Abstract
The sensitivity of testing BRCA1 and BRCA2 remains unresolved as the frequency of deep intronic splicing variants has not been defined in high-risk familial breast/ovarian cancer families. This variant category is reported at significant frequency in other tumour predisposition genes, including NF1 and MSH2. We carried out comprehensive whole gene RNA analysis on 45 high-risk breast/ovary and male breast cancer families with no identified pathogenic variant on exonic sequencing and copy number analysis of BRCA1/2. In addition, we undertook variant screening of a 10-gene high/moderate risk breast/ovarian cancer panel by next-generation sequencing. DNA testing identified the causative variant in 50/56 (89%) breast/ovarian/male breast cancer families with Manchester scores of ≥50 with two variants being confirmed to affect splicing on RNA analysis. RNA sequencing of BRCA1/BRCA2 on 45 individuals from high-risk families identified no deep intronic variants and did not suggest loss of RNA expression as a cause of lost sensitivity. Panel testing in 42 samples identified a known RAD51D variant, a high-risk ATM variant in another breast ovary family and a truncating CHEK2 mutation. Current exonic sequencing and copy number analysis variant detection methods of BRCA1/2 have high sensitivity in high-risk breast/ovarian cancer families. Sequence analysis of RNA does not identify any variants undetected by current analysis of BRCA1/2. However, RNA analysis clarified the pathogenicity of variants of unknown significance detected by current methods. The low diagnostic uplift achieved through sequence analysis of the other known breast/ovarian cancer susceptibility genes indicates that further high-risk genes remain to be identified.
Introduction
Functional variants in BRCA1 and BRCA2 account for 2–3% of all cases of breast cancer and 10–15% of epithelial ovarian cancer.1, 2 While the relationship between the two genes and cancer predisposition has been known for over 20 years and variant screening is offered routinely by a number of diagnostic laboratories worldwide, the sensitivity of variant detection is unresolved. This is mainly due to the heterogeneity of breast cancer with BRCA1/2 contributing only about 15–20% of the hereditary component.3 Loss of function variants in two genes, RAD51C4 and RAD51D,5 confer a high risk of ovarian cancer; however, their contribution to breast cancer risk is less clear.5, 6 Until the discovery of functional variants in these two genes, it was assumed that the majority, if not all, of the inherited link between breast and ovarian cancer was due to variants in BRCA1 or BRCA2.7 Ninety-five percent of families with the Breast Cancer Linkage Consortium (BCLC) criteria of at least four individuals with breast/ovarian cancer, with the breast cancers diagnosed <60 years of age and at least one case of ovarian cancer in the family, were predicted to have a variant in either BRCA1 or BRCA2. This increased to 100% for families with two or more women with ovarian cancer. Similarly, 95% of male breast cancer families fulfilling BCLC criteria were due to variants in these genes (80% BRCA2 and 15% BRCA1).7 We have previously shown that variants in RAD51C and RAD51D make little contribution to very high-risk BRCA1/2-negative breast ovarian cancer families with only one BCLC criteria family having a variant.8
We have reported that families with Manchester score of 40+ are extremely likely to harbour a pathogenic BRCA1/2 variant.8, 9, 10 In probands with female breast cancer, only the variant detection rates are 2–12% lower in absolute terms than in families in which a proband with ovarian cancer or breast and ovarian cancer has been tested. This difference is likely due to the higher rate of phenocopies in breast cancer accounting for a reduction in sensitivity of about 6%.11
The reduced sensitivity of testing for BRCA1/2 variants could be attributed to variants in other as yet unidentified genes, which increase both breast and ovarian cancer risk. However, a small increase in the sensitivity of testing of BRCA1/2 would have more clinical application than identifying another gene with a similar variant frequency and risk profile to RAD51C/D. Deep intronic splicing variants contribute about 2–3% of the mutational spectrum in neurofibromatosis 1 (NF1)12 and we have reported similar rates for neurofibromatosis 2.13 Sensitivity of variant detection in both genes is at least 95% with RNA sequence analysis,12, 13 and we have detected variants in 97% of individuals fulfilling NF1 diagnostic criteria including 10/347 (2.9%) with deep intronic splicing mutations. There has been no reported comprehensive analysis of BRCA1 and BRCA2 using RNA sequencing to identify functional splicing isoforms secondary to deep intronic variants.
For families with multiple affected members with breast/ovary cancer, it is vital to identify the causative gene variant. Such information allows accurate clarification of risk in other family members, and therefore the need for screening or risk reducing surgery. In addition, it may predict sensitivity to novel therapies, including PARP inhibitors. Therefore, we aimed to establish the utility of RNA analysis to clarify the pathogenicity of variants of uncertain significance predicted to alter splicing. We aimed to determine how many of the unresolved cases of familial breast/ovarian cancer or male breast cancer were due to BRCA1/2 deep intronic splicing variants and what proportion may be due to variants in other known or unknown genes.
Materials and Methods
Affected individuals from families with breast and/or ovarian cancer have been screened in Manchester mainly using Sanger sequencing of DNA for all exons and intron–exon boundaries as well as a test for large rearrangements using Multiple Ligation Dependent Amplification (MLPA) as previously described.8, 9, 10 DNA had been extracted from lymphocytes. We have ascertained 37 breast/ovary/male breast cancer families (Tables 1 and 2) with no identified clearly functional variant in BRCA1/2 in at least one fully tested affected individual. These included six families with males affected by breast cancer including one family with two affected by ovarian cancer. Another family included a 37-year-old affected male with breast cancer with five female relatives <50 years with breast cancer and a family with three males with breast cancer and no BRCA1/2 variant were also available. There were 31 non-male breast cancer families containing at least one non-mucinous epithelial ovarian cancer with a combined Manchester score (Table 3)9 of 25 or greater. Twelve of these families have had at least two affected family members fully tested by Sanger sequencing of DNA and MLPA. An additional eight female only breast cancer families were selected on the basis of a known variant of uncertain significance (VUS) or BRCA-like features (eg, two sisters with high-grade triple-negative breast cancer <35), lymphoblastoid cell lines were available on all families. Variants, functional or not identified in this study, were named according to HGVS guidelines using the following reference sequences; LRG_292t1 (BRCA1); LRG_293t1 (BRCA2); LRG_135_t1 (ATM) and LRG_516_t1 (RAD51D).
Table 1. RNA testing of high-risk families without a clear functional BRCA1/2 variant and subsequent gene panel testing.
| Number | Manchester score | Type | Ratio | BC | BC age1 | BC age2 | OC age | BRCA1/2 | BCLC criteria | VUS | Other gene | Panel positive | RNA positive |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G22836 | 65 | Breast/ovary | 109 | Yes | 32 | 39 | - | BRCA1 | Yes | BRCA1 c.5152+4A>G | No | No | Yes |
| fh0028 | 59 | Breast/ovary | 107 | Yes | 45 | - | No | Yes | No | No | No | ||
| C0344/fh2061 | 56 | Breast/ovary | 302 | No | - | No | Yes | BRCA2 c.8488-5T>C | No | No | No | ||
| g31973 | 55 | Breast/ovary | 303 | Yes | 32 | - | No | Yes | RAD51D c.556C>T,p.(Arg186*) | Yes | No | ||
| C2298 | 55 | Breast/ovary | 305 | Yes | 76 | 76 | No | Yes | No | No | No | ||
| FH5563 | 53 | Breast/ovary | 302 | Yes | 42 | - | No | Yes | No | No | No | ||
| g41478 | 51 | Breast/ovary | 204 | Yes | 53 | - | No | Yes | No | No | No | ||
| C1048 | 49 | Breast/ovary | 302 | Yes | 42 | - | BRCA1 | Yes | BRCA1 c.4868C>G, p.(Ala1623Gly) | No | No | Yes | |
| G71619/FH7849 | 48 | Breast/ovary | 303 | No | 60 | No | Yes | No | No | No | |||
| C0658/fh2755 | 43 | MBC | 204 | Yes | 31 | - | No | Yes | BRCA2 c.1395A>C, p.( =) | No | No | No | |
| FH0221 | 42 | Breast/ovary | 203 | Yes | 49 | 61 | - | No | Yes | No | No | No | |
| FH1670 | 41 | Breast/ovary | 204 | No | 60 | No | Yes | No | No | No | |||
| MBC14 | 40 | MBC | Yes | 54 | - | No | No | No | No | ||||
| g53650 | 40 | Ovary | 300 | No | 30 | No | No | No | No | ||||
| g79535 | 40 | FBC only | Yes | 45 | 48 | - | No | No | No | No | |||
| FH2910 | 39 | MBC | Yes | 37 | - | No | Yes | CHEK2 c.1263delT, p.(Ser422Valfs*15) | Yes | No | |||
| g61338 | 39 | MBC | Yes | 36 | - | No | Yes | No | No | No | |||
| g33876 | 36 | Breast/ovary | 103 | Yes | 31 | 43 | No | Yes | No | No | No | ||
| g8510 | 36 | Ovary | 300 | No | 68 | No | No | No | No | ||||
| FH6284 | 36 | Breast/ovary | 102 | Yes | 46 | - | No | BRCA2 c.4614T>C, p.(=) | No | No | No | ||
| G56432 | 36 | FBC only | Yes | 35 | 35 | - | No | No | No | No | |||
| g31074 | 35 | Breast/ovary | 202 | Yes | 33 | - | No | Yes | No | No | No | ||
| g39632/fh4173 | 35 | MBC | Yes | 39 | - | No | BRCA2 c.6725A>T, p.(Asp2242Val) | No | No | No | |||
| fh5674 | 35 | Breast/ovary | 105 | Yes | 67 | - | No | No | No | No | |||
| FH4584 | 35 | Breast/ovary | 104 | Yes | 45 | 56 | No | BRCA1: c.4676-7C>T | ATM c.7271T>G, p.(Val2424Gly) | Yes | No | ||
| MBC18 | 35 | MBC | Yes | 29 | - | No | No | No | No | ||||
| g37126 | 33 | Breast/ovary | 103 | Yes | 43 | - | No | Yes | BRCA2 c.9082G>C, p.(Ala3028Pro) | No | No | No | |
| g33630 | 33 | Breast/ovary | 103 | Yes | 37 | 43 | No | Yes | No | No | No | ||
| c1922/g53571 | 33 | Breast/ovary | 103 | Yes | 32 | 37 | No | Yes | No | No | No | ||
| g67282 | 32 | Breast/ovary | 106 | Yes | 58 | - | No | Yes | BRCA2, c.8488-5T>C | No | No | No | |
| C1346 | 31 | Breast/ovary | 103 | Yes | 36 | - | No | Yes | No | No | No | ||
| g53030 | 31 | Breast/ovary | 103 | Yes | 46 | 49 | No | No | No | No | |||
| FH5206 | 31 | Breast/ovary | 104 | Yes | 46 | - | No | Yes | No | No | No | ||
| g78035/fh1887 | 30 | Breast/ovary | 103 | Yes | 47 | - | No | Yes | No | No | No | ||
| g46192 | 29 | Breast/ovary | 103 | Yes | 52 | - | No | Yes | No | No | No | ||
| X1611879 | 29 | Breast/ovary | 105 | Yes | 50 | 55 | - | No | No | No | |||
| FH0722 | 28 | FBC only | Yes | 37 | - | No | BRCA2 c.7540A>G, p.(Lys2514Glu) | No | |||||
| G78690 | 27 | Breast/ovary | 102 | Yes | 34 | - | No | BRCA2 c.5319G>C, p.(Glu1773Asp) | No | No | No | ||
| g45393 | 26 | Breast/ovary | 103 | Yes | 42 | 66 | - | No | No | No | |||
| G79865 | 26 | Breast/ovary | 105 | Yes | 46 | 65 | 73 | No | BRCA2 c.6750A>G, p.(=) | No | No | No | |
| FH3252 | 24 | FBC only | Yes | 41 | - | No | No | No | No | ||||
| FH4522 | 20 | FBC only | Yes | 48 | - | No | BRCA2 c.7939C>G, p.(Leu2647Val) | No | No | No | |||
| X1908593 | 20 | FBC only | Yes | 25 | - | No | No | No | No | ||||
| 200907192 | 20 | FBC only | Yes | 33 | - | No | No | No | No | ||||
| g82840 | 19 | FBC only | Yes | 28 | - | No | No | No | No |
Abbreviations: FBC, female breast cancer; MBC, male breast cancer; OC, ovarian cancer. Ovary breast ratio: 103 is one ovarian and 3 breast cancers; 302 three ovarian and 2 breast cancers. Reference sequences used: LRG_292t1 (BRCA1); LRG_293t1 (BRCA2).
Table 2. Detection rates for BRCA1 and BRCA2 variants in breast/ovarian and male breast cancer families by Manchester score.
| Manchester score 50+ | Manchester score 40–49 | Manchester score 35–39 | Manchester score 30–34 | Manchester 25–29 | Total | |
|---|---|---|---|---|---|---|
| Breast/ovary | 44/51 (87%) | 46/60 (77%) | 38/53 (66%) | 40/87 (43%) | 44/146 (30%) | 212/397 (53%) |
| MBC | 5/5 (100%) | 5/8 (62.5%) | 7/12 (58%) | 8/13 (61.5%) | 2/16 (12.5%) | 27/54 (50%) |
| Total | 49/56 (87.5%) | 51/68 (75%) | 45/65 (69%) | 48/100 (48%) | 46/162 (28%) | 239/451 (53%) |
| RNA analysis in negative | 7/7 | 7/17 | 10/20 | 8/62 | 5/116 | 37/222 |
| RNA positive | 1/7 | 1/7 | 0/11 | 0/8 | 0/5 | 2/37 |
| Panel testing | 6/6 | 7/16 | 11/20 | 8/62 | 5/116 | 37/220 |
| Panel positive | 1x RAD51D of 6 | 0/7 | 1x ATM 1x CHEK2 of 11 | 0/8 | 0/5 (0/5 <25) | 3/42 |
| Female breast only | 4/5 (80%) | 11/19 (58%) | 21/37 (57%) | 34/69 (49%) | ||
| RNA FBC | 1 | 1 | 2 (4-16-20) | 0/8 | ||
| Panel testing | 0/1 | 0/1 | 0/1 (0/1 20) | 0/3 |
Abbreviations: FBC, female breast cancer; MBC, male breast cancer.
Table 3. Manchester scoring system to identify likelihood of a BRCA1/2 mutation.
| Case of relevant cancer and age at diagnosis | BRCA1 | BRCA2 |
|---|---|---|
| FBC <30 | 6 | 5 |
| FBC 30–39 | 4 | 4 |
| FBC 40–49 | 3 | 3 |
| FBC 50–59 | 2 | 2 |
| FBC >59 | 1 | 1 |
| MBC <60 | 5 if BRCA2 tested | 8 |
| MBC >59 | 5 if BRCA2 tested | 5 |
| Ca ovary <59 | 8 | 5 if BRCA1 tested |
| Ca ovary >60 | 5 | 5 if BRCA1 tested |
| Ca pancreas | 0 | 1 |
| Ca prostate <60 | 0 | 2 |
| Ca prostate >59 | 0 | 1 |
Each cancer in a direct lineage is scored including contralateral breast cancers. The highest lineage score is used.
Previously identified variants in BRCA1/2
Currently, VUS in BRCA1/2 have been identified on DNA sequencing in 13 of 45 families (10/38 breast/ovary/male families) (Table 1) with two families with BRCA2 c.8488-5T>C a potential splicing variant. Four of the twelve different variants are likely polymorphic, but eight may affect splicing and RNA analysis will confirm this and define pathogenicity. The variant BRCA1 c.4868C>G, p.(Ala1623Gly) is the only one with a high likelihood of being functional based on the previous work.
RNA analysis
Although we could have whole genome sequencing at the DNA level we chose RNA analysis as would provide evidence of a functional effect of any variant identified, whereas identification of a deep intronic variant on genome sequencing would still require assessment at the RNA level. RNA was obtained from a fresh blood sample from an affected family member least likely to be a phenocopy or from cell lines already available. This was one with breast and ovarian cancer, ovarian cancer or one with an affected offspring. Sanger sequencing of the cDNA of BRCA1/2 was carried out to identify whether any changes in the transcript suggestive of deep intronic splice variants were present. We have experience of identifying these in NF1 and NF2. Any unexplained allelic imbalance in RNA prompted promoter sequencing and microarray analysis to identify centromeric deletions such as those seen in EPCAM that can cause methylation of the MSH2 promoter. RNA was extracted from blood collected in PAXgene blood tubes using the PAXgene blood RNA kit (PreAnalytiX GmbH, Hombrechtikon, Switzerland). Where lymphoblastoid cell lines were available, RNA was extracted using Qiagen QIAshredder (Qiagen, Hilden, Germany) and Qiagen BioRobot EZ1 (Qiagen) following the manufacturer's protocols. RNA was reverse transcribed to cDNA using the Life Technologies High Capacity RNA-to-cDNA kit (Life Technologies, Carlsbad, CA, USA). Overlapping primer pairs were designed to produce amplicons of around 1500 bp, covering the whole of the BRCA1 and BRCA2 genes. PCRs were performed using Promega GoTaq Hot Start Green master mix (Promega, Madison, WI, USA) and run on Applied Biosystems Veriti thermal cyclers (Life Technologies). Sequencing PCRs were carried out using Life Technologies BigDye Terminator reagents and run on an ABI 3730 analyser (Life Technologies).
In individuals where there was no heterozygosity of common variants, a selection of variants (BRCA1 hg19.chr17:g.41245466G>A (rs1799949), hg19.chr17:g.41245237G>A (rs16940), hg19.chr17:g.41245936G>A (rs799917), hg19.chr17:g.41244435T>C (rs16941), hg19.chr17:g.41244000T>C (rs16942) and BRCA2 hg19.chr13:g.32911888A>G (rs1801406), hg19.chr13:g.32912299T>C (rs543304), hg19.chr13:g. 32973012A>C (rs15869), hg19.chr13:g. 32889792A>G (rs206118), hg19.chr13:g. 32906729A>C (rs144848), hg19.chr13:g. 32929232A>G (rs1799955)) was Sanger sequenced using DNA previously extracted to rule out hemizygosity of the transcript.
Panel testing
Panel testing was carried out by next-generation sequencing of 12 genes known to confer increased risk of breast, ovarian or both breast/ovarian cancer. These were BRCA1, BRCA2, TP53, CDH1, STK11, PTEN, PALB2, BRIP1, RAD51C, RAD51D, ATM and CHEK2. Enrichment was performed using the Illumina 94 gene TruSight Cancer Sequencing Panel according to the manufacturer's protocol. Quantified libraries were sequenced on the Illumina MiSeq platform using the paired end (dual indexed) 2 × 150 bp configuration to a depth of at least 30 × . Novoalign (v3.01.00, Novocraft Technologies, Selangor, Malaysia) was used to align to the human genome build hg19. Reads were converted to a bam file, sorted and indexed using samtools (version 0.1.19; https://urldefense.proofpoint.com/v2/url?u=https-3A__samtools.github.io_&d=BQIFAgc=bMxC-A1upgdsx4J2OmDkk2Eep4PyO1BA6pjHrrW-ii0&r=1DJkrySuN1oA897_rRkN9LSRniqquwAY7jkl3rFxkFI&m=9kERfOVGZ_H1gIDwKU-VsZPrszMX2cM1tUtHJaWQhsc&s=4N50QNjl5exOvObLZYfTfza27wUgHyZzZvHGMu98hD8&e=). The Genome Analysis Toolkit (GATK) (v2.7-1; https://urldefense.proofpoint.com/v2/url?u=https-3A__www.broadinstitute.org_gatk_&d=BQIFAg&c=bMxC-A1upgdsx4J2OmDkk2Eep4PyO1BA6pjHrrW-ii0&r=1DJkrySuN1oA897_rRkN9LSRniqquwAY7jkl3rFxkFI&m=9kERfOVGZ_H1gIDwKU-VsZPrszMX2cM1tUtHJaWQhsc&s=MTfE6NILD2cmEGLvb7UlldDtcpS_vy8T4Atgh5XkUkU&e=) was used to quality score and recalibrate reads and duplicate reads were removed using the MarkDuplicates tool from Picard (v1.97; https://urldefense.proofpoint.com/v2/url?u=http-3A__broadinstitute.github.io_picard_&d=BQIFAg&c=bMxC-A1upgdsx4J2OmDkk2Eep4PyO1BA6pjHrrW-ii0&r=1DJkrySuN1oA897_rRkN9LSRniqquwAY7jkl3rFxkFI&m=9kERfOVGZ_H1gIDwKU-VsZPrszMX2cM1tUtHJaWQhscs=7PCxOado_kCuVoNqyv1D86Yo6IaaQ9Kp_1JI8ou-baU&e=). Variants were called using both HaploTypeCaller from the GATK (v2.7-1) and Pindel (v0.2.5a1; https://urldefense.proofpoint.com/v2/url?u=http-3A__gmt.genome.wustl.edu_packages_pindel_&d=BQIFAg&c=bMxC-A1upgdsx4J2OmDkk2Eep4PyO1BA6pjHrrW-ii0&r=1DJkrySuN1oA897_rRkN9LSRniqquwAY7jkl3rFxkFI&m=9kERfOVGZ_H1gIDwKU-VsZPrszMX2cM1tUtHJaWQhsc&s=9Fmu9iWox_DJ5kYoj8xFsdZu3L-0rya10mY9lYHDndo&e=) for more complex variants. Variants were annotated using the Table_annovar.pl script from the annovar package (v2013-06-21). All variants classified as likely functional or functional were validated by Sanger sequencing.
This study was approved by Central Manchester Research Ethics Committee reference number 10/H1008/24, 11 July 2013.
Results
RNA sequencing of samples from 45 individuals at high risk of a functional BRCA1/2 variant but with no clearly functional variant on DNA analysis has been carried out between 2012 and 2015 (Tables 1 and 2). This included 24 samples from breast/ovarian or male breast cancer families with Manchester scores >35 and 13 with scores between 25 and 34. A further eight samples from affected individuals from families with female breast cancer only were analysed.
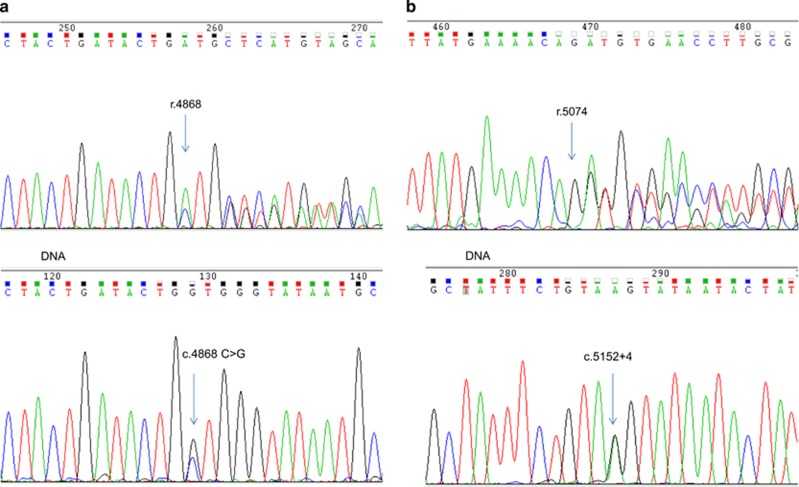
Abnormal splicing was demonstrated in two families with putative splicing variants identified on standard variant analysis. These were BRCA1 c.4868C>G p.(Ala1623Gly) and a novel splice donor variant in intron 17 of BRCA1 (c.5152+4A>G), which to our knowledge has not been reported previously with RNA data as a functional variant does appear on dbSNP as rs397509232 (hg19.chr17:g.41215887T>C) with no reported frequency (Figures 1a and b). BRCA1 c.4868C>G p.(Ala1623Gly) leads to heterozygous deletion of the 3' end of exon 15 (r.4868_4686del) predicted to result in p.Ala1623Aspfs*16. The other variant BRCA1 c.5152+4A>G led to heterozygous deletion of exon 17 (r.5075_5152del) predicted to result in p.Asp1692_Trp1718delinsGly.
Figure 1.
(a) Chromatogram of cDNA showing abnormal splicing in BRCA1. Heterozygous deletion of 3' end of exon 15 (r.4868_4686), caused by c.4868C>G predicted to result in p.Ala1623Aspfs*16. (b) Chromatogram of cDNA showing abnormal splicing in BRCA1. Heterozygous deletion of exon 17 (r.5075_5152del) caused by c.5152+4 A>G in intron 17, predicted to result in p.Asp1692_Trp1718delinsGly.
No abnormal splicing was found secondary to the BRCA2 variant c.7939C>G p.(Leu2647Val), which has been reported to be functional. Two further candidate splicing variants were found not to affect splicing BRCA2 c.8488-5T>C and BRCA1 c.4676-7C>T (Table 4).
Table 4. Predicted effect of 12 VUS in BRCA1 and BRCA2 and effects at RNA level.
| Variant | Predicted effect In silico | Predicted effect Alamut | VUS classification before RNAa | Evidence to support classification | Abnormal splicing on RNA | Effect on RNA |
|---|---|---|---|---|---|---|
| BRCA1c.4676-7C>T | No significant change predicted | N/A | Class 2 | Published data based on a multifactorial likelihood-ratio model predicts it to be neutral | No | None |
| BRCA1 c.4868C>G p.(Ala1623Gly) | Strong effect predicted (creation of cryptic novel high ranking splice-acceptor site) | Amino acid not evolutionary conserved | Class 5 | From previous published evidence of effect on splicing | Yes | Heterozygous deletion in exon 15 r.4868_4986del (NM_007294.3 on Ensembl) |
| BRCA1 c.5152+4A>G | Strong effect predicted (significant reduction in splice donor ranking plus creation of novel splice donor and acceptor) | N/A | Class 3 | Yes | Heterozygous deletion of exon 17 r.5075_5152del (NM_007294.3 on Ensembl) | |
| BRCA2c.1395A>C, p.(=) | No significant change predicted (loss of cryptic splice acceptor) | N/A | Class 1 | Range of published evidence including no effect on splicing | No | None |
| BRCA2c.4614T>C, p.(=) | No significant change predicted | N/A | Class 1 | Good co-occurrence data | No | None |
| BRCA2c.5319G>C, p.(Glu1773Asp) | No significant change predicted | Amino acid not evolutionary conserved | Class 3 | No | None | |
| BRCA2c.6725A>T, p.(Asp2242Val) | No significant change predicted | Amino acid conserved, change not predicted as disruptive | Class 3 | No | None | |
| BRCA2 c.7540A>G, p.(Lys2514Glu) | Moderate effect (3/5 predict new splice donor) | Amino acid conserved change predicted disruptive | Class 3 | No | None | |
| BRCA2c.7939C>G, p.(Leu2647Val) | Moderate effect (3/5 predict new splice donor) | Amino acid conserved, change predicted disruptive (2/3) | Class 3 | No | None | |
| BRCA2c.8488-5T>C | Moderate effect (reduction in ranking of splice acceptor) | N/A | Class 3 | No | None | |
| BRCA2c.6750A>G, p.(=) | No significant change predicted | N/A | Class 3 | No | None | |
| BRCA2c.9082G>C, p.(Ala3028Pro) | No significant change predicted | Amino acid not evolutionary conserved, change not predicted disruptive | Class 3 | No | None |
Reference sequences used: LRG_292t1 (BRCA1); LRG_293t1 (BRCA2).
Submission to LOVD: http://databases.lovd.nl/shared/genes/BRCA1
BRCA1 c.4676-7C>T Submission of screening #0000063401.
BRCA1 c.4868C>G p.(Ala1623Gly) Submission of screening #0000063402.
BRCA1 c.5152+4A>G Submission of screening #0000063403.
BRCA2 c.1395A>C, p.(=) Submission of screening #0000063404.
BRCA2 c.4614T>C, p.(=) Submission of screening #0000063405.
BRCA2 c.5319G>C, p.(Glu1773Asp) Submission of screening #0000063406.
BRCA2 c.6725A>T, p.(Asp2242Val) Submission of screening #0000063407.
BRCA2 c.7540A>G, p.(Lys2514Glu) Submission of screening #0000063414.
BRCA2 c.7939C>G, p.(Leu2647Val) Submission of screening #0000063417.
BRCA2 c.8488-5T>C Submission of screening #0000063422.
BRCA2 c.6750A>G, p.(=) Submission of screening #0000063427.
BRCA2 c.9082G>C, p.(Ala3028Pro) Submission of screening #0000063428.
No evidence for deep intronic splicing variants was identified in any of the 45 samples. In all, 13 samples appeared to be homozygous for all intragenic variants in BRCA1 and 6 were homozygous in BRCA2. These samples were known to be heterozygous at the DNA level for all exons on previous MLPA testing. The remaining 32 samples were heterozygous for at least one marker in BRCA1 and 39 in BRCA2 on RNA testing. All the DNA samples from the 10 BRCA1 and 6 BRCA2 cases were also homozygous ruling out apparent hemizygosity from loss of constitutional heterozygosity.
Assessing against BCLC criteria only 125/159 (78.5%) of samples with proven non-mucinous epithelial ovarian cancer meeting these criteria had identifiable BRCA1/2 variants after DNA and RNA analysis. Of the 34 without proven BRCA1/2 variants 16 had panel testing with only the RAD51D variant identified. Of families with at least two proven ovarian cancers meeting BCLC criteria, 10/69 (14.5%) did not have a variant in BRCA1/2 with 9/10 not having a variant in RAD51D or RAD51C either. Thus, 9/69 (13%) could be due to other genes (Table 1).
Panel testing was carried out on 42 samples subjected to RNA analysis (Tables 1 and 2). This included the previously identified RAD51D c.556C>T, p.(Arg186*) variant.10 The only other clearly functional actionable variant from previous work identified was ATM c.7271T>G, p.(Val2424Gly) in a breast ovarian family. The woman tested had breast cancer aged 45 years and a granulosa cell tumour of the ovary aged 56 years. There were three further female breast cancers in the family with a Manchester score of 35 points and the variant has been found in two further affected family members with breast cancer. A second family had a CHEK2 c.1263delT, p.(Ser422Valfs*15) variant that is presumed functional. This has been seen six times in the ExAc database (http://exac.broadinstitute.org/) equivalent to about 1 in 10 000 people. The variant was carried by a 37-year-old female with breast cancer and her brother who developed breast cancer at the same age, samples from three further close female relatives with breast cancer aged 44, 48 and 49 were not available. No additional functional variants were identified and in particular no variant in PALB2. Importantly, the family with RAD51D which had previously shown clear linkage to BRCA1 also was negative on RNA analysis of BRCA1 but also heterozygous for a known SNP in RNA hg19.chr17:g.41246481C>T (rs1799950) ruling out loss of BRCA1 RNA function.
The median Manchester score in the 38 breast/ovary/male breast cancer families tested with RNA and panel testing was 36 compared to a median score of only 16 points for 2820 other samples from breast/ovarian cancer patients that had tested negative through the Manchester service. The predicted detection rate for BRCA1/2 on DNA testing is 10% for samples with 16 points and >50% for samples with a combined score of 36.
Discussion
We have carried out the first comprehensive testing of BRCA1/2 using RNA analysis in a large series of families with extremely high a priori likelihood of a BRCA1/2 variant but without a definite identified functional variant. The concentration on families with a very high Manchester score with 38 samples having a breast/ovarian or male breast cancer phenotype should have provided the ideal sample set to assess added sensitivity of RNA analysis. Indeed, the sample selection with high likelihood of BRCA1/2 and prescreening with sequencing and MLPA should have identified four mutations if 3% of BRCA1/2 involvement was due to deep intronic mutations. Unlike NF1 and NF2 RNA analysis does not appear to add any additional sensitivity as no deep intronic splicing variants were identified. A deep intronic variant (intron 12, c.6937+594T>G) has been found on RNA analysis in a BRCA2 family previously,14 but introduction of comprehensive RNA testing does not appear necessary for BRCA1/2 on the basis of the current results, as the contribution of deep intronic functional variants is probably below 1% of all functional variants. Nonetheless, RNA analysis does add value to Sanger or next-generation sequencing by confirming pathogenicity as demonstrated for two BRCA1 variants in the present report and by showing that other candidate splicing variants are almost certainly benign variants. While another study has assessed RNA allelic imbalance, this concentrated only on 54 samples that had an identified VUS and did not carry out full assessment on RNA.15 Although one sample was found to have allelic imbalance this was due to a missed exon 12–13 deletion in genomic DNA. We found no evidence for allelic imbalance in our set of families.
Table 2 shows that DNA-based testing has very high sensitivity for BRCA1/2 as evidenced by the identification of variants in 49/56 (87.5%) breast/ovary/male breast cancer samples with a Manchester score of 50+ with RNA confirming another variant boosting sensitivity to 50/56 (89%). Including the panel testing only identifies one further variant (RAD51D). Overall panel testing of breast/ovary/male breast cancer samples with Manchester scores of 40+ only boosts identification of actionable variants from 128/150 (85%) by a further 1/13 of the missing samples. Assuming a similar detection rate in the remaining 9 samples panel testing for the 10 additional genes would identify only 2 variants compared with 128 for BRCA1/2 only 2/150 (1.3%) overall with sensitivity similar to use of RNA to confirm candidate splicing variants in BRCA1/2. Importantly, PALB2 that has recently been reported to confer high risk of breast cancer16 appears to contribute little from this UK-based sample set. While the very high rate of identification in Manchester score families with scores over 50 do not leave a great deal of room for other high-risk breast/ovarian genes, the fact that 21% of BCLC families do not have variants suggests that there may well be other as yet unidentified genes that predispose to breast and ovarian and male breast cancer. Certainly, the original families in the BCLC consortium are likely to have been extremely high risk equivalent to a Manchester score above 50 and although the original assessments for families meeting these criteria with at least 2 ovarian cancers were close to 100%,7 these estimates now need revising downwards in view of the present research.
Although testing for the RAD51 genes may have clinical utility, the overall contribution to high-risk breast/ovarian cancer families is low. In families with Manchester scores of 40+ or in BCLC families that have tested negative, testing RAD51D in particular may be worthwhile. However, in families with lower probabilities detection rates fall <1% for both genes combined. Nonetheless, there remains a minority of very high-risk breast/ovary families that are unaccounted for.
Panel testing did identify one new probable actionable functional variant. The ATM variant p.Val2424Gly has been shown to confer high risk of breast cancer and segregate with breast cancer in families.17 Interestingly, the proband identified with the variant in the present report also had a granulosa cell tumour of the ovary. Granulosa cell tumours unlike other germ cell tumours have been associated with BRCA1/2 variants.9 We have now found variants in BRCA1 and BRCA2 in 2 (1 each) of 10 index cases screened for variants and in both (1 each) affected first-degree relatives tested in families with a known variant. As such granulosa cell tumours should not be overlooked like other germ cell tumours and mucinous tumours in assessing high-risk breast/ovarian families.9 Although the CHEK2 variant identified in a male breast cancer family is likely to have contributed to the risk, it is highly unlikely that it accounts for the full pattern in the family as a similar variant 1100delC only confers around a 30% lifetime risk17 and we do not propose offering this as a presymptomatic test in this family.
In conclusion, the present report confirms high sensitivity for current techniques of DNA sequencing and MLPA in BRCA1/2 with a minimum sensitivity of 89%, but more likely sensitivity nearer to 100% with the remaining 11% accounted for by phenocopies or other genes. RNA testing did not identify any deep intronic splicing variants and there was no evidence of loss of RNA expression, thus RNA testing is probably best reserved for problem-solving variants that may affect splicing detected on DNA analysis. Current panel testing adds very little to BRCA1/2 testing in high-risk breast/ovarian families, but given the ease and lack of expense of testing multiple genes this is likely to continue to be offered in addition to standard BRCA1/2 testing. Finally, the present report confirms that there may still be rare breast/ovarian and male breast cancer high-risk genes that are yet to be identified.
Acknowledgments
We acknowledge funding from the Genesis Breast Cancer Prevention Appeal. DGE and WN designed the study and obtained funding. HB, EvV, YW, NB and KR carried out the experiments. DGE, AW, HB, and WN interpreted the data, DGE and HB drafted the manuscript. All authors commented and approved the final version.
The authors declare no conflict of interest.
References
- Lalloo F, Evans DG: Familial breast cancer. Clin Genet 2012; 82: 105–114. [DOI] [PubMed] [Google Scholar]
- Alsop K, Fereday S, Meldrum C et al: BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 2012; 30: 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles SA, Aboagye EO, Ali S et al: Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res 2013; 15: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Hellebrand H, Wiek C et al: Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010; 42: 410–414. [DOI] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ramsay E et al: Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 2011; 43: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ruark E et al: Germline RAD51C mutations confer susceptibility to ovarian cancer. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 1998; 62: 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Gifford FL, Lalloo F, Newman WG, Evans DG: High sensitivity for BRCA1/2 mutations in breast/ovarian kindreds: are there still other breast/ovary genes to be discovered? Breast Cancer Res Treat 2012; 134: 895–897. [DOI] [PubMed] [Google Scholar]
- Evans DG, Lalloo F, Cramer et al: Addition of pathology and biomarker information significantly improves the performance of the Manchester scoring system for BRCA1 and BRCA2 testing. J Med Genet 2009; 46: 811–817. [DOI] [PubMed] [Google Scholar]
- Evans DGR, Young K, Bulman M, Shenton A, Lalloo F: Probability of BRCA1/2 mutation varies with ovarian histology: results from screening 442 ovarian cancer families. Clin Genet 2008; 73: 338–345. [DOI] [PubMed] [Google Scholar]
- Smith A, Moran A, Boyd MC et al: The trouble with phenocopies: are those testing negative for a family BRCA1/2 mutation really at population risk? J Med Genet 2007; 44: 10–15.17079251 [Google Scholar]
- Messiaen LM, Callens T, Mortier G et al: Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat 2000; 15: 541–555. [DOI] [PubMed] [Google Scholar]
- Evans DGR, Ramsden RT, Shenton et al: Mosaicism in NF2 an update of risk based on uni/bilaterality of vestibular schwannoma at presentation and sensitive mutation analysis including. MLPA J Med Genet 2007; 44: 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Buisson M, Léoné M et al: BRCA2 deep intronic mutation causing activation of a cryptic exon: opening toward a new preventive therapeutic strategy. Clin Cancer Res 2012; 18: 4903–4909. [DOI] [PubMed] [Google Scholar]
- Caux-Moncoutier V, Pagès-Berhouet S, Michaux D et al: Impact of BRCA1 and BRCA2 variants on splicing: clues from an allelic imbalance study. Eur J Hum Genet 2009; 17: 1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Casadei S, Heikkinen T et al: Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgar DE, Healey S, Dowty JG et al: Rare variants in the ATM gene and risk of breast cancer. Breast Cancer Res 2011; 13: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pharoah PD, Antoniou AC et al: Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015; 372: 2243–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles DM, Easton D, Foulkes WD et al: Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008; 29: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis Y, Payne S, McAnulty C et al: Practice Guidelines for the Evaluation of Pathogenicity and the Reporting of Sequence Variants in Clinical Molecular Genetics, Available from http://www.acgs.uk.com/media/774853/evaluation_and_reporting_of_sequence_variants_bpgs_june_2013_-_finalpdf.pdf.