Abstract
Whole-exome sequencing of a patient with intellectual disability and without recognisable phenotype yielded a mutation in the intron20 splice donor site of CREBBP. Mutations at different positions within the same intron20 splice donor site were observed in three patients clinically suspected as having Rubinstein-Taybi syndrome (RSTS). All mutations were de novo and likely disease-causing. To investigate a putative difference in splicing between the patient without RSTS phenotype and the three patients with the RSTS phenotype, we analysed the effects of these mutations on splicing of the pre-mRNA of CREBBP. As no RNA of patients was available, we generated a new and improved exon-trap vector, pCDNAGHE, and tested the effect of the various mutations on splicing in vitro. All mutations lead to skipping of exon20. In one of the patients with an RSTS phenotype, there was also some normal splicing detectable. We conclude that the splicing pattern obtained by exon-trapping cannot explain the difference in phenotype between the patient without the RSTS phenotype and the patients with clinical RSTS. Patient or tissue-specific splice effects as well as modifying genes likely will explain the difference in phenotype.
Introduction
Rubinstein-Taybi syndrome (RSTS1, OMIM #180849; RSTS2, #613684) is an autosomal, dominantly inherited syndrome characterised by intellectual disability, microcephaly, postnatal growth retardation, broad thumbs and halluces, and an unusual face, consisting of hypertelorism, down-slanted palpebral fissures, long eyelashes, prominent nose with convex nasal ridge and mal-positioned ears.1 RSTS is caused by mutations in CREBBP (RSTS1)2 and less frequently by mutations in EP300 (RSTS2).3 The mutations found in RSTS1 can be insertions and deletions of variable sizes, inversions and translocations, but typically are frameshift, nonsense, missense and splice-site mutations.4 The effect of sequence alterations, especially splice mutations, is not always predictable. Pre-mRNA splicing is a high-precision process and depends on the strength of the donor (5′) and acceptor (3′) sites, the branch point, poly-pyrimidine tract structure, binding sites of the spliceosome snRNPs as well as on auxiliary splicing-factor RNA-binding proteins that can enhance or suppress alternative splicing by interfering with it.5
Analysis of mutations affecting splicing is performed preferably on RNA of the patient using RT-PCR; however, RNA cannot always be obtained. In those instances, in vitro analysis using an exon-trap vector construct or a mini-gene construct is possible.6, 7 To best mimic canonical splicing, it is important that the vector contains as much of the splicing-influencing factors as possible.
Here we describe a new and improved exon-trap vector that we used to analyse the effect of four mutations in the same splice site in CREBBP, that is, in the intron20 splice donor site. The mutations were identified in three patients in whom the diagnosis of RSTS was clinically suspected, and in a patient with intellectual disability and a phenotype not resembling RSTS.
Materials and methods
Patients
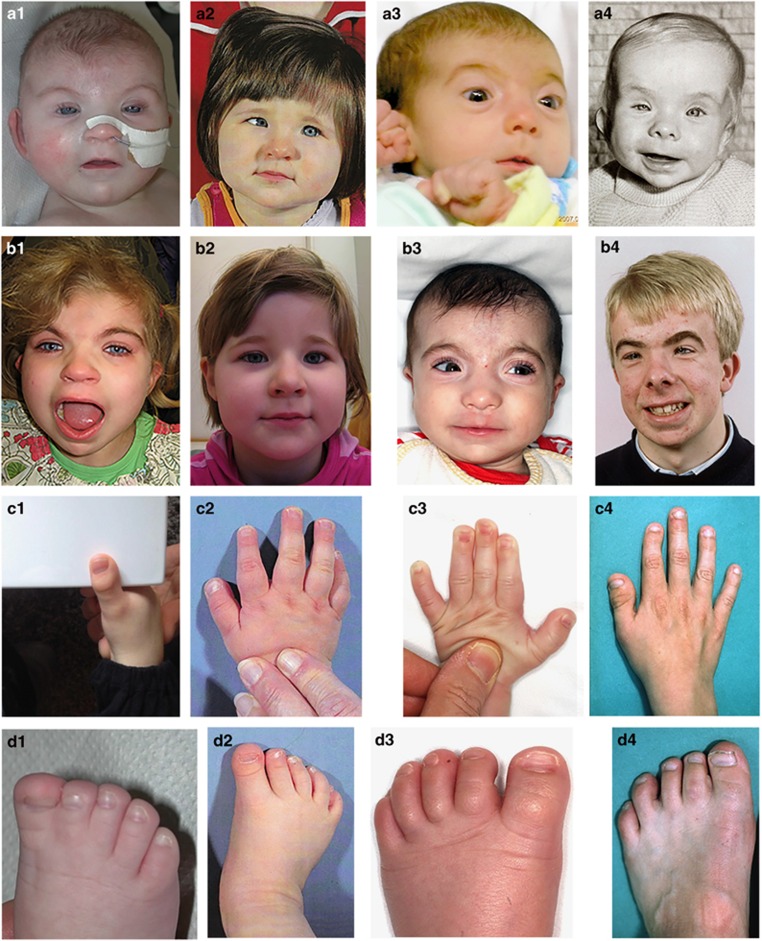
The phenotype of the patients is provided in Table 1 and illustrated in Figure 1. Patients 2, 3 and 4 show a phenotype fitting RSTS including broad thumbs and big toes, although not all facial characteristics are present. The phenotype of patient 1 differs markedly from the classical RSTS phenotype, and the long neatly pencilled eyebrows, long eyelashes, long philtrum, thin upper vermillion and downturned corners of the mouth show some resemblances to Cornelia de Lange syndrome (CdlS). The resemblance to the latter syndrome, however, did not extend to the shape of the nose or the philtrum. This patient does not show broad thumbs or big toes but did have almost complete cutaneous syndactyly between toes 1 and 2. The CREBBP mutation was identified using exome-sequencing. DNA of this patient and her parents was analysed on a CytoScan HD Array (Affymetrix, Santa Clara, CA, USA); no de novo CNVs were observed. RNA of the patients was not available. Informed consent was obtained for all patients.
Table 1. Main signs and symptoms of the patients with a mutation in the CREBBP intron20 splice donor site.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Growth retardation <3rd centile | + | − | + | − |
| Intellectual disability | Severe | Mild | Severe | Mild |
| Highly arched, broad eyebrows | + | − | + | + |
| Long eyelashes | + | + | + | + |
| Hypertelorism | + | + | + | + |
| Downward slanted palpebral fissures | − | − | − | − |
| Convex nasal ridge | − | − | − | − |
| Nasal septum below alae nasi | − | + | − | + |
| Long philtrum | + | + | − | − |
| Thin upper vermillion | + | − | + | + |
| Down-turned corners of mouth | + | − | − | − |
| Highly arched palate | − | − | + | − |
| Grimacing smile | − | + | + | + |
| Broad/angulated thumbs | − | +/− | +/− | +/− |
| Broad terminal phalange fingers | − | + | − | + |
| Broad, angulated big toes | − | +/+ | +/− | −/− |
| CREBBP mutation | c.3779+1G>A | c.3779+2T>C | c.3779+3A>T | c.3779+5G>C |
| De novo (paternity and maternity tests) | y | y father deceased | y | y |
| Other | Syndactyly toes 1–3 | Pectus excavatum | Vesicoureteral reflux | − |
| Short middle phalange fingers |
Mutations submitted to www.LOVD.nl/CREBBP (patient IDs 00166, 00112, 00098, 00035).
CREBBP reference sequence NM_004380.2; exons were numbered like in NG_009873.1.
Figure 1.
Facial and distal limb phenotype in the presently described four patients with an intron20 splice donor site in CREBBP. Patients are shown as infant (a1–4) and children (b1–4). Note the difference in phenotype between patient 1 and patients 2–4, and resemblance with Cornelia de Lange syndrome in shape and position of eyebrows, eyelashes and downturned corners of the mouth in patient 1, especially as an infant (a1). Please note that none of the depicted patients has downturned palpebral fissures, all have hypertelorism, none has a convex nasal ridge and patients 2 and 4 demonstrate their nasal septum being low positioned. The thumbs are broad in patients 2–4, and the halluces are broad in patients 2 and 3.
Construction of the exon-trap vector pCDNAGHE
The sCOGH6 vector was previously described.8 See Supplementary Methods for details of the pCDNAGHE construction.
Generation of the CREBBP constructs
Mutations c.3779+1G>A, c.3779+2T>C, c.3779+3A>T and c.3779+5G>C (www.LOVD.nl/CREBBP (patient IDs 00166, 00112, 00098, 00035) were introduced in a 13.7 kb EcoRI fragment, starting in intron17 and ending in intron22 of the CREBBP gene (CREBBP reference sequence NM_004380.2; exons were numbered like in NG_009873.1). Wild type (wt), inverted wt and mutated fragments were cloned into sCOGH6 and in pCDNAGHE. For details see Supplementary Methods.
Transfection of the constructs
For the transient transfection experiments, V79 (lung fibroblast from Chinese hamster, reference CCL-93, ATCC, Manassas, VA, USA), cells were seeded in six-well plates the day before transfection in DMEM/F12, glutaMAX, supplemented with 10% fetal calf serum (heat-inactivated), 2 μM MEM sodium pyruvate, 0.5 U/ml penicillin and 0.5 μg/ml streptomycin (Thermo Fisher Scientific, Breda, The Netherlands). The cells were transfected with DNA of the constructs; 4 μg DNA was diluted in serum-free DMEM/F12 and was complexed to 10 μl lipofectamin2000 (Thermo Fisher Scientific) according to the manufacturer's instructions. The cells were incubated with the transfection complexes for 4 h before changing to complete medium, and were harvested after an additional 20 h.
RNA isolation, RT-PCR and Sanger sequencing
RNA of the cells was isolated 24 h after transfection, using the High Pure RNA Isolation Kit (Roche Diagnostics, Almere, the Netherlands) according to the manufacturer's instructions. RNA (1 μg) was used in the transcriptor first-strand cDNA synthesis kit (Roche Diagnostics) with random hexamer primers at 55 °C for 30 min for cDNA synthesis. PCR was performed with a combination of CREBBP- and GH1-specific primers (Supplementary Table I). PCR fragments were analysed on a 2.5% agarose gel. Fragments were excised from gel, and DNA was extracted using the Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA, USA). Sanger sequencing was carried out by Macrogen (Macrogen Europe, Amsterdam, the Netherlands).
Results and Discussion
Exome-sequencing in patient 1 yielded a CREBBP intron20 splice donor site mutation, c.3779+1G>A. Additional variants observed in this patient have been listed in Supplementary Table II. Sanger sequencing confirmed the mutation and showed that it was de novo (Table 1). As the patient did not show the classical RSTS phenotype, we searched for other patients with a CREBBP mutation in the same intron, and found three patients in whom the clinical diagnosis of RSTS had been made (Figure 1) and a mutation had been found using Sanger sequencing. Their mutations were c.3779+2T>C, c.3779+3A>T and c.3779+5G>C (Table 1). Subsequently, we studied whether there was a correlation between the effect on splicing and the variation in phenotype using exon-trapping.
For these experiments we generated an upgraded vector, pCDNAGHE, for which we adapted the previously published sCOGH6 construct (Supplementary Methods and Results).8 The system is based on the insertion of multiple exons and introns surrounding the mutation in an intron of the complete GH1 gene (Supplementary Figure 1a).
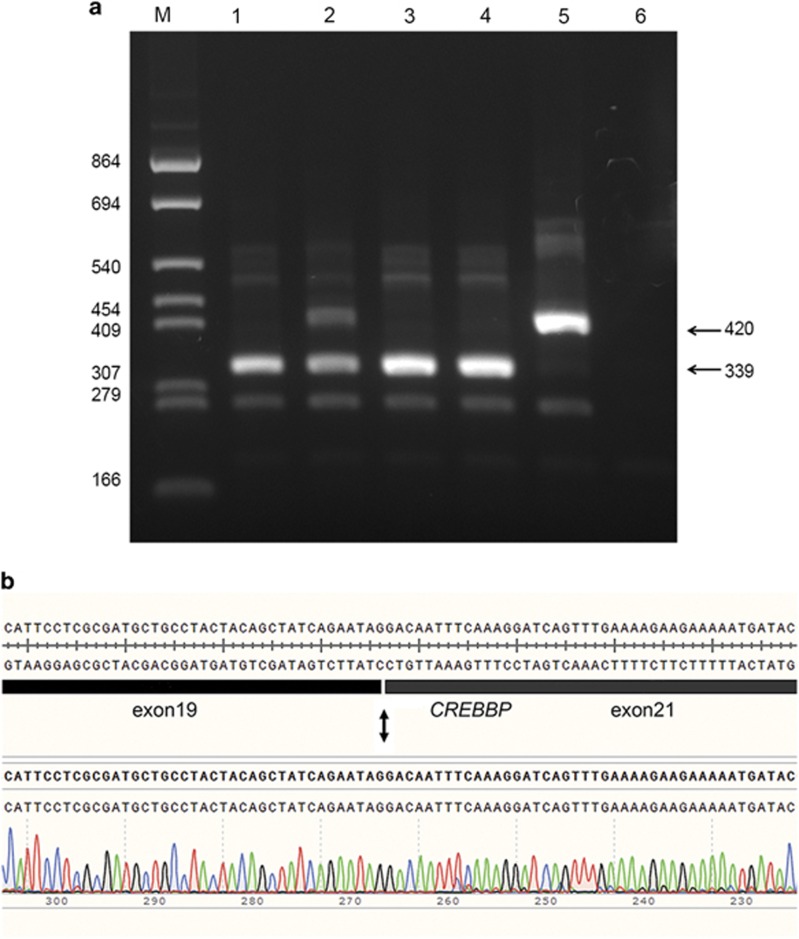
We generated the six constructs containing the normal 13.7 kb EcoRI fragment of CREBBP intron17 to intron22 (pCDNAGHE137), the inverted normal fragment and the four splice-site mutations, c.3779+1G>A, c.3779+2T>C, c.3779+3A>T and c.3779+5G>C, and transfected these into V79 cells. Using primers located in CREBBP exon19 and GH1 exon5 (Supplementary Table I), the normal construct showed a band of the expected 420 bp, whereas the mutant constructs showed only a smaller band of 339 bp, except for the construct of patient 2, which showed both the wt and the smaller band, indicating partial normal splicing in this patient. As expected, no bands were seen for the inverted construct (Figure 2a). Sequence analysis of the 420 and 339 bp fragments showed the normal sequence and a deletion of exon20 (Figure 2b), respectively. Other primer combinations demonstrated the same pattern (data not shown); the whole experiment was repeated three times, giving the same outcome.
Figure 2.
Alternative splicing of CREBBP intron20 splice donor site mutations demonstrated by exon-trapping with the pCDNAGHE vector. (a) A 2.5% agarose gel showing the results of a RT-PCR with primers in CREBBP exon19 and GH1 exon5 on samples (1) pCDNAGHE137pat1, (2) pCDNAGHE137pat2, (3) pCDNAGHE137pat3, (4) pCDNAGHE137pat4, (5) pCDNAGHE137, (6) pCDNAGHE137inv. A 339 bp fragment is seen in all patients corresponding with a deletion of exon20, with only patient 2 showing additionally a wild-type band of 420 bp, which is also seen in the normal control. No bands can be seen for the inverted insert of pCDNAGHE137inv. (b) Sequence of the 339 bp fragment (patient 1) shows a CREBBP exon20 deletion.
Therefore, all splice mutations resulted in a skip of exon20, and in the c.3779+2T>C mutation construct additionally the normally spliced product was produced, indicating less disturbed splicing. As in the splice-site consensus sequence only the +1G and the +2T are found in 100% of the splice donor sites, the position of the nucleotide involved could not explain the partial normal splicing seen for the c.3779+2T>C mutation.9
Skipping of exon20 leads to the in-frame deletion of 81 bp in the CREBBP gene. At the protein level the deletion of 27 amino acids partially deletes the DUF902 domain. This domain of unknown function has been found in several transcriptional co-activators, including in the C terminus of the bromodomain in CREBBP, and is localised in the histone acetyltransferase (HAT) domain. As most missense mutations found in RSTS1 are clustered in the HAT domain, it was suggested that loss of HAT activity may be sufficient to cause RSTS1.4, 10, 11 This, together with the fact that all mutations are de novo (Table 1), makes it likely that the splice mutations presented in this study are pathogenic.
The preferred method to study the effect of mutations on pre-mRNA splicing is the analysis of patient-derived mRNA obtained from blood or preferably from relevant tissue(s). Frequently, patient material is difficult to obtain, particularly when young and anxious patients are involved, as was the case in this study. Therefore, we applied exon-trapping, which enables the comparison of mutations in the same cellular environment. Using the new pCDNAGHE vector we successfully showed splicing of wt and mutant CREBBP constructs. By analysing a fragment containing multiple exons and introns surrounding the mutated splice site, the chance to miss splicing because of non-sequential splicing is reduced. This is of importance as recent studies have shown that some parts of pre-mRNAs splice non-sequential and affect the splicing of multiple exons.12 Our data indicate that in vitro there is no correlation between altered splicing and the phenotypes of these patients. We cannot rule out, however, that in vivo such a correlation exists. This could be related to cell types and developmental stage and/or the influence of variations in splicing associated factors in the different patients.
The phenotypical differences most likely can be explained by the influence of other modifying genes. Mutations in those genes may disturb a biological balance, thereby increasing or decreasing the expressivity of certain symptoms.13 In recent years, it became increasingly evident that many malformation/intellectual disability syndromes not only share clinical characteristics but also overlap in genes that are mutated, suggesting a connection and convergence of functional pathways. For example, CdlS (OMIM #122470, #300590, #610759, #614701 and #300882), Wiedemann–Steiner syndrome (WDSTS, OMIM #605130), Floating-Harbor syndrome (FLHS, OMIM #136140), Coffin-Siris syndrome (CSS, OMIM #135900) and RSTS share overlapping clinical features. Previously, a mutation in EP300 (RSTS2) has been reported in a patient with symptoms resembling CdlS, and also patient 1 in this report shows overlapping clinical characteristics fitting CdlS.14 Although the large majority of CdlS patients carry mutations in genes that encode subunits or regulators of the cohesin complex, these proteins also have a role in the regulation of transcription. Even more, mutations in the methyl-transferases KMT2A and TAF6, genes involved in epigenetic modification and chromatin remodelling, have been reported as well.13 Altogether, global transcriptional disturbances may underlie CdlS and related syndromes such as RSTS and FLHS. In addition, a mutation in the synaptonemal complex component SYCP2 has also been shown to act as a disease modifier in two SMC1A mutation carriers diagnosed with WDSTS, supporting the hypothesis that the mutational load may affect the biological balance and lead to occurrence or aggravation of the disease.13
In conclusion, we describe four patients carrying a mutation in the same splice donor site: three patients in whom the diagnosis of RSTS was clinically suspected and one patient with intellectual disability and a phenotype not resembling RSTS. The phenotypic differences could not be explained by effects on splicing and suggests the involvement of modifying genes, altering the expressivity of the disease phenotypes.
Acknowledgments
We would like to thank the patients and their families for their participation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Rubinstein JH, Taybi H: Broad thumbs and toes and facial abnormalities. A possible mental retardation syndrome. Am J Dis Child 1963; 105: 588–608. [DOI] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG et al: Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature 1995; 376: 348–351. [DOI] [PubMed] [Google Scholar]
- Roelfsema JH, White SJ, Ariyürek Y: Genetic heterogeneity in Rubinstein–Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet 2005; 76: 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema JH, Peters DJ: Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med 2007; 20: 1–16. [DOI] [PubMed] [Google Scholar]
- de Klerk E, 't Hoen PA: Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet 2015; 31: 128–139. [DOI] [PubMed] [Google Scholar]
- Burn TC, Connors TD, Klinger KW et al: Increased exon-trapping efficiency through modifications to the pSPL3 splicing vector. Gene 1995; 161: 183–187. [DOI] [PubMed] [Google Scholar]
- Baralle D, Lucassen A, Buratti E: Missed threads. The impact of pre-mRNA splicing defects on clinical practice. EMBO Rep 2009; 10: 810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datson NA, van de Vosse E, Dauwerse HG et al: Scanning for genes in large genomic regions: cosmid-based exon trapping of multiple exons in a single product. Nucleic Acids Res 1996; 15: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM: A catalogue of splice junction sequences. Nucleic Acids Res 1982; 22: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorry EK, Keddache M, Lanphear N et al: Genotype-phenotype correlations in Rubinstein-Taybi syndrome. Am J Med Genet A 2008; 146A: 2512–2519. [DOI] [PubMed] [Google Scholar]
- Murata T, Kurokawa R, Krones A et al: Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum Mol Genet 2001; 10: 1071–1076. [DOI] [PubMed] [Google Scholar]
- Pulyakhina I, Gazzoli I, 't Hoen PA et al: SplicePie: a novel analytical approach for the detection of alternative, non-sequential and recursive splicing. Nucleic Acids Res 2015; 43: e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Pehlivan D, Karaca E et al: Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest 2015; 125: 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods SA, Robinson HB, Kohler LJ et al: Exome sequencing identifies a novel EP300 frame shift mutation in a patient with features that overlap Cornelia de Lange syndrome. Am J Med Genet A 2014; 164A: 251–258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.