Abstract
Analysis of chromosomal rearrangements has been highly successful in identifying genes involved in many congenital abnormalities including hearing loss. Herein, we report a subject, designated DGAP242, with congenital hearing loss (HL) and a de novo balanced translocation 46,XX,t(1;5)(q32;q15)dn. Using multiple next-generation sequencing techniques, we obtained high resolution of the breakpoints. This revealed disruption of the orphan receptor ESRRG on chromosome 1, which is differentially expressed in inner ear hair cells and has previously been implicated in HL, and disruption of KIAA0825 on chromosome 5. Given the translocation breakpoints and supporting literature, disruption of ESRRG is the most likely cause for DGAP242's phenotype and implicates ESRRG in a monogenic form of congenital HL, although a putative contributory role for KIAA0825 in the subject's disorder cannot be excluded.
Introduction
Hearing loss (HL) is one of the most common birth defects, affecting over one in a thousand newborns. Over half of these cases can be attributed to a genetic etiology.1 Chromosomal abnormalities have been implicated in several genetic forms of congenital HL including Down, Smith-Magenis and Branchio-Oto-Renal syndromes.2 In cases of congenital HL accompanied with balanced chromosomal abnormalities (BCAs), high resolution of chromosomal breakpoints can reveal genes that have been disrupted, implicating them in auditory pathology.3 By employing this strategy, the Developmental Genome Anatomy Project (DGAP, dgap.harvard.edu) has successfully resolved the genetic etiology of HL in several cases of BCAs.4, 5, 6 Herein, we report the high-resolution breakpoints from a de novo translocation discovered in a 6-year-old female with congenital bilateral sensorineural hearing loss (SNHL) accompanied with a developmental disorder of speech and language.
Materials and methods
The subject, referred to as DGAP242, was recruited into DGAP after identification of a BCA and congenital HL. Informed consent, medical records and blood samples were obtained through the DGAP protocol approved by the Partners HealthCare Systems Institutional Review Board.
Clinical report
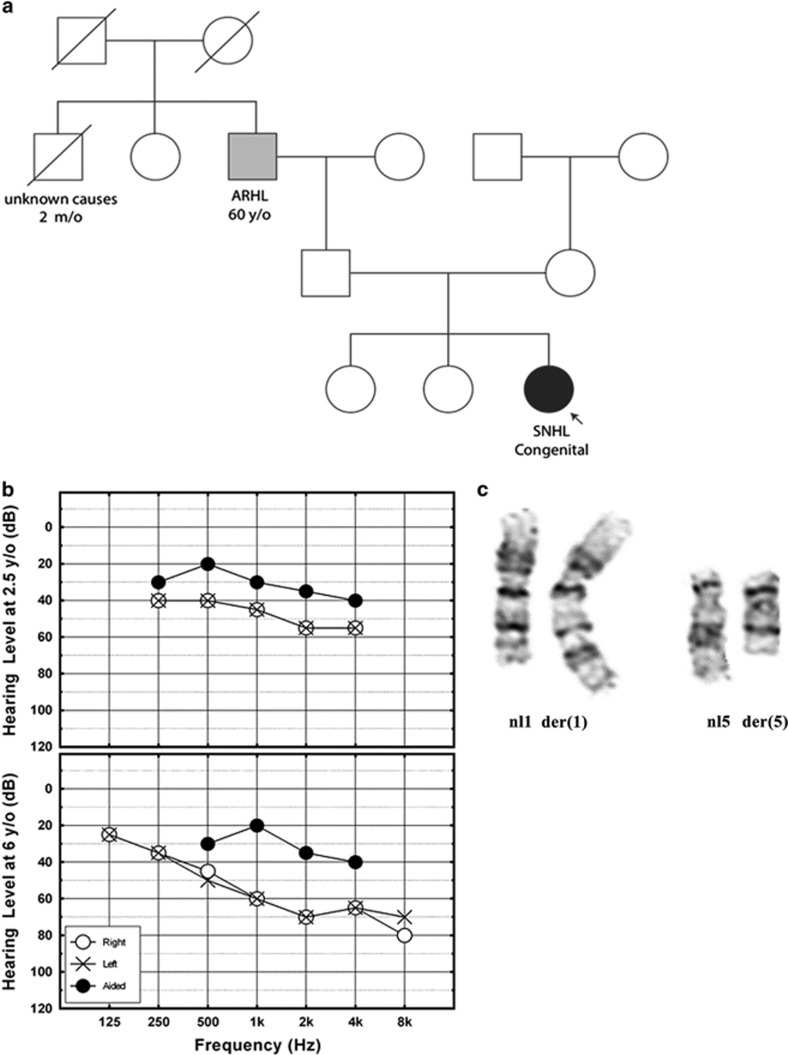
DGAP242 was delivered at 40 weeks gestation with a birth weight of 3.36 kg (50th percentile) to a 40-year-old G3P3 mother after elective cesarean section. The pregnancy was remarkable for gestational diabetes, hypothyroidism and high blood pressure. There were no prenatal risk factors for HL, including no known family history of congenital or childhood HL. The paternal grandfather started wearing hearing aids after age 60 years with hearing impairment attributed to age-related hearing loss (ARHL) and occupational noise exposure (Figure 1a).
Figure 1.
(a) DGAP242 pedigree (SNHL, sensorineural hearing loss; ARHL, age-related hearing loss). (b) Audiometric evaluation of both ears simultaneously, using play audiometry in sound field, shows downward sloping mild-to-moderate hearing loss in DGAP242 at age 2.5 years (top). Audiometric evaluation by pure tone audiometry shows bilateral, symmetrical and moderate SNHL in DGAP242 at age six years. (c) DGAP242 partial karyotype of normal (left) and derivative (right) GTG-banded chromosomes 1 and 5.
DGAP242 did not pass her newborn hearing screening and was referred for diagnostic audiological evaluation. Absent otoacoustic emissions, automated auditory brainstem response and auditory steady state response measurements indicated congenital, bilateral SNHL. GJB2 testing was negative for pathogenic variants. DGAP242 demonstrated hypotonia and motor delay, standing at 16 months and walking at 20 months of age. At this time, chromosome analysis and array-based comparative genomic hybridization of the proband and her parents revealed a de novo apparently balanced translocation between chromosomes 1 and 5: 46,XX,t(1;5)(q32;q15)dn.
Upon enrollment in DGAP 6 months later, several features were noted. DGAP242 had delayed speech, slightly posteriorly rotated ears and mild epicanthal folds. Play audiometry indicated mild-to-moderate high-frequency SNHL (Figure 1b, top). She started receiving speech therapy at age three years, and started wearing hearing aids regularly at age four. In a follow-up at 6 years of age, DGAP242 had bilateral, symmetrical, moderate SNHL and receptive language disorder (Figure 1b, bottom) and significant problems in listening comprehension and in understanding complex sentence structures. Additional observations included difficulty with abstract visual reasoning and balance tasks, but normal visual acuity and appropriate gross and fine motor skills (Table 1). DGAP242's clinical description and refined translocation breakpoints are deposited in DECIPHER (decipher.sanger.ac.uk/patient/317883).7
Table 1. HPO-coded phenotypes for DGAP242.
| Phenotype | Code |
|---|---|
| Bilateral sensorineural hearing impairment | HP:0008619 |
| Congenital sensorineural hearing impairment | HP:0008527 |
| Infantile muscular hypotonia | HP:0008947 |
| Motor delay | HP:0001270 |
| Expressive language delay | HP:0002474 |
| Poor speech | HP:0002465 |
| Posteriorly rotated ears | HP:0000358 |
| Epicanthus | HP:0000286 |
| Receptive language delay | HP:0010863 |
Lymphoblastoid cell line
Epstein–Barr virus transformation of lymphocytes, library generation, sequencing and bioinformatic analysis were performed at the Genomics and Technology Core in the Center for Human Genetic Research at Massachusetts General Hospital (Boston, MA, USA). Confirmatory GTG-banded karyotyping was performed on the Epstein–Barr virus-transformed lymphoblastoid cell line at the Brigham and Women's Hospital CytoGenomics Core Laboratory (Boston, MA, USA).
Whole-genome jumping library
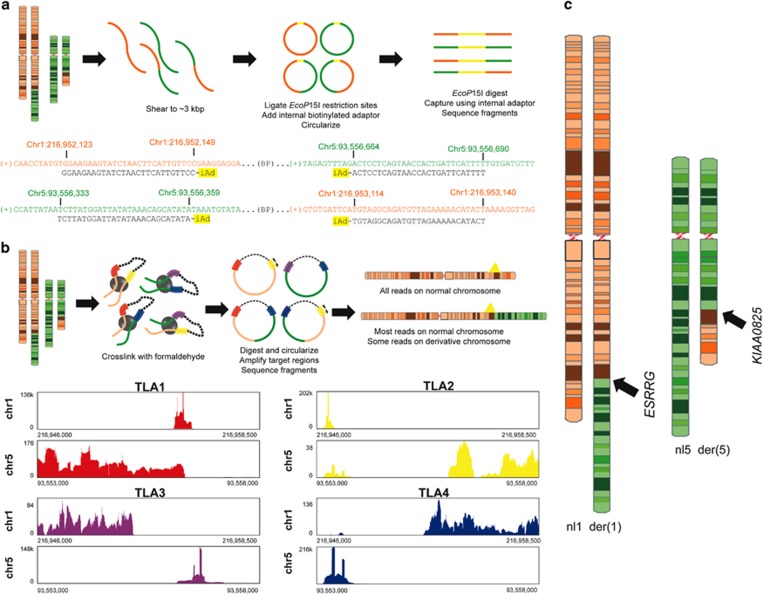
Large-insert (‘jumping library') whole-genome sequencing was performed as previously described.8, 9 In brief, genomic DNA was sheared to ~3 kbp, end-ligated with EcoP15I restriction adapters, size-selected and then circularized about an internal biotinylated adapter. Circular DNAs were EcoP15I restriction digested and purified using internal adapter pull-down, resulting in short DNA fragments whose ends reflect ~3 kbp of physical separation in the human reference genome. Fragments were ligated to Illumina TruSeq multiplexing adapters and amplified by PCR. The jumping library was used for next-generation sequencing (NGS) on an Illumina HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) using PE25 chemistry (2 × 25 bp reads). Data processing and analysis were performed as previously described (Figure 2a).10, 11
Figure 2.
(a) Diagram of jumping library methods and nearest aligned sequences adjacent to the translocation breakpoints (iAd, internal adapter for circularization). (b) Diagram of targeted locus amplification (TLA) library and histogram of nearest aligned reads adjacent to breakpoints. (c) Ideograms illustrate a simple q-q translocation disrupting ESRRG on chr1 and KIAA0825 on chr5.
Targeted locus amplification library
A targeted locus amplification library was prepared as previously described.12 In brief, genomic DNA was crosslinked, enzymatically digested and then re-ligated to create circularized DNA fragments. After de-crosslinking, breakpoint regions were amplified using PCR with anchored primers designed to target regions upstream and downstream of the presumed breakpoints. PCR products were sheared by sonication, and then barcode adapters were added. After amplification, the library was used for NGS on an Ion Torrent PGM with a 316v2 chip (Figure 2b).
Results
Fine-resolution mapping of DGAP242's translocation breakpoints was performed using sequential high-resolution methods. First, metaphase chromosome analysis confirmed the band-level resolution of the clinical karyotype indicating a balanced translocation, t(1;5)(q32;q15)dn (Figure 1c). Large-insert whole-genome sequencing was then performed substantiating the reciprocal translocation of chromosomes 1 and 5, and resolving the breakpoints to a 965 bp window on chr1 (216 952 149 to 216 953 114) and a 305 bp window on chr5 (93 556 359 to 93 556 664; Figure 2a).8, 9 Breakpoints of the translocation resulted in a revision of the karyotype to t(1;5)(q41;q15)dn. The chr1 breakpoint region includes a large ~500 bp AT-rich region that precluded identifying the precise junction to nucleotide resolution using standard polymerase-based amplification and Sanger sequencing.
In an effort to improve resolution of the breakpoints, a secondary NGS approach was pursued by constructing a targeted locus amplification library. Using four primer sets, regions were targeted either upstream or downstream of the putative breakpoints discovered in the jumping library (Supplementary Table 1). All primer sets had reads that aligned to both the targeted chromosome and to an untargeted chromosome (Figure 2b). These reads confirmed the t(1;5), refining the breakpoint region further to an 816 bp window on chr1 (216 952 298 to 216 953 114) and a 153 bp window on chr5 (93 556 359 to 93 556 512).
On the basis of these orthogonal NGS approaches, we mapped the breakpoints to sub-kilobase resolution. The karyotypic description of the chromosome rearrangement is given according to Next-Generation Cytogenetic Nomenclature13 and newly proposed recommendations by the Human Genome Variation Society (www.hgvs.org/mutnomen/comments004.html):
Next Generation Cytogenetic Nomenclature (NGCN)
46,XX,t(1;5)(q32;q15)dn.seq[GRCh37/hg19]
t(1;5)(1pter->1q41(216,952,298~216,953,114)::
5q15(93,556,359~93,556,512)->5qter;
5pter->5q15(93,556,359~93,556,512)::
1q41(216,952,298~216,953,114)->1qter)dn
Human Genome Variation Society (HGVS)
46,XX,t(1;5)(q32;q15)dn.seq[GRCh37] t(1;5)(q41;q15)dn
g.[chr1:pter_cen_(216952298_216953114)::
chr5:(93556359_93556512)_qter]
g.[chr5:pter_cen_(93556359_93556512)::
chr1:(216952298_216953114)_qter]
The t(1;5) disrupts two genes, ESRRG and KIAA0825, suggesting a single copy loss for both genes (Figure 2c). Both genes have low haploinsufficency scores according to DECIPHER (KIAA0825, %HI=7.51; ESRRG, %HI=0.51), further arguing that these disruptions could be pathogenic.14 Of the two genes, only ESRRG is significantly enriched in cochlear hair cells according to inner ear expression data from the Shared Harvard Inner Ear Laboratory Database (SHIELD, shield.hms.harvard.edu; fold enrichment=4.55, FDR=8.20E−03).15 This indicates that loss of ESRRG is more likely to have a HL phenotype than KIAA0825 (Supplementary Table 2). Sanger sequencing of coding regions and flanking exon–intron boundaries of ESRRG did not identify any additional pathogenic variant, further supporting a haploinsufficiency mechanism of pathogenicity. In addition to disrupted genes, potentially dysregulated genes within the topologically associating domains containing the breakpoints were assessed, due to possible changes in regional regulators of transcription (Supplementary Figure 1).16
Discussion
In individuals with congenital anomalies and a BCA with normal array-based comparative genomic hybridization results, investigation of the chromosomal breakpoints frequently elucidates causal genes that have been disrupted.3 Directly disrupted genes can be associated with a haploinsufficiency mechanism of pathogenicity, as one allele is lost due to interruption of the coding sequence. In this report, we analyzed genes disrupted by an apparently balanced de novo chromosomal translocation in a subject with congenital SNHL.
High-resolution analysis of the t(1;5) in DGAP242 illuminated the possible genetic etiology in two predicted haploinsufficient genes: KIAA0825 and ESRRG. Although little is known about KIAA0825, which limits assessment of its contribution to DGAP242's phenotype, its lack of enrichment in inner ear cells does not support its involvement in DGAP242's HL. However, disruption of ESRRG, which is preferentially expressed in inner ear hair cells, could explain DGAP242's HL. ESRRG encodes estrogen-related receptor gamma (ERRγ), a dimeric orphan nuclear transcription factor that is a paralog to ERRβ, a known HL gene.17, 18, 19 In addition, ESRRG has been shown by a candidate gene association study to be associated with age-related HL in females, a finding that has been supported by hair cell-specific expression of Esrrg in the adult mouse inner ear, and significantly elevated hearing thresholds in Esrrg−/− mice relative to their wild-type littermates.20 These data suggest that disruption of ESRRG may reduce its expression in hair cells and result in congenital HL in humans.
ESRRG is hypothesized to have a role in DGAP242's congenital, bilateral SNHL: ESRRG is predicted to be haploinsufficient, is differentially expressed in the inner ear, and has been formerly associated with HL. To our knowledge, this is the first report of a heterozygous disruption of ESRRG implicated in human HL. This study supports the candidate gene association study and Esrrg−/− mouse studies that previously implicated ESRRG in age-related hearing loss.20 Given these findings, evaluation of ESRRG in other cases of unexplained congenital bilateral SNHL may be warranted.
Acknowledgments
We thank the patient and her family for their participation in our study. This research was supported by the National Institute of General Medical Sciences GM061354 (CCM, JFG), the National Institute of Mental Health MH095867 (MET), the National Institute on Deafness and Other Communication Disorders R03DC013866 (JS) and F32DC012466 (BBC) and the National Science Foundation DGE1144152 (SLPS). Any opinion, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding institutions. This study makes use of data generated by the DECIPHER community. A full list of centers that contributed to the generation of the DECIPHER data is available from http://decipher.sanger.ac.uk and via E-mail from decipher@sanger.ac.uk. Funding for DECIPHER was provided by the Wellcome Trust.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Morton CC, Nance WE: Newborn hearing screening—a silent revolution. N Engl J Med 2006; 354: 2151–2164. [DOI] [PubMed] [Google Scholar]
- Giersch ABS, Morton CC: Cytogenetics and cochlear expressed sequence tags (ESTs) for identification of genes involved in hearing and deafness. In: Keats BJB, Popper AN, Fay RR (eds): Genetics and Auditory Disorders. New York: Springer, 2002, ppxii, 322 p. [Google Scholar]
- Higgins AW, Alkuraya FS, Bosco AF et al: Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet 2008; 82: 712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger GJ, Crocker S, McKenzie K et al: X-linked deafness-2 (DFNX2) phenotype associated with a paracentric inversion upstream of POU3F4. Am J Audiol 2014; 23: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KK, Reiss JA, Crow K et al: Deletion of an enhancer near DLX5 and DLX6 in a family with hearing loss, craniofacial defects, and an inv(7)(q21.3q35). Hum Genet 2010; 127: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RE, Darrow KN, Michaud S et al: Methylthioadenosine phosphorylase (MTAP) in hearing: gene disruption by chromosomal rearrangement in a hearing impaired individual and model organism analysis. Am J Med Genet A 2007; 143 A: 1630–1639. [DOI] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Mungall CJ et al: The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014; 42: D966–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscom C, Talkowski M: Design of large-insert jumping libraries for structural variant detection using illumina sequencing. Curr Protoc Hum Genet 2014; 80: 7 22 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Ernst C, Heilbut A et al: Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Human Genet 2011; 88: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C et al: Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 2012; 44: 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand H, Collins RL, Hanscom C et al: Paired-duplication signatures mark cryptic inversions and other complex structural variation. Am J Hum Genetics 2015; 97: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vree PJ, de Wit E, Yilmaz M et al: Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat Biotechnol 2014; 32: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Ordulu Z, Wong KE, Currall BB et al: Describing sequencing results of structural chromosome rearrangements with a suggested next-generation cytogenetic nomenclature. Am J Hum Genet 2014; 94: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME: Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 2010; 6: e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer DI, Shen J, Corey DP, Chen ZY: Gene expression by mouse inner ear hair cells during development. J Neurosci 2015; 35: 6366–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V et al: Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015; 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin RW, Kalay E, Tariq M et al: Mutations of ESRRB encoding estrogen-related receptor beta cause autosomal-recessive nonsyndromic hearing impairment DFNB35. Am J Hum Genet 2008; 82: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Yang L, Stallcup MR: Hormone-independent transcriptional activation and coactivator binding by novel orphan nuclear receptor ERR3. J Biol Chem 1999; 274: 22618–22626. [DOI] [PubMed] [Google Scholar]
- Horard B, Vanacker JM: Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol 2003; 31: 349–357. [DOI] [PubMed] [Google Scholar]
- Nolan LS, Maier H, Hermans-Borgmeyer I et al: Estrogen-related receptor gamma and hearing function: evidence of a role in humans and mice. Neurobiol Aging 2013; 34: e2071–e2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.