develop progressive walking difficulties and frequent falls around age 5 and PME with slow and sharp bilateral waves of 3 to 4 cycles/s on EEG around age 7(refs. 6, 16). The disease is progressive and causes recurrent lung infections and respiratory failure, resulting in death in the teenage years.
Despite the very severe phenotype, symptoms in SMA-PME appear to be restricted to the nervous system and the disease course is less dramatic than that observed in the allelic disorder FL, which is a very rare autosomal recessive condition resulting from a profound reduction or complete lack of acid ceramidase activity.3 FL manifests most commonly between 2 weeks and 4 months of age and is clinically characterized by painful and progressively deformed joints, subcutaneous nodules (lipogranulomata), hoarseness due to laryngeal dysfunction and variable involvement of liver, spleen, lungs, heart and nervous system.
A genomic ASAH1 variant (c.125C>T; p.(Thr42Met)) has been previously reported in homozygosis in five children from two SMA-PME families and in association with a deletion of the whole gene in a third SMA-PME family.5 This study showed that the acid ceramidase activity of the mutant c.125C>T cDNA, transiently expressed into fibroblast cell line from a Farber disease-affected individual, was 32% of the wild-type cDNA.5
It is generally accepted that a marked reduction or absence of acid ceramidase activity leads to FL, whereas a milder reduction of enzyme activity causes the later-onset and nervous system-restricted SMA-PME phenotype.3, 5
Inefficient breakdown of ceramides and impaired production of its breakdown products are thought to have a role in the nerve cell damage by causing defects of motor neuron axonal branching and an increase in apoptosis in the spinal cord.2, 5, 6 However, the exact pathogenic mechanism is still unknown.
Our study describes for the first time the association between an ASAH1 variant and an adult ‘pure' SMA phenotype.
The novel c.124A>G, p.(Thr42Ala) variant caused a ASAH1-linked SMA variant with a mild and slow clinical course: no death in teenage years, no myoclonic epilepsy and no serious pulmonary involvement. It must be noted, even though unexplained, that both our patients retained tendon reflexes.
In order to rule out other causative genetic factors in association with the peculiar patient's phenotype, next generation sequencing analysis of a neuromuscular gene panel was also performed excluding the presence of variants of possible pathological significance.
Interestingly, the c.124A>G nucleotide change, causing the amino acid substitution p.(Thr42Ala), occurred very close to the c.125C>T change affecting the same codon (Thr42), previously described in association with the classical ASAH1-linked SMA phenotype.5
Biochemical studies on fibroblasts from the patient #1 bearing the c.124A>G change showed 10% of ceramidase activity and a twofold increase ceramide accumulation of normal control without specific changes in the molecular species. The same experiments, conducted in parallel on Farber fibroblasts, demonstrated 2% of ceramidase activity and a fivefold increase in ceramide (compared with the control), this suggesting that the molecular defect underlying the SMA phenotype of patient #1, led to a higher acid ceramidase residual activity and subsequently to a lower accumulation of ceramide than Farber patient.
Our patient showed a more marked reduction of enzyme activity than the previously described SMA-PME patients.5 Although these data seem to suggest that the extent of reduction of enzyme activity and accumulation of ceramide are not the only ‘players' in disease pathogenesis, the different experimental approaches used here compared with previous studies may make the results scarcely comparable.
In conclusion, the evidence of an ASAH1 linked SMA adult phenotype with no myoclonic epilepsy nor signs of FL suggests to extend the molecular testing to ASAH1 gene even in classic SMA patients without SMN1 gene alterations.
Acknowledgments
The patient's samples were obtained from the ‘Cell Line and DNA Biobank from patients affected by Genetic Diseases' (Istituto Giannina Gaslini), member of Telethon Network of Genetic Biobanks (project no. GTB12001). This work was partially supported by unrestricted grants from ‘Cinque per mille e Ricerca Corrente, ‘Ministero della Salute' to MiF, SL and RM. All human studies have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All persons gave their informed consent prior to their inclusion in the study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Bernardo K, Hurwitz R, Zenk T et al: Purification, characterization, and biosynthesis of human acid ceramidase. J Biol Chem 1995; 270: 11098–11102. [DOI] [PubMed] [Google Scholar]
- Li CM, Park JH, He X et al: The human acid ceramidase gene (ASAH): structure, chromosomal location, mutation analysis, and expression. Genomics 2000; 62: 223–231. [DOI] [PubMed] [Google Scholar]
- Moser HW, Moser AB, Chen WW, Schram AW. Ceramidase deficiency: Farber lipogranulomatosis In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic Basis of Inherited Disease Vol. II 6th ed. McGraw-Hill: New York, USA, 1989, pp 1645–1654. [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN: The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tawk M, Tiziano FD et al: Spinal muscular atrophy associated with progressive myoclonic epilepsy is caused by mutations in ASAH1. Am J Hum Genet 2012; 91: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment DA, Sell E, Vanstone MR et al: Evidence for clinical, genetic and biochemical variability in spinal muscular atrophy with progressive myoclonic epilepsy. Clin Genet 2014; 86: 558–563. [DOI] [PubMed] [Google Scholar]
- Haliloglu G, Chattopadhyay A, Skorodis L et al: Spinal muscular atrophy with progressive myoclonic epilepsy: report of new cases and review of the literature. Neuropediatrics 2002; 33: 314–319. [DOI] [PubMed] [Google Scholar]
- Filocamo M, Mazzotti R, Corsolini F et al: Cell line and DNA biobank from patients affected by genetic diseases. Open J Bioresources 2014; 1: e2. [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR: An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 2006; 15: 2490–2508. [DOI] [PubMed] [Google Scholar]
- Wang M, Marin A: Characterization and prediction of alternative splice sites. Gene 2006; 366: 219227. [DOI] [PubMed] [Google Scholar]
- Ben-Yoseph Y, Gagné R, Parvathy MR, Mitchell DA, Momoi T: Leukocyte and plasma N-laurylsphingosine deacylase (ceramidase) in Farber disease. Clin Genet 1989; 36: 38–42. [DOI] [PubMed] [Google Scholar]
- Scandroglio F, Venkata JK, Loberto N: Lipid content of brain, brain membrane lipid domains, and neurons from acid sphingomyelinase deficient mice. J Neurochem 2008; 107: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni L, Bassi R, Sonnino S, Tettamanti G: Formation of free sphingosine and ceramide from exogenous ganglioside GM1 by cerebellar granule cells in culture. FEBS Lett 1992; 300: 188–192. [DOI] [PubMed] [Google Scholar]
- Valsecchi M, Mauri L, Casellato R et al: Ceramide and sphingomyelin species of fibroblasts and neurons in culture. J Lipid Res 2007; 48: 417–424. [DOI] [PubMed] [Google Scholar]
- Rubboli G, Veggiotti P, Pini A et al: Spinal muscular atrophy associated with progressive myoclonic epilepsy: A rare condition caused by mutations in ASAH1. Epilepsia 2015; 56: 692–698. [DOI] [PubMed] [Google Scholar]
- Giráldez BG, Guerrero-López R, Ortega-Moreno L et al: Uniparental disomy as a cause of spinal muscular atrophy and progressive myoclonic epilepsy: phenotypic homogeneity due to the homozygous c.125C>T mutation in ASAH1. Neuromuscul Disord 2015; 25: 222–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.