Abstract
Concomitant with the evolution of biological diversity must have been the evolution of mechanisms that facilitate evolution, because of the essentially infinite complexity of protein sequence space. We describe how evolvability can be an object of Darwinian selection, emphasizing the collective nature of the process. We quantify our theory with computer simulations of protein evolution. These simulations demonstrate that rapid or dramatic environmental change leads to selection for greater evolvability. The selective pressure for large-scale genetic moves such as DNA exchange becomes increasingly strong as the environmental conditions become more uncertain. Our results demonstrate that evolvability is a selectable trait and allow for the explanation of a large body of experimental results.
Darwin was obsessed with variation. His books, considered as an ensemble, devote much more attention to variation than to natural selection, because he knew that no satisfactory theory of evolutionary change could be constructed until the causes of variation and the empirical rule of its form and amount had been elucidated (1).
Whether the propensity to evolve, or evolvability (2–4), can be an object of Darwinian natural selection is a topic of interest. Causality would suggest not because of the apparently anticipatory nature of evolvability (5, 6). Many within the field of evolutionary biology are uncomfortable with the concept that evolvability is a selectable trait. A growing body of experimental data, however, would be explained if evolvability were a selectable trait (7–15).
Higher organisms cannot evolve, or adapt, by germ-line mutation to an environmental change within their own lifetime. Does this mean that lineages and individuals cannot be under selection for evolvability? Although viability is the selection criterion, the genotype that determines the viability arises in a mutated, evolved way from that of the previous generation as a result of base substitution, recombination, transposition, and horizontal gene transfer. These mutational processes are the driving forces of evolution, and their rates fundamentally determine evolvability. The perspective we offer here is that the evolvability of an organism is defined by the rates of genetic change, that genetic change is not always deleterious, and that these rates of genetic change are not fixed and are under selective pressure. That is, the mechanisms that define the rates of change are encoded in the genotype, and so they are selectable. An analogy with thermodynamics illuminates the issue: How is free energy minimized in a physical system of particles despite the difficulty in defining the entropy of a given configuration of the particles? An ensemble of particle configurations allows the definition of free energy and the approach to thermodynamic equilibrium just as a population of evolving organisms allows the definition of and selection for evolvability.
Within the framework of point mutation, base substitution, and recombination, correlations of adaptation with function have been observed. It is known that immunoglobins have evolved such that the mutation rates in complementary determining regions, in which mutation is most likely to generate useful variants, are much higher than those in framework regions (14, 16). Recent data point to a role for DNA polymerases in regulating the somatic hypermutation rate of immunoglobin genes (13, 17). Similarly, codon usage within the influenza hemagglutinin protein seems to be biased to favor more rapid antigenic drift (14). Furthermore, in HIV-1 protease, the probability of mutation is not randomly distributed within the structure but rather concentrated at sites that alter the geometry of the protein-binding domain, conferring significant propensity for antigenic drift (18). Such behavior is not mere curiosity and has widespread implications for drug design and the evolution of drug resistance (19). Stressful conditions may generally provoke activation of error-prone polymerases, triggering a large increase in adaptive rates (20). Not only point mutation but also recombination are widely appreciated to confer increased evolvability (9, 21, 22). Recombination among the hemagglutinin and neuraminidase proteins, for example, is believed to be a significant mechanism leading to the emergence of new virulent strains of influenza (23). Computational and theoretical studies have also shown the persistence under selection of evolvability-enhancing moves in the context of point mutation and recombination evolutionary dynamics (24–29).
The selective forces that lead to the evolution and maintenance of mechanisms for rearrangement, deletion, transfer, and transposition of genetic material, inasmuch as they lead to even greater evolution than point mutation and recombination alone, are of great interest. For example, the development of antibiotic resistance in bacteria has evolved mainly through the swapping of DNA pieces between the evolving bacteria (8, 30). Similarly, the evolution of Escherichia coli from Salmonella is thought to have occurred exclusively from DNA swapping (31). It has been proposed that the success of bacteria as a group stems from a capacity to acquire genes from a large and diverse range of species (32). It would seem, then, that large genetic moves are pervasive and crucial to evolutionary dynamics (6, 8, 10–12, 15, 30, 31, 33–39). Concomitantly, evolvability is enhanced by these larger moves, as shown experimentally for the case of DNA shuffling (32, 40–44). A key question is whether selection for evolvability fosters the husbandry of these moves.
We address here, from a theoretical point of view, selection of evolvability in the presence of large-scale genetic moves. Although the use of the term evolvability has only recently come into vogue in the scientific community, investigations into the evolution of adaptation go back several decades (45–47). Prominent from a theoretical perspective are works in population genetics (48, 49) and game theory (50–52). Despite the insights that these studies give as to the origin and maintenance of evolvability, evolution of and selection for evolvability remains a contested issue primarily because of the causality principle (5, 6). We show here that evolvability is selected for, notwithstanding the constraints imposed by causality, when a system is subject to a constant, random environmental change. This selection for evolvability occurs even when viability as a function of genotype is an extremely complex function, with exponentially many optima, and when the evolving system is unable to reach the global optimum of viability in any one instance of the environment. We demonstrate our results by using computer simulations of protein molecular evolution that incorporate selection in a varying environment. The genotype of a protein molecule is mapped to a complex phenotype by using a generalized NK model in which all assumptions and relevant parameters are known. The selective pressure for evolvability is shown to be greater for larger rates of environmental change. Interestingly, a generalized susceptibility of the system correlates with the fluctuations in the environment, albeit not as a result of generalized linear response theory (53). The addition of selection for evolvability as a phenomenological law to the toolbox of evolutionary theory allows for the explanation of a large body of experimental results.
The Generalized Block NK Model
Whether evolvability is selectable has been a difficult question to answer, primarily because observations in evolutionary biology tend to be correlative in nature and difficult on which to make mechanistic conclusions. Therefore, we consider here the dynamics of evolvability in a well defined theoretical model of protein evolution (54). Within this model of protein structure and function, we have a fixed population of proteins, which we take to be 1,000. We have a microscopic selection criterion, which we take to be the folding and binding of a protein to a substrate. And we have a means of inducing constant, random environmental change.
We model the molecular evolution of protein systems by using a generalization of the NK (55–57) and block NK (58) models that has been used previously to study protein molecular evolution strategies (54) and the immune-system response to vaccination and disease (59). The model includes a population of sequences, upon which selection acts and in which occur genetic mutations. The mutational hierarchy includes both point mutations and large-scale swapping moves, akin to transposition or translocation events. Although the model does not include recombination, such inclusion is not expected to change the results because swapping can be viewed as a powerful form of recombination (54). For example, linkage effects are mitigated even more rapidly by swapping in our model than they would be by recombination. The selection for greater swapping rates in more rapidly changing environments observed in our model parallels results found in studies of the evolution of sex, for which adaptation and variation in a heterogeneous environment is well researched (60).
In the generalized block NK model, each individual evolving protein sequence has an energy that is determined by secondary structural subdomain energies, Usd, subdomain–subdomain interaction energies, Usd–sd, and chemical binding energies, Uc. This energy is used as the selection criteria in our studies and is given by
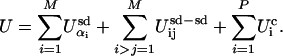 |
[1] |
Within our generalized block NK model, each protein molecule is composed of M = 10 secondary structural subdomains of N = 10 aa in length. We consider five chemically distinct amino acid classes (negative, positive, polar, hydrophobic, and other) and L = 5 different types of subdomains (helices, strands, loops, turns, and others). We therefore have L different subdomain energy functions of the NK form
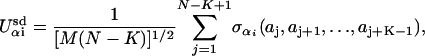 |
[2] |
where aj is the amino acid type of the jth amino acid in the subdomain, and αi is the type of the ith subdomain. As in previous studies, we consider the case in which the range of the interactions within a subdomain is specified by K = 4 (54, 59). Here σαi is a quenched Gaussian random number with zero mean and a variance of unity, and it is different for each value of its argument for each of the L subdomain types, αi. The interaction energy between secondary subdomain structures is given by
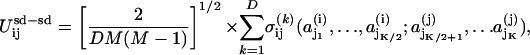 |
[3] |
where we consider D = 6 interactions between secondary structures (54, 59). The zero-mean, unit-variance Gaussian 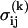 and the interacting amino acids, j1,..., jK, are selected at random for each interaction (i, j, k). In our model, P = 5 aa contribute directly to a binding event, as in a typical pharmacophore, where the chemical binding energy of each amino acid is given by
and the interacting amino acids, j1,..., jK, are selected at random for each interaction (i, j, k). In our model, P = 5 aa contribute directly to a binding event, as in a typical pharmacophore, where the chemical binding energy of each amino acid is given by
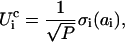 |
[4] |
where the zero-mean, unit-variance Gaussian σi and the contributing amino acid, i, are chosen at random.
System Evolution and Environmental Change
Our model system maintains a constant population of 1,000 proteins, each protein of 100 aa in length and initially distinct in sequence. The system evolves through the base substitution of single amino acids and through DNA swapping of amino acid subdomains from structural pools representing the five different subdomain types, each containing 250 low-energy subdomain sequences. These moves represent the small-scale adaptation and the large-scale, dramatic evolution that occur in nature. For protein i, nmut(i) point mutations occur per sequence per round of selection. In addition, for protein i, subdomain sequences are replaced randomly with sequences from the same-type low-energy pools with probability pswap(i).
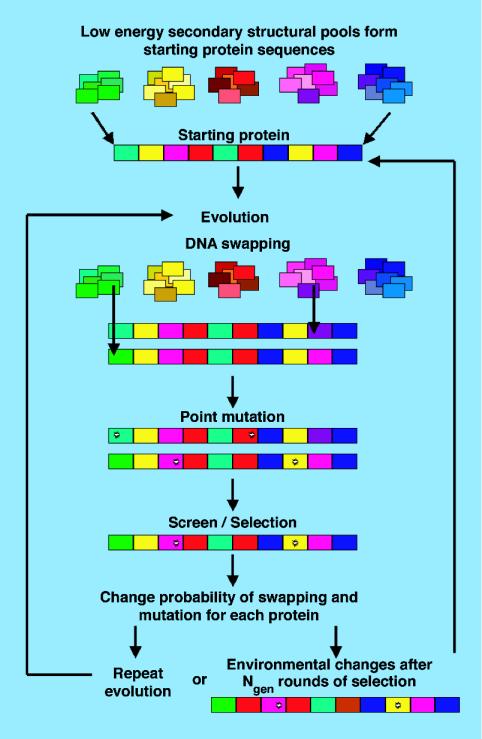
After pool swapping and point mutations, selection occurs, and the 20% lowest-energy protein sequences are kept and amplified to form the population of 1,000 proteins for the next round of selection. The parameters pswap(i) and nmut(i) are allowed to take a log-Gaussian random walk for each protein sequence. This process is repeated for Ngen rounds of selection, after which an environmental change is imposed on the system with a severity that is characterized by the parameter p (59). The parameter p is the probability of (i) changing the type of each of the 10 subdomains in the protein sequences, αi in Eq. 2, (ii) changing the amino acids and energies that are involved in subdomain–subdomain interactions, jk and σ(k)ij in Eq. 3, and (iii) changing the amino acids and energies that are involved in the chemical binding, i and σi in Eq. 4. We repeat the process for a total of 100 environmental changes and average our results over 1,000 instances of the ensemble. For each system studied, a steady state in nmut, pswap, and the average energies at the beginning, 〈U〉start, and end, 〈U〉end, of the dynamics in a single instance of the environment is reached after <80 environmental changes in all cases. We average the data over the last 20 environmental changes. We study how the frequency of environmental change, 1/Ngen, and the severity of environmental change, p, affect the evolvability of the protein sequences. A schematic diagram showing the molecular evolution of our protein system can be seen in Fig. 1.
Fig. 1.
Schematic diagram showing the evolution of the protein system.
Selection for Evolvability
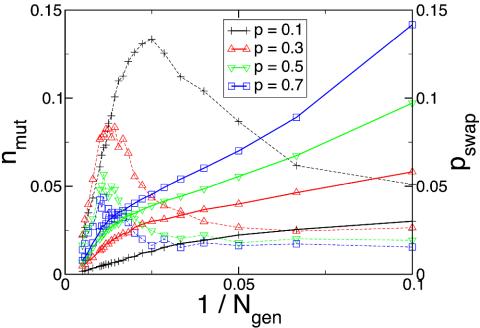
Shown in Fig. 2 are the steady-state values of pswap and nmut that our protein system selects as a function of imposed frequency of environmental change, 1/Ngen, and severity of environmental change, p. The DNA swapping moves that we propose have a high capacity for evolutionary change, because a significant number of amino acids may be altered in a protein sequence in one swap move. It is clear that our systems select for higher probabilities of DNA swapping moves, and thus evolvability, as the frequency and severity of environmental change increases. We stress the importance of this result. Mainstream evolutionary theory does not recognize a need for the selection of evolvability. More generally, we see that only in the limit of little or no environmental change, pswap → 0, do large-scale changes tend to be disfavored. The role of base substitution in our evolving system is more complex. For more severe environmental changes and for higher frequencies of environmental change, the system depends more on DNA swapping than on point mutation to produce low-energy proteins. In these cases, because the protein must make large changes to its sequence to adapt to the environmental change, selection results in high values of pswap, with base substitution having only a small effect on the energy of the protein. For less severe environmental changes and lower frequencies of environmental change, base substitution is sufficient to achieve the small modifications in protein sequence that are required for adaptation to the environmental change. Thus, we observe the higher dependence on nmut and lower dependence on pswap for small p. In addition, as 1/Ngen → 0, nmut → 0, because mutations tend to be deleterious in stable systems with no environmental fluctuations.
Fig. 2.
nmut (dashed lines) and pswap (solid lines) as a function of the frequency of environmental change, 1/Ngen, for different values of the severity of environmental change, p. The statistical errors in the results are smaller than the symbols on the figure.
Evolvability is intimately related to the diversity of a population. At short times, evolvability can be quantified by the diffusion coefficient in protein sequence space, D0, which is given by the combined diffusion due to swapping of the subdomains and the point mutation of individual amino acids (61):
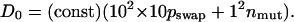 |
[5] |
The overwhelming contribution to D0 comes from the swapping step, because the swapping move far more dramatically changes the sequence. The short-time diffusion rate selected for reflects, as a function of environmental change, a balance between staying within a favorable basin of attraction, or niche, and adaptation to a newly created, superior niche. As Fig. 2 shows, greater environmental change favors greater local diffusion, as indicated by the monotonic increase of pswap with p.
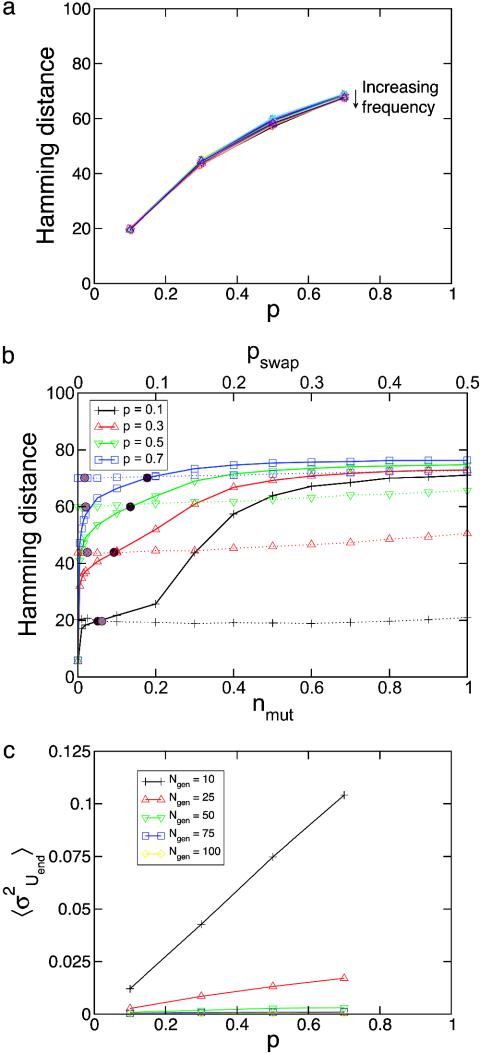
It is useful to regard base substitution as a means of fine tuning the protein sequences, whereas DNA swapping can be considered a source of more substantial evolutionary change. This hierarchy within the space of evolutionary moves becomes more apparent when studying the difference between starting and ending protein sequences within one environment as a function of p, pswap, and nmut. The distance between protein sequences is characterized by the Hamming distance between the respective amino acid sequences. For a given p, the Hamming distance decreases only slightly as the frequency of environmental change, 1/Ngen, increases, but it has a very strong dependence on the severity of the environmental change, p, as shown in Fig. 3a. The sensitivity of the Hamming distance also shows markedly different behavior to pswap and nmut, as shown in Fig. 3b. For state points with fixed nmut, 1/Ngen, and p, the Hamming distance strongly depends on the value of pswap. However, for state points with fixed pswap, 1/Ngen, and p, the Hamming distance displays little or no variation with nmut. The Hamming distance is a long-time measure of the evolvability of the system. The long-time diffusion coefficient can be defined as the square of the Hamming distance multiplied by the frequency of environmental change. As Fig. 3a implies, the long-time evolvability, as measured by the long-time diffusion coefficient, increases with both the severity and frequency of environmental change.
Fig. 3.
Hamming distance and average variance. (a) Hamming distance as a function of the severity of environmental change, p, for the state points shown in Fig. 2. (b) Hamming distance as a function of nmut (dashed lines) and pswap (solid lines) for fixed Ngen = 15 and for different severities of environmental change, p. In displaying the Hamming distance dependence on nmut (pswap), we fix pswap (nmut) to the selected values from Fig. 2. The selected values of nmut and pswap at each state point are shown by light and dark circles, respectively. (c) Average variance, σ2Uend, of the energy of a population at the end of an evolution, Uend, as a function of the severity of environmental change, p, for different frequencies of environmental change, 1/Ngen.
Due to the roughness of viability as a function of sequence, the exploration performed by any particular individual is limited to a local basin of attraction defined by the short-time mutation rates, and thus more independent traces through sequence space allow for more thorough evolution. In other words, the more diverse the starting population of individuals, the greater potential there is for evolution. Fig. 3c shows the average variance of the energy values at the end of the dynamics within a single instance of the environment as a function of the severity and frequency of environmental change. It is clear that the diversity increases monotonically with p and 1/Ngen.
As we have seen, evolvability is quantifiable at any point in time through measurement of diversity and the local mutation rates. For this reason, causality does not prevent selection for evolvability. Because evolvability is an observable property, it can be selected for.
Susceptibility
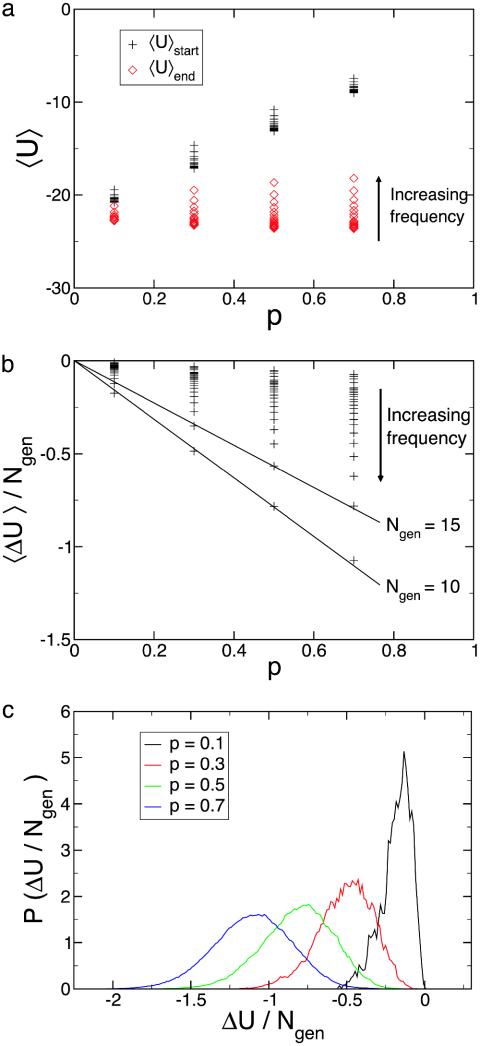
A further measure of long-time evolvability is the response, or susceptibility, of the system to environmental change. In Fig. 4a we plot the average energy at the start, 〈U〉start, and end, 〈U〉end, of the dynamics within a single instance of the environment. This quantity is shown as a function of the severity, p, and frequency, 1/Ngen, of environmental change. It is apparent that at low frequencies of environmental change, populations with greater diversity and variation, which are more evolvable, have slightly lower values of 〈U〉end. There is also a clear increasing trend in 〈U〉start as a function of p, which is a feature of the generalized NK model. Considering the ending energy of a protein molecule within one instance of the environment to be roughly the sum of n Gaussian terms from the generalized NK model,
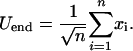 |
[6] |
The starting energy of this protein molecule after an environmental change is given by
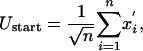 |
[7] |
where
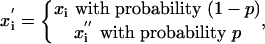 |
[8] |
and where 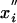 are random Gaussian variables with zero mean (
are random Gaussian variables with zero mean (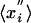 = 0), whereas xi are evolved variables that are better than random and typically negative. Thus, the average starting energy of this protein molecule is
= 0), whereas xi are evolved variables that are better than random and typically negative. Thus, the average starting energy of this protein molecule is
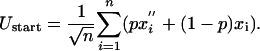 |
[9] |
Thus, averaging over the values in the new environment
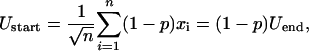 |
[10] |
or, averaging over many environmental changes
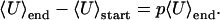 |
[11] |
Fig. 4.
Average energy, average change in energy, and probability distribution. (a) Average energy immediately after, 〈U〉start, and immediately before, 〈U〉end, an environmental change as a function of the severity of environmental change, p, for different frequencies of environmental change. (b) Average change in energy, 〈ΔU〉, multiplied by the frequency of environmental change, 1/Ngen, as a function of the severity of environmental change, p. (c) Probability distribution of the susceptibility for different values of the severity of environmental change, p, for a fixed frequency of environmental change, 1/Ngen = 0.1.
This average reduction in the energy is a measure of the susceptibility of a system, 〈ΔU〉/Ngen = (〈U〉end – 〈U〉start)/Ngen. In Fig. 4b we plot the susceptibility of our system as a function of the severity of environmental change, p. For a fixed frequency of environmental change, the susceptibility is a linear function of the severity of environmental change, as in Eq. 11. This simple analysis captures the essence of the dynamics that occurs in the correlated, generalized NK model. Fig. 4c shows that the probability distribution of the susceptibility is Gaussian in shape. Note also that the variance of the susceptibility increases with p in Fig. 4c, and thus the linearity of the susceptibility in Fig. 4b is not simply the result of a generalized fluctuation–dissipation theorem.
Implications for Evolution
Our results have implications for evolutionary theory. In our model system, populations of protein molecules that are subject to greater environmental change select for higher rates of evolvability. The selection criterion that we use is not a measure of evolvability in any way, yet the system selects for evolvability based on the implicit energetic benefits of adaptation to environmental change. In addition, there is no reason to assume that selection is optimal. In fact, systems optimal for one environment tend to have too little evolvability and tend to be selected against when faced with the inevitability of change.
Given our results, we propose that it is not mere chance that highly evolvable species tend to be found in rapidly changing environments or that an environmental crisis can trigger an increase in the rate of the evolution of a species. Indeed, selection for evolvability allows for the explanation of many data: the existence of somatic hypermutation in the immune system (13, 14, 16, 17), the evolution of drug resistance in species of bacteria (8, 30), and the occurrence and success of transpositional events in bacterial evolution (10, 31, 36). A recently studied example from mammals is the San Nicolas Island fox, which is a highly endangered species and the most monomorphic sexually reproducing animal known. This species, however, is found to have high levels of genetic variation within the major histocompatibility complex loci (62) that allows for increased pathogen resistance.
We believe that our results are of relevance to the field of vaccine and drug design. Currently, the design of new vaccines and drugs is largely based on the assumption that pathogens evolve by local space searching in response to therapeutic and immune selection. However, it is clear that we must anticipate the evolutionary potential of large DNA swapping events in the development of viruses, parasites, bacteria, and cancers if we are to engineer effective methods of treating them. How evolvability correlates with treatment strategy, and how to drive pathogens into regions of low evolvability where they are eradicated most easily, is of importance to efforts for vaccine and drug engineering.
Specific pathogenic examples of evolvability include the emergence of new influenza strains by a novel hemagglutinin neuraminidase recombination, followed by antigenic drift to a highly infectious strain (23); emergence of many new HIV strains with the spread of the disease from its site of origin in Africa (63, 64); and the increased emergence of new infectious diseases associated with modern, post-World War II travel (65). Additionally, a recent study of the dynamics of HIV-1 recombination suggests that HIV-1 may have evolved high recombination rates to foster rapid diversification and further its survival (66).
Note that evolvability is not simply the observation that new strains occur; rather, it is the underlying probability with which new strains are created by genetic modification. These new strains may proliferate and be observed, or they may fail and not be observed to an appreciable extent. Fundamental study of evolvability, then, requires an appreciation of these underlying rates of genetic change. These underlying rates, such as polymerase error rates, recombination rates, and transposition rates, are what selection for increased evolvability may modulate (67). These underlying rates of change are inheritable and can be altered by mutation. Study of these rates of genetic change, deconvoluted from observed rates of evolution, which are these rates multiplied by a probability of survival, is of fundamental interest.
It is intriguing that we find that at low frequencies of environmental change, populations that are subject to more severe environmental changes can produce lower-energy individuals than populations that are not subject to environmental changes (Fig. 4a). Thus, under some conditions, adaptability can provide global benefits. This finding can be contrasted to the more customary expectation that specialists are better than generalists (68). In experimental studies of Chlamydomonas, generalists that were evolved in alternating light and dark conditions were found to be better than their ancestors in both light and dark conditions but less good than specialists that had evolved exclusively in one of the environmental conditions (69). Studies of the evolution of E. coli at constant and alternating temperatures produced similar results (70, 71). The nature of the environmental change in these studies is not completely random as in our model. In addition, the number of rounds of selected evolution under each environmental condition is perhaps better defined within our model. These experiments do point to possible tests of our theory. For a species that is capable of DNA swapping evolutionary moves, a systematic study of competency as a function of the frequency of a random environmental change would be of interest. We predict that under some conditions, certain frequencies of environmental change will produce better individuals, after a given number of rounds of evolution and selection, than would be produced by evolution in a constant environment. Different severities of environmental change could also be imposed by altering the change in environmental variables between samples, such as temperature, food concentrations, light conditions, and exposure to disease. With regard to susceptibility, we would expect the rate of change of viability within an environment to be higher in systems with more frequent and harsher environmental changes because of greater evolvability.
Summary
Not only has life evolved, but life has evolved to evolve. That is, correlations within protein structure have evolved, and mechanisms to manipulate these correlations have evolved in tandem. The rates at which the various events within the hierarchy of evolutionary moves occur are not random or arbitrary but are selected by Darwinian evolution. Sensibly, rapid or extreme environmental change leads to selection for greater evolvability. This selection is not forbidden by causality and is strongest on the largest-scale moves within the mutational hierarchy.
Many observations within evolutionary biology, heretofore considered evolutionary happenstance or accidents, are explained by selection for evolvability. For example, the vertebrate immune system shows that the variable environment of antigens has provided selective pressure for the use of adaptable codons and low-fidelity polymerases during somatic hypermutation. A similar driving force for biased codon usage as a result of productively high mutation rates is observed in the hemagglutinin protein of influenza A. Selection for evolvability explains the prevalence of transposons among bacteria and recombination among higher organisms. We suggest that therapeutics also confer selective pressure on the evolvability of pathogens, and that this driving force for antigenic drift should be considered in drug- and vaccine-design efforts.
Acknowledgments
We thank Kevin R. Foster for a careful reading of the manuscript. This research is supported by the National Institutes of Health.
References
- 1.Gould, S. J. (1983) Hen's Teeth and Horse's Toes (Norton, New York).
- 2.Kirschner, M. & Gerhart, J. (1998) Proc. Natl. Acad. Sci. USA 95, 8420–8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins, R. (1989) in Artificial Life, ed. Langton, C. G. (Addison–Wesley, New York), pp. 201–220.
- 4.Radman, M., Matic, I. & Taddei, F. (1999) Ann. N.Y. Acad. Sci. 870, 146–155. [DOI] [PubMed] [Google Scholar]
- 5.Chicurel, M. (2001) Science 292, 1824–1827. [DOI] [PubMed] [Google Scholar]
- 6.Partridge, L. & Barton, N. H. (2000) Nature 407, 457–458. [DOI] [PubMed] [Google Scholar]
- 7.Kidwell, M. G. (1997) Proc. Natl. Acad. Sci. USA 94, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro, J. A. (1997) Trends Genet. 13, 98–104. [DOI] [PubMed] [Google Scholar]
- 9.Barton, N. H. & Charlesworth, B. (1998) Science 281, 1986–1990. [PubMed] [Google Scholar]
- 10.Fedoroff, N. (2000) Proc. Natl. Acad. Sci. USA 97, 7002–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro, J. A. (2002) J. Biol. Phys. 28, 745–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro, J. A. (2002) Ann. N.Y. Acad. Sci. 981, 111–134. [DOI] [PubMed] [Google Scholar]
- 13.Storb, U. (2001) Nat. Immunol. 2, 484–485. [DOI] [PubMed] [Google Scholar]
- 14.Plotkin, J. B. & Dushoff, J. (2003) Proc. Natl. Acad. Sci. USA 100, 7152–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporale, L. H. (2003) Am. Sci. 91, 234–241. [Google Scholar]
- 16.Kepler, T. B. (1997) Mol. Biol. Evol. 14, 637–643. [DOI] [PubMed] [Google Scholar]
- 17.Friedberg, E. C., Feaver, W. F. & Gerlach, V. L. (2000) Proc. Natl. Acad. Sci. USA 97, 5681–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freire, E. (2002) Nat. Biotechnol. 20, 15–16. [DOI] [PubMed] [Google Scholar]
- 19.Kepler, T. B. & Perelson, A. S. (1998) Proc. Natl. Acad. Sci. USA 95, 11514–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenberg, S. M. (2001) Nat. Rev. Genet. 2, 504–515. [DOI] [PubMed] [Google Scholar]
- 21.Pepper, J. W. (2003) BioSystems 69, 115–126.12689725 [Google Scholar]
- 22.Colegrave, N. (2002) Nature 420, 664–666. [DOI] [PubMed] [Google Scholar]
- 23.Frank, S. A. (2002) Immunology and Evolution of Infectious Disease (Princeton Univ. Press, Princeton). [PubMed]
- 24.Wagner, G. P. & Altenberg, L. (1996) Evolution (Lawrence, Kans.) 50, 967–976. [DOI] [PubMed] [Google Scholar]
- 25.Lenski, R. E., Ofria, C., Pennock, R. T. & Adami, C. (2003) Nature 423, 139–144. [DOI] [PubMed] [Google Scholar]
- 26.Blasio, F. V. D. (1999) Phys. Rev. E 60, 5912–5917. [Google Scholar]
- 27.Travis, J. M. J. & Travis, E. R. (2002) Proc. R. Soc. London Ser. B 269, 591–597. [Google Scholar]
- 28.Siegal, M. L. & Bergman, A. (2002) Proc. Natl. Acad. Sci. USA 99, 10528–10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergman, A. & Siegal, M. L. (2003) Nature 424, 549–552. [DOI] [PubMed] [Google Scholar]
- 30.Shapiro, J. A. (1992) Genetica 86, 99–111. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, J. G. (1997) Trends Microbiol. 5, 355–359. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y.-X., Perry, K., Vinci, V. A., Powell, K., Stemmer, W. P. C. & del Cardayre, S. B. (2002) Nature 415, 644–646. [DOI] [PubMed] [Google Scholar]
- 33.Pennisi, E. (1998) Science 281, 1131–1134. [DOI] [PubMed] [Google Scholar]
- 34.Gilbert, W. (1978) Nature 271, 501. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert, W., DeSouza, S. J. & Long, M. (1997) Proc. Natl. Acad. Sci. USA 94, 7698–7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duret, L., Marais, G. & Biemont, C. (2000) Genetics 156, 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonnig, W.-E. & Saedler, H. (2002) Annu. Rev. Genet. 36, 389–410. [DOI] [PubMed] [Google Scholar]
- 38.Levin, B. R. & Bergstrom, C. T. (2000) Proc. Natl. Acad. Sci. USA 97, 6981–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doolittle, W. F. (2000) Sci. Am. 282 (2), 90–95. [DOI] [PubMed] [Google Scholar]
- 40.Stemmer, W. P. C. (1994) Nature 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 41.Crameri, A., Raillard, S. A., Bermudez, E. & Stemmer, W. P. C. (1998) Nature 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, J.-H., Dawes, G. & Stemmer, W. P. C. (1997) Proc. Natl. Acad. Sci. USA 94, 4504–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. C., Jin, H.-M., Kuchner, O. & Arnold, F. H. (1997) J. Mol. Evol. 272, 336–347. [DOI] [PubMed] [Google Scholar]
- 44.Lutz, S. & Benkovic, S. J. (2000) Curr. Opin. Biotechnol. 11, 319–324. [DOI] [PubMed] [Google Scholar]
- 45.Clarke, B. C. (1979) Proc. R. Soc. London Ser. B 205, 453–474. [DOI] [PubMed] [Google Scholar]
- 46.Dawkins, R. & Krebs, J. R. (1979) Proc. R. Soc. London Ser. B 205, 489–512. [DOI] [PubMed] [Google Scholar]
- 47.Gould, S. J. & Lewontin, R. C. (1979) Proc. R. Soc. London Ser. B 205, 581–598. [DOI] [PubMed] [Google Scholar]
- 48.Gillespie, J. H. (1991) The Causes of Molecular Evolution (Oxford Univ. Press, Oxford).
- 49.Frank, S. A. & Slatkin, M. (1990) Am. Nat. 136, 244–260. [Google Scholar]
- 50.Smith, J. M. (1979) Proc. R. Soc. London Ser. B 205, 475–488. [DOI] [PubMed] [Google Scholar]
- 51.Smith, J. M. (1982) Evolution and the Theory of Games (Cambridge Univ. Press, Cambridge, U.K.).
- 52.Sasaki, A. & Ellner, S. (1995) Evolution (Lawrence, Kans.) 49, 337–350. [DOI] [PubMed] [Google Scholar]
- 53.Sato, K., Ito, Y., Yomo, T. & Kaneko, K. (2003) Proc. Natl. Acad. Sci. USA 100, 14086–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogarad, L. D. & Deem, M. W. (1999) Proc. Natl. Acad. Sci. USA 96, 2591–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kauffman, S. & Levin, S. (1987) J. Theor. Biol. 128, 11–45. [DOI] [PubMed] [Google Scholar]
- 56.Kauffman, S. A. (1993) The Origins of Order (Oxford Univ. Press, New York).
- 57.Kauffman, S. A. & MacReady, W. G. (1995) J. Theor. Biol. 173, 427–440. [DOI] [PubMed] [Google Scholar]
- 58.Perelson, A. S. & Macken, C. A. (1995) Proc. Natl. Acad. Sci. USA 92, 9657–9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deem, M. W. & Lee, H. Y. (2003) Phys. Rev. Lett. 91, 068101. [DOI] [PubMed] [Google Scholar]
- 60.Michod, R. E. & Levin, B. R., eds. (1988) The Evolution of Sex (Sinauer, Sunderland, MA).
- 61.Chandrasekhar, S. (1943) Rev. Mod. Phys. 15, 1–89. [Google Scholar]
- 62.Aguilar, A., Roemer, G., Debenham, S., Binns, M., Garcelon, D. & Wayne, R. K. (2004) Proc. Natl. Acad. Sci. USA 101, 3490–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, T. F., Korber, B. T., Nahmias, A. J., Hooper, E., Sharp, P. M. & Ho, D. D. (1998) Nature 391, 594–597. [DOI] [PubMed] [Google Scholar]
- 64.Gao, F., Bailes, E., Robertson, D. L., Chen, Y. L., Rodenburg, C. M., Michael, S. F., Cummins, L. B., Arthur, L. O., Peeters, M., Shaw, G. M., et al. (1999) Nature 397, 436–441. [DOI] [PubMed] [Google Scholar]
- 65.Lederberg, J., Shope, R. E. & S. C. Oaks, J., eds. (1992) Emerging Infections: Microbial Threats to Health in the United States (Natl. Acad. Press, Washington, DC). [PubMed]
- 66.Levy, D. N., Aldrovandi, G. M., Kutsch, O. & Shaw, G. M. (2004) Proc. Natl. Acad. Sci. USA 101, 4204–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan, T., Bogarad, L. D. & Deem, M. W. (2004) J. Mol. Evol., in press. [DOI] [PubMed]
- 68.Elena, S. F. & Lenski, R. E. (2003) Nat. Rev. Genet. 4, 457–469. [DOI] [PubMed] [Google Scholar]
- 69.Reboud, X. & Bell, G. (1997) Heredity 78, 507–514. [Google Scholar]
- 70.Bennett, A. F. & Lenski, R. E. (1993) Evolution (Lawrence, Kans.) 47, 1–12. [DOI] [PubMed] [Google Scholar]
- 71.Leroi, A. M., Lenski, R. E. & Bennett, A. F. (1994) Evolution (Lawrence, Kans.) 48, 1222–1229. [DOI] [PubMed] [Google Scholar]