Abstract
Agonist-mediated degradation of estrogen receptor α (ERα) has been associated with its transcriptional activity. However, the mechanism by which ERα is targeted for degradation and whether there is a direct functional link between ERα stability and ERα-mediated transactivation have not been elucidated. Here we provide evidence that the p160 coactivator, AIB1, uniquely mediates agonist-induced, but not antagonist-induced, ERα degradation. We show that AIB1 recruitment by ERα is not only necessary but also sufficient to promote degradation. Suppression of AIB1 levels leads to ERα stabilization in the presence of 17β-estradiol and, despite increased ERα levels, reduced recruitment of ERα to endogenous target gene promoters. In addition, association of RNA polymerase II with ERα target promoters is lost when AIB1 is suppressed, leading to inhibition of target gene transcription. AIB1 thus plays a dual role in regulating ERα activity, one in recruiting transcription factors including other coactivators involved in gene activation and the other in regulating ERα protein degradation mediated by the ubiquitin–proteosome machinery.
Estrogen plays a central role in the control of development, sexual behavior, and reproductive functions, and its effects have been linked to the progression of the majority of human breast cancers. The diverse biological effects of estrogen are mediated by two estrogen receptors (ERs), ERα and ERβ, which are members of the nuclear hormone receptor superfamily (1, 2). Upon 17β-estradiol (E2) binding, ER undergoes a major conformational change, binds to its cognate DNA response element (ERE) located in the promoter/enhancer regions of target genes, and regulates gene transcription (3). It is recognized that ER-mediated transcription is a highly complex process involving a multitude of coregulatory factors and “cross talk” among distinct signaling pathways (reviewed in ref. 4). The cofactors can be broadly divided into coactivators, which augment functions of activated receptors, and corepressors, which sustain the inactivated state of receptors (reviewed in ref. 5). The coactivators enhance receptor activity by modulating chromatin state through recruitment of ATP-dependent chromatin remodeling complexes and modification of the histones by methyltransferases and acetyltransferases. The p160 family of coactivators serves as platforms to recruit factors that have intrinsic enzymatic activities, whereas the DRIP/TRAP/SMCC complexes are “mediator” complexes bridging the receptor to the basal transcription machinery. We and others have demonstrated that these coactivators become recruited to target gene promoters by agonist-bound ERα in a dynamic fashion after a sequential order cycling on and off the promoter (6, 7).
Cellular responses to E2 are highly controlled, involving regulation of the ERα level through transcriptional, posttranscriptional, and posttranslational mechanisms (8–10). E2 binding accelerates receptor degradation, reducing the half-life of the ERα protein from ≈5 days to ≈3–4 h (11). In addition to E2, the ERα-pure antagonist fulvestrant (Faslodex or ICI 182,780, AstraZeneca, Wilmington, DE) inhibits ERα activity by inducing rapid down-regulation of the receptor (12). The selective ER modulator (SERM) GW5638 has also been shown to induce ERα degradation (13). In contrast, another SERM, 4-hydroxytamoxifen (OHT), stabilizes ERα (14). In common with many short-lived transcription factors, ERα degradation is through the ubiquitin–proteasome pathway because treatment with specific inhibitors of proteasome function can block E2-mediated ERα degradation (15–17). Evidence from a number of investigators has suggested that the ERα down-regulation is linked to its transcriptional activity (16, 17). Mutant ERα that has lost its transcriptional activity and association with coactivators is not down-regulated in the presence of E2. Conversely, inhibiting proteasome activity abrogates ERα transactivation and immobilizes ERα on the nuclear matrix (18). Recent work by Reid et al. (17) further strengthened the notion that down-regulation of ERα by E2 is an integral part of the transcriptional activity of the receptor. By using chromatin immunoprecipitation (ChIP) assays, they demonstrated that in the presence of MG132, RNA polymerase II (Pol II) was never associated with ERα target gene promoters after E2 treatment, and that the length of ERα chromatin-association cycle was considerably extended. Reciprocally, inhibition of transcription by inhibitors prevented proteasome-mediated degradation of ERα.
Despite the strong indication that regulation of ERα level via the proteasome pathway is essential for ERα-mediated transactivation, the mechanism by which E2-activated ERα is ubiquitinated and subsequently targeted for proteasomal degradation remains to be determined. Ubiquitination of proteins involves the action of three classes of enzymes, namely the ubiquitin-activating enzyme (UBA), the ubiquitin-conjugating enzyme (UBC), and the ubiquitin ligase (19). Interestingly, several steroid receptor-interacting proteins have been identified as components of the ubiquitin–proteasome degradation system, including SUG1/Trip1, an ATPase subunit of the 26S proteasome complex (20); uba3, the catalytic subunit of the NEDD8-activating enzyme (21); UBC9 (22); and RSP5/RPF1 and E6-AP, both ubiquitin ligases (23, 24). Not only do these proteins interact with the steroid receptors, they also modulate the receptor activity, suggesting that mechanisms targeting receptor for degradation are linked to those regulating receptor activation.
Here we demonstrate that AIB1 (SRC3/ACTR/RAC3/p/CIP/NCoA3), a member of the p160 coactivator family and encoded by a gene frequently amplified in breast cancer, is required for agonist-stimulated ERα degradation. Knockdown of the AIB1 protein, but not other members of the p160 coactivator family, leads to ERα protein stabilization in the presence of E2. Further, E2- and fulvestrant- or GW5638-induced ERα degradation appear to be mediated through distinct pathways because reduced AIB1 expression affects ERα degradation only in the presence of E2. Despite increasing ERα levels, suppression of AIB1 results in decreased ERα-mediated transactivation through the reduction of recruitment of ERα and Pol II to target gene promoters. These findings suggest that the coactivator AIB1 may be a modulator of both ERα transcriptional activity and protein stability, thus directly linking these two molecular processes.
Materials and Methods
Cell Lines and Reagents. MCF-7 cells were cultured in hormone-free condition for 3 days before the hormone treatment. The ERα, progesterone receptor (PR), and SRC-1 Abs were purchased from Neomarkers (Fremont, CA), the Pol II and hemagglutinin (HA)-tagged Abs from Covance (Princeton), and the calnexin Ab from Stressgen Biotechnologies (Victoria, Canada). The AIB1 Ab was raised against GST-AIB1 (amino acids 695–933) (25).
Short Interfering RNA (siRNA) Transfections. The 19-nt RNA oligonucleotides with a 3′-dTdT overhang were designed for each target gene, then synthesized and annealed to generate siRNA (Dharmacon, Lafayette, CO). For transient transfections, siRNAs were transfected into cells by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At 48 h posttransfection, cells were treated with either vehicle (ethanol) or hormone and harvested later for RNA and protein analyses.
Western Blot and Coimmunoprecipitation Analyses. Whole-cell lysates were prepared in RIPA lysis buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). For Western blotting, 20 μg of the lysates was resolved by SDS/PAGE and transferred to a nitrocellulose membrane, and proteins that reacted with the Abs were detected with a chemiluminescent substrate. For coimmunoprecipitation, 500-μg samples of lysates were immunoprecipitated with either 1 μg of rabbit IgG or 1 μg of E6-AP polyclonal Ab. The immunoprecipitated proteins were then subjected to Western blot analysis.
Quantitative RT-PCR Analysis. The expression of mRNA or heterogeneous nuclear RNA was analyzed by real-time RT-PCR. Optimal DNA primers were designed with the program primer express (Applied Biosystems). For the analysis of heterogeneous nuclear RNA, DNA primers were designed that spanned the intron–exon boundary. Quantitative RT-PCRs were conducted in 96-well plates by using the SYBR Green PCR kit (Applied Biosystems), and samples were amplified with the ABI Prism 7700 Sequence Detector (Applied Biosystems).
ChIP. Cells were plated on 15-cm culture dishes under hormone-free conditions and transfected with either siLuc or siAIB1. On the third day posttransfection, cells were treated with 100 nM E2 for various periods. After treatment, ChIP was performed as described in ref. 6.
In Vitro GST Binding Assay. Immobilized GST fused to different fragments of E6-AP was preincubated at 4°C for 1 h in the binding buffer containing 1 mg/ml BSA. The fusion proteins were then incubated with 100,000 cpm of 35S-labeled in vitro transcribed translated AIB1 in the binding buffer at 4°C for 1 h. The Sepharose beads were then washed three times with the same binding buffer, resuspended in 25 μlof2× SDS sample buffer, and boiled, and the eluted proteins were analyzed by electrophoresis.
Results
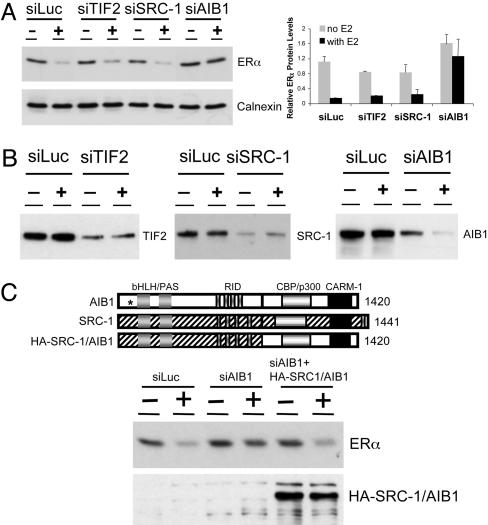
AIB1 Mediates ERα Down-Regulation by E2. In an effort to delineate the link between ERα degradation and its transcriptional activity and to determine whether the coactivators, in addition to enhancing the receptor activity, also play a role in E2-induced degradation of ERα, we used RNA interference (RNAi) to knock down the expression of specific coactivators and asked whether reduction in the level of any of the coactivators influences ERα stability. Studies by Lonard et al. (16) have demonstrated that ERα mutants that have lost or have reduced binding to the p160 coactivators become stabilized in the presence of E2. We thus decided to focus primarily on this family of coactivators. There are three members of the p160 coactivator family, SRC-1 (NCoA-1) (26), TIF2 (NCoA-2, GRIP1) (27, 28), and AIB1 (29, 30). Despite significant sequence homology among p160 members, several lines of evidence suggest that in addition to some degree of functional redundancy they have unique functions as well (31–33). To address whether these three coactivators have distinct or similar functions in mediating the ERα action, siRNAs were designed to target specific mRNA sequences of TIF2, SRC-1, or AIB1 and then transfected into ERα-positive human breast carcinoma MCF-7 cells. To evaluate the regulation of ERα protein level by hormone after transfection, the cells were maintained in hormone-free conditions for 3 days before being treated with vehicle or 100 nM E2 for 16 h. In cells transfected with control siRNA targeting the luciferase gene (siLuc), E2 treatment led to >80% down-regulation of ERα. This down-regulation was also observed in cells transfected with either siTIF2 or siSRC-1. In contrast, knockdown of AIB1 completely blocked E2-induced down-regulation of ERα (Fig. 1A).
Fig. 1.
Suppression of AIB1 by RNAi leads to ERα protein stabilization in the presence of E2. (A) Suppression of AIB1 inhibits E2-induced ERα degradation. MCF-7 cells were seeded in hormone-free condition before siRNA transfection. Forty-eight hours posttransfection, cells were treated with either vehicle (–) or 100 nM E2 (+) for 16 h. The ERα protein expression was analyzed by Western blotting (Left). The expression of calnexin protein was included as a loading control. (Right) Quantitative analyses of the ERα expression relative to the calnexin level in three independent experiments are represented. (B) siRNA specific to each of the p160 coactivators leads to efficient inhibition of the target protein. siRNA targeting the luciferase gene was used as a negative control. (C) The carboxyl-terminal region of AIB1 is necessary to mediate the ERα degradation by E2. (Top) Schematic diagram of AIB1, SRC-1, and the HA-tagged SRC-1/AIB1 is shown; the site in AIB1 targeted by siAIB1 is indicated by an asterisk. (Middle and Bottom) Western blots.
To verify the silencing efficiency and specificity of RNAi, we examined the expression level of each coactivator in cells transfected with either siLuc or specific siRNA. siRNA transfection led to a significant suppression in the levels of protein and mRNA of the target p160 coactivator but not of the two other members of the family (Fig. 1B and data not shown). Interestingly, we consistently observed that the expression of AIB1 was down-regulated by E2 in both siLuc- and siAIB1-transfected cells (Fig. 1B and Fig. 2). This observation is in line with previous reports demonstrating that E2 can repress AIB1 at mRNA and protein levels, whereas the antiestrogens fulvestrant and OHT up-regulate the expression of AIB1 (34, 35).
Fig. 2.
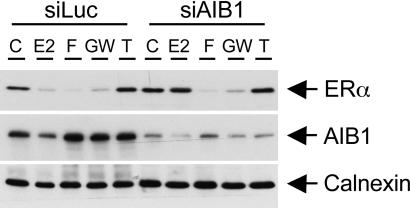
Suppression of AIB1 inhibits E2-induced ERα degradation but does not affect the ERα stability in the presence of OHT, fulvestrant, or GW5638. Solutions used in the experiments were vehicle control (C), 100 nM E2, 10 nM fulvestrant (F), 1 μM GW5638 (GW), and 1 μM OHT (T).
To further demonstrate that AIB1 is necessary for ERα down-regulation in the presence of E2, we used a HA-tagged chimeric construct that contains the amino-terminal portion of SRC-1 fused to the carboxyl-terminal region of AIB1. This chimera, not targeted by the specific siAIB1 used, was coexpressed with siAIB1 and tested for the ability to rescue ERα down-regulation (Fig. 1C). Western blot analysis showed that siAIB1 transfection in MCF-7 cells led to ERα stabilization in the presence of E2. However, the ERα stabilization could be reversed when siAIB1 was cotransfected with the chimeric HA-SRC-1/AIB1, demonstrating that AIB1, specifically the region between residues 875 and 1420 encompassing the CBP/p300 and CARM-1 binding domains, is necessary to mediate the effect of AIB1 on ERα protein turnover.
AIB1 Is Necessary for Agonist-Induced but Not Antagonist-Induced ERα Degradation. Several studies have demonstrated that the ERα protein degradation is mediated through the ubiquitin–proteasome pathway. We confirmed in MCF-7 cells that ERα was degraded in the presence of either E2 or fulvestrant. This effect was ligand-specific, because OHT stabilized ERα protein levels. Both E2- and fulvestrant-induced ERα degradation was found to be mediated by the ubiquitin–proteasome pathway because it could be reversed by the treatment with the proteasome inhibitor MG132 (data not shown). Because knockdown of AIB1 blocked ERα degradation induced by E2 (Fig. 1A), we next asked whether it had a similar effect on ERα stability in the presence of other ligands. MCF-7 cells were transfected with siLuc or siAIB1, cultured in hormone-free condition for 3 days, and then treated with various ligands for 16 h. The expression of ERα and AIB1 in these cells was analyzed by Western blotting (Fig. 2). E2 or fulvestrant treatment resulted in degradation of ERα, whereas OHT stabilized the receptor. When the AIB1 level was reduced by RNAi, ERα was stabilized in the presence of E2. Conversely, this stabilization was not observed in the presence of fulvestrant, because suppression of AIB1 had no effect on ERα degradation induced by fulvestrant. We also evaluated the role of AIB1 in mediating ERα degradation induced by GW5638, a SERM that has properties distinct from OHT or fulvestrant (36, 37). Similar to fulvestrant-induced ERα degradation, GW5638-induced ERα degradation was not affected by the inhibition of AIB1 (Fig. 2). These findings suggest that although ERα degradation induced by E2, fulvestrant, or GW5638 requires the proteasome as the ultimate step, distinct pathways mediated by different intermediary factors are involved.
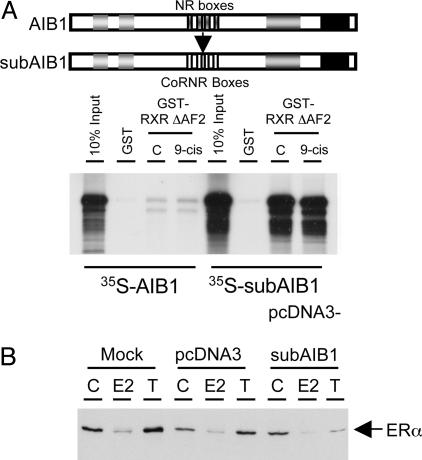
AIB1 Is Sufficient to Mediate Ligand-Induced ERα Degradation. To test whether the recruitment of AIB1 by ERα is sufficient for ERα degradation, we made a “reversed pharmacology” mutant of AIB1 in which the nuclear receptor interacting sequences (NR boxes) identified for coactivator–receptor interaction were replaced with the CoRNR boxes, which have been shown to mediate the interaction between corepressors and receptors in the absence of ligand or presence of antagonists (6, 38) (Fig. 3A, schematic diagram). The CoRNR box-substituted AIB1, termed subAIB1, would be predicted to interact with ERα in the presence of OHT instead of E2. If the recruitment of AIB1 to ERα is sufficient to induce the receptor degradation, expression of subAIB1 should be sufficient to reverse the effect of OHT, resulting in OHT-induced ERα degradation instead of stabilization. subAIB1 was recruited to the AF2 domain deleted form of retinoid X receptor (RXR ΔAF2) in a GST pull-down assay, whereas the WT AIB1 was not, confirming its reversed specificity (Fig. 3A). We then overexpressed the vector alone or subAIB1 in MCF-7 cells, and the level of ERα was assessed in the absence or presence of either E2 or OHT (Fig. 3B). In mock- or vector-transfected cells, ERα was degraded in the presence of E2 and stabilized by OHT. In contrast, the ERα level was diminished in the presence of OHT when subAIB1 was overexpressed in cells, demonstrating that the recruitment of AIB1 is sufficient to induce ERα degradation.
Fig. 3.
AIB1 binding to ERα is sufficient to induce its degradation. (A Upper) Schematic diagram showing the replacement of the nuclear receptor interacting sequences in AIB1 with CoRNR boxes in subAIB1. (A Lower) Interaction of 35S-labeled in vitro translated AIB1 and subAIB1 with GST or GST-RXR ΔAF2 in the absence or presence of 9-cis-retinoic acid was assessed by GST pull-down assay. (B) Protein expression of ERα was examined by Western blot analysis in MCF-7 cells mock-transfected or transfected with vector alone or subAIB1 in the presence of vehicle (C), E2, or OHT (T).
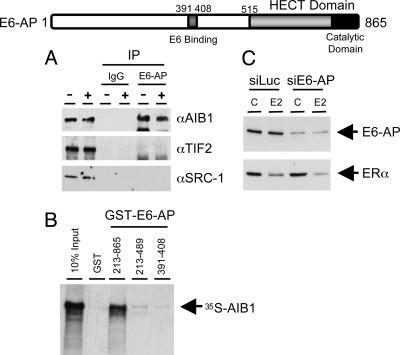
AIB1 Interacts with a Ubiquitin Ligase, E6-AP. In an effort to delineate the mechanisms by which AIB1 mediates ERα degradation, we investigated whether AIB1 could interact with components of the ubiquitin-proteasome pathway. Several proteins within the proteasome pathway have been implicated in modulating steroid hormone receptor activity, one of which is E6-AP. E6-AP was originally identified as a ubiquitin protein ligase mediating the degradation of the tumor suppressor p53 by the E6 protein of the human papillomavirus (39). A previous report (24) has shown that E6-AP can interact with and potentiate the transcriptional activity of several steroid hormone receptors, including ERα. This finding prompted us to examine the interaction between AIB1 and E6-AP. We performed coimmunoprecipitation assays to test the interaction between E6-AP and each of the p160 proteins. Total cellular extracts were prepared from MCF-7 cells treated with or without E2, immunoprecipitated with either rabbit IgG or rabbit anti-human E6-AP, and then immunoblotted with Ab against AIB1, TIF2, or SRC-1 (Fig. 4A). Of the p160 coactivators, only AIB1 was found in complex with E6-AP, and this interaction was independent of the presence of E2. GST pull-down assays were carried out to further delineate the basis of the E6-AP–AIB1 interaction (Fig. 4B). These studies demonstrated that AIB1 can directly interact with E6-AP in vitro, and that the interaction domain may reside within the carboxyl terminus of the E6-AP between residues 489 and 865.
Fig. 4.
AIB1 interacts with the ubiquitin ligase E6-AP both in vivo and in vitro. (Top) Diagram showing the structure of E6-AP. The HECT and catalytic domains are as indicated. (A) AIB1 interacts with E6-AP in vivo as determined by coimmunoprecipitation. (B) AIB1 interacts with E6-AP directly in vitro as assessed by GST pull-down assay. (C) E6-AP is not necessary for E2-induced ERα degradation.
To address the question whether the ability of AIB1 to target ERα for degradation requires the E6-AP ligase, we inhibited the expression of E6-AP by siRNA and examined whether loss of E6-AP would block ER degradation. Contrary to what we found with AIB1, reduction of E6-AP levels did not stabilize ERα in the presence of E2 (Fig. 4C), suggesting E6-AP is not necessary to mediate ERα degradation.
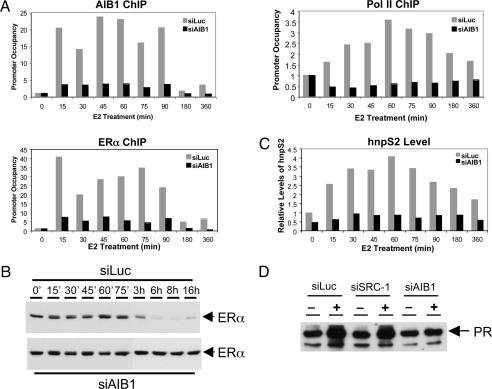
AIB1 Is Required for Optimal Binding of ERα to Target Gene Promoters and Transactivation. We performed ChIP assays to assess the effect of AIB1 knockdown on the recruitment of ERα, AIB1, and Pol II to the promoter of an endogenous ERα target gene, pS2. MCF-7 cells were transiently transfected with siLuc or siAIB1, cultured in hormone-free conditions for 3 days, and then treated with 100 nM E2 for various times (Fig. 5). To quantify E2-induced recruitment of ERα and other factors to the promoter, coprecipitated DNA was analyzed by quantitative real-time PCR, and the fold enrichment after immunoprecipitation, relative to the untreated control, was presented as promoter occupancy (Fig. 5A). The knockdown of the AIB1 level by siRNA was verified by Western blot (data not shown). Consistently, ChIP assay demonstrated that in siAIB1-transfected cells, very little AIB1 was recruited to the pS2 promoter as compared with the robust and cyclic recruitment of AIB1 to the promoter observed in control siLuc-transfected cells (Fig. 5A Upper Left). ERα recruitment followed a similar pattern: in siLuc-transfected cells, E2 stimulation rapidly led to a >40-fold increase in promoter occupancy by ERα after 15 min of treatment, and it declined after 30 min, followed by another round of promoter association (Fig. 5A Lower Left). Similar to that of AIB1, the ligand-induced recruitment of ERα to the promoter appeared to be transient as it decreased to the basal level with extended E2 treatment after 3 h. Interestingly, in cells in which expression of AIB1 was suppressed, nearly 6-fold less ERα was associated with the promoter, demonstrating that AIB1 binding is required for optimal association of ERα with the promoter. In parallel, we examined the expression level of ERα after E2 treatment (Fig. 5B). In cells transfected with siLuc, ERα levels were unchanged for the first 75 min of E2 treatment, fell >50% after 90 min, and were barely detectable after 6 h (Fig. 5B Upper and data not shown). In contrast, in siAIB1-transfected cells, ERα levels were stable in the presence of E2 for >16 h (Fig. 5B Lower).
Fig. 5.
AIB1 is required for optimal association of ERα with target gene promoter and ERα transactivation. (A) The association of ERα, AIB1, and Pol II with the pS2 promoter upon E2 stimulation was determined by ChIP analyses in MCF-7 cells. (B) The protein levels of ERα in cells after E2 treatment at different time points were determined by Western blotting. (C) Inhibition of the AIB1 expression abolishes the E2-dependent transactivation of the pS2 gene. The levels of hnpS2 were measured by quantitative RT-PCR. (D) Suppression of AIB1 has a more marked effect on the E2-inducible PR up-regulation than does SRC-1. The protein levels of PR were determined by Western blotting.
To determine the transcriptional activity of ERα in control cells vs. cells in which the level of AIB1 had been decreased by RNAi, we first examined Pol II recruitment to the pS2 promoter by ChIP (Fig. 5A Right). In siLuc-transfected cells, the association of Pol II with the promoter followed a pattern distinct from that of ERα and AIB1: it increased less rapidly, reached a maximum by 60 min, and gradually declined with longer treatment. The transcriptional activation of the pS2 gene, as measured by the quantitative RT-PCR analysis of the unprocessed heterogeneous nuclear pS2 RNA (hnpS2), coincided with the time course of the Pol II recruitment to the promoter (Fig. 5C). In line with much reduced recruitment of ERα and AIB1 in siAIB1-transfected cells, Pol II in these cells failed to bind to the promoter, and consequently very little transcription of pS2 was detected. These results suggest that AIB1 binding to ERα target gene promoters is essential for the transcriptional activation of the target gene by facilitating the recruitment of ERα and Pol II to the promoter. After transactivation, AIB1 plays an additional role in mediating ERα degradation, which further contributes to receptor regulation. ERα degradation after E2 stimulation occurs at a time subsequent to maximal transcriptional initiation, indicating that the E2-dependent transcriptional activation of the receptor precedes the degradation of the receptor. More-detailed ChIP and Western blot analyses at different time points revealed that the ERα receptor level started to decrease after 90 min, when strong pS2 promoter occupancy was still observed with ERα, AIB1, and Pol II (Fig. 5 and data not shown), suggesting that the receptor degradation occurs before the cessation of ERα-mediated gene transactivation.
We have shown that AIB1, but not SRC-1 or TIF2, mediates E2-induced ERα degradation. We then asked whether the p160 coactivators would have different effects on ERα transactivation. We compared the E2-dependent up-regulation of the PR in MCF-7 cells that had either suppressed AIB1 or SRC-1 expression (Fig. 5D). Western blot analysis showed that the E2-induced expression of PR was retained in siSRC-1-transfected MCF-7 cells. In contrast, suppression of AIB1 abolished this induction (Fig. 5D). This result was not due to differences in knockdown because SRC-1 and AIB1 were reduced to similar levels by Western blot (data not shown). This finding supports a unique role for AIB1 in ERα function.
Discussion
Gene expression responding to hormonal stimulation in eukaryotic cells is rigorously controlled. In addition to modulation of the intracellular hormone levels, other mechanisms limit the transactivation by nuclear hormone receptors. Hormone-induced transcriptional initiation is a rapid process, as is the cessation of transcription after hormone withdrawal. The binding of multicomponent coactivator complexes to the liganded receptor would make the passive disassembly of the complexes and release of the ligand from the receptor quite inefficient. Three mechanisms have been proposed that may contribute to a more rapid termination of transcription: (i) binding of molecular chaperones to the responsive DNA promoters to facilitate the disassembly of the transcription regulatory complexes (40), (ii) binding of histone deacetylases and SWI/SNF complexes, which remodels the chromatin structure and allows the dissociation of the regulatory complexes (7), and (iii) recruitment of ubiquitin ligases and components of the 19S proteasome regulatory subunit onto the promoters, leading to polyubiquitination of the receptor, which is subsequently translocated to active sites of degradation (17, 41). Here, we identify a mechanism that mediates the E2-dependent degradation of ERα, carried out by one of the coactivators within the regulatory complexes, AIB1. Our experimental data suggest that AIB1 uniquely mediates the agonist- but not antagonist-induced ERα degradation. More importantly, suppression of AIB1 leads to reduced recruitment of ERα to its target gene promoter, loss of Pol II recruitment, and correspondingly, inhibition of transcription. Although the ubiquitin ligase E6-AP can interact directly with AIB1, it alone is not necessary for ERα degradation, as loss of E6-AP does not affect ERα stability. However, it may play a role in the stability of other components of the complex. We propose that AIB1 has a dual function in regulating ERα signaling: first as a coactivator by recruiting cofactors that have enzymatic activities such as histone acetyltransferases and histone methyltransferases, and thus increasing ERα transcriptional activity by modulating chromatin, and second by the posttranslational modification of ERα through recruitment of components of the ubiquitin–proteasome pathway, which then target ERα for proteasomal degradation. Loss of AIB1 will thus affect ERα-mediated signaling by both directly inhibiting transcriptional initiation and blocking ERα turnover, which may further compromise transcriptional regulation by the receptor. This model is consistent with previous findings that the ERα turnover is an integral part of the estrogen signaling and blockade of ERα degradation by proteasome inhibitors results in transcriptional inactivation. By using RNAi that specifically inhibits the expression of a target gene, our studies demonstrate that the nuclear receptor coactivator AIB1 has a unique role in modulating ERα stability and thus provides a direct link between the two pathways regulating ERα activity.
AIB1-mediated ERα degradation is agonist-specific, as decreased expression of AIB1 does not alter the ERα stability in the presence of partial or full antagonists but only in the presence of E2 (Fig. 2). Binding of antagonists and SERMs to ERα induces conformational changes of the receptor distinct from the change induced by E2 binding. In the presence of OHT, helix 12 of the ERα ligand-binding domain is in a position that prevents the formation of the coactivator binding surface but favors the association of corepressors (42, 43). Fulvestrant, on the other hand, elicits an unusual organization of the receptor ligand-binding domain and may be recognized by the cell as a misfolded protein, targeting it for destruction (44). Thus, fulvestrant induces ERα degradation through a distinct mechanism, independent of coactivator association. A recent study suggested that the NEDD8 conjugation pathway might be involved in mediating fulvestrant-induced ERα degradation (45). GW5638, a SERM, appears to exhibit some functional properties similar to fulvestrant because it induces a unique structural change in ERα distinct from that observed in the presence of OHT, and it inhibits the growth of tamoxifen-resistant breast tumors (37). However, given a chemical structure that more closely resembles OHT than fulvestrant, it remains to be determined whether the mechanism by which GW5638 induces ERα degradation is shared with fulvestrant or is distinct. Nonetheless, our findings clearly show that the mechanism underlying ERα degradation induced by E2 is distinct from that by fulvestrant or GW5638 because association of the coactivator AIB1 with ERα is essential for E2-induced degradation.
The ubiquitin ligase E6-AP was recently found to be able to interact directly with PR in a ligand-dependent fashion and enhance the transcriptional activity of several steroid hormone receptors. In addition, E6-AP, the ubiquitin ligase MDM2, and the SUG1/Rpt6 protein, a component of the 19S regulatory subunit of the proteasome, have been reported to cyclically associate with the ERα target gene promoter (17). We demonstrate that AIB1 can interact with E6-AP both in vivo and in vitro in a ligand-independent fashion (Fig. 4 A and B). However, E6-AP is not uniquely required to mediate the ERα degradation because repression of the E6-AP expression by RNAi does not affect ERα stability (Fig. 4C). In addition to degradation, E2 binding leads to other posttranslational modifications of the receptor, such as phosphorylation and acetylation, and these modifications may be intrinsically associated with receptor degradation. Mutation of the mitogen-activated protein kinase (MAPK) phosphorylation site in PR prevents progestin-dependent PR degradation (46). We have reported that AIB1 is also a target of the MAPK pathway. AIB1 phosphorylation by MAPK enhances the ERα activity partly by facilitating its interaction with CBP/p300 (25). It is conceivable that AIB1 mediates ERα degradation by other molecular changes of the receptor induced by E2 binding. It will be necessary to determine the mechanism by which these events contribute to the ERα turnover and how distinct and/or overlapping signaling pathways collaborate to modulate the disassembly of the ERα–coregulator complexes from target promoters.
Our ChIP analyses demonstrate that the inhibition of AIB1 recruitment to the promoter results in a reduced promoter association by ERα, suggesting that ERα binds to the promoter DNA more efficiently when complexed with the AIB1 coactivator (Fig. 5A). This result is consistent with the role of AIB1 as a scaffold protein that brings in histone acetyltransferases and histone methyltransferases, which alter the local chromatin structure allowing the formation of stable ERα–coactivator complexes on the promoter. The profile of Pol II recruitment to the pS2 promoter differs from that of ERα and AIB1 in control cells because its E2-induced promoter association occurs at a later time and reaches its maximum more gradually. This finding may be explained by the binding of other transactivating factors, including the subunits within the SWI/SNF complex, histone acetyltransferases, histone methyltransferases, and components of the preinitiation complex, that occurs before the recruitment of Pol II (6, 7). The finding that suppression of AIB1 leads to a complete loss of the Pol II recruitment and target gene transactivation is striking. Given the large number of regulatory factors recruited to the promoter by ligand, one might expect that these other factors could compensate for the inhibition of AIB1. We hypothesize that AIB1 has an essential role in mediating ERα activity. This hypothesis is supported by the illustration that inhibition of the AIB1 protein expression has a more detrimental effect on ERα target gene regulation than the inhibition of another p160 protein, SRC-1 (Fig. 5D). The dynamics of the coregulator complex assembly on the target gene promoter needs to be systematically evaluated by ChIP in MCF-7 cells that have a specific factor suppressed by RNAi to determine whether each of these cofactors has a different or similar role in modulating the ERα target gene transcription.
AIB1 was originally identified on the basis of its frequent amplification in breast and ovarian cancers (29). Overexpression of AIB1 mRNA and protein has been revealed in >60% and ≈10% of the breast tumors, respectively (47, 48). High levels of both AIB1 and HER2 protein in breast tumors have been recently associated with tamoxifen resistance (49). Given the unique functions of AIB1 in regulating the ERα activity, it is not surprising that abnormal expression of AIB1 is associated with malignancies in estrogen target tissues. The finding that GW5638, a SERM with a chemical structure similar to tamoxifen, is able to induce ERα degradation in an AIB1-independent fashion raises the possibility that it or compounds like it might prove to be useful in the treatment of tamoxifen-resistant breast cancers.
Acknowledgments
We thank Dr. P. Howley for kindly providing the E6-AP Ab and Dr. M. Stallcup for the TIF2 Ab. This work was supported in part by the Dana–Farber/Harvard Cancer Center Specialized Programs of Research Excellence in Breast Cancer, a Dana–Farber Women's Cancers Program grant, and National Institutes of Health Grants P01CA080111 (to M.B.) and DK48807 (to D.P.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ERα, estrogen receptor α; E2, 17β-estradiol; ChIP, chromatin immunoprecipitation; HA, hemagglutinin; OHT, 4-hydroxytamoxifen; Pol II, RNA polymerase II; PR, progesterone receptor; RNAi, RNA interference; SERM, selective estrogen receptor modulator; siRNA, short interfering RNA.
References
- 1.Green, S., Walter, P., Greene, G., Krust, A., Goffin, C., Jensen, E., Scrace, G., Waterfield, M. & Chambon, P. (1986) J. Steroid Biochem. 24, 77–83. [DOI] [PubMed] [Google Scholar]
- 2.Kuiper, G. G. J. M., Enmark, E., Pelto-Huikko, M., Nilsson, S. & Gustafsson, J.-A. (1996) Proc. Natl. Acad. Sci. USA 93, 5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klinge, C. M. (2001) Nucleic Acids Res. 29, 2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall, J. M., Couse, J. F. & Korach, K. S. (2001) J. Biol. Chem. 276, 36869–36872. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld, M. G. & Glass, C. K. (2001) J. Biol. Chem. 276, 36865–36868. [DOI] [PubMed] [Google Scholar]
- 6.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. (2000) Cell 103, 843–852. [DOI] [PubMed] [Google Scholar]
- 7.Metivier, R., Penot, G., Hubner, M. R., Reid, G., Brand, H., Kos, M. & Gannon, F. (2003) Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- 8.Saceda, M., Lippman, M. E., Chambon, P., Lindsey, R. L., Ponglikitmongkol, M., Puente, M. & Martin, M. B. (1988) Mol. Endocrinol. 2, 1157–1162. [DOI] [PubMed] [Google Scholar]
- 9.Read, L. D., Greene, G. L. & Katzenellenbogen, B. S. (1989) Mol. Endocrinol. 3, 295–304. [DOI] [PubMed] [Google Scholar]
- 10.Pink, J. J. & Jordan, V. C. (1996) Cancer Res. 56, 2321–2330. [PubMed] [Google Scholar]
- 11.Eckert, R. L., Mullick, A., Rorke, E. A. & Katzenellenbogen, B. S. (1984) Endocrinology 114, 629–637. [DOI] [PubMed] [Google Scholar]
- 12.Dauvois, S., Danielian, P. S., White, R. & Parker, M. G. (1992) Proc. Natl. Acad. Sci. USA 89, 4037–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wijayaratne, A. L. & McDonnell, D. P. (2001) J. Biol. Chem. 276, 35684–35692. [DOI] [PubMed] [Google Scholar]
- 14.Wijayaratne, A. L., Nagel, S. C., Paige, L. A., Christensen, D. J., Norris, J. D., Fowlkes, D. M. & McDonnell, D. P. (1999) Endocrinology 140, 5828–5840. [DOI] [PubMed] [Google Scholar]
- 15.Nawaz, Z., Lonard, D. M., Dennis, A. P., Smith, C. L. & O'Malley, B. W. (1999) Proc. Natl. Acad. Sci. USA 96, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonard, D. M., Nawaz, Z., Smith, C. L. & O'Malley, B. W. (2000) Mol. Cell 5, 939–948. [DOI] [PubMed] [Google Scholar]
- 17.Reid, G., Hubner, M. R., Metivier, R., Brand, H., Denger, S., Manu, D., Beaudouin, J., Ellenberg, J. & Gannon, F. (2003) Mol. Cell 11, 695–707. [DOI] [PubMed] [Google Scholar]
- 18.Stenoien, D. L., Patel, K., Mancini, M. G., Dutertre, M., Smith, C. L., O'Malley, B. W. & Mancini, M. A. (2001) Nat. Cell Biol. 3, 15–23. [DOI] [PubMed] [Google Scholar]
- 19.Hartmann-Petersen, R., Seeger, M. & Gordon, C. (2003) Trends Biochem. Sci. 28, 26–31. [DOI] [PubMed] [Google Scholar]
- 20.vom Baur, E., Zechel, C., Heery, D., Heine, M. J. S., Garnier, J. M., Vivat, V., LeDouarin, B. & Losson, R. (1996) EMBO J. 15, 110–124. [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, M., Long, X., Bailey, J. A., Reed, C. A., Osborne, E., Gize, E. A., Kirk, E. A., Bigsby, R. M. & Nephew, K. P. (2002) Mol. Endocrinol. 16, 315–330. [DOI] [PubMed] [Google Scholar]
- 22.Poukka, H., Aarnisalo, P., Karvonen, U., Palvimo, J. J. & Janne, O. A. (1999) J. Biol. Chem. 274, 19441–19446. [DOI] [PubMed] [Google Scholar]
- 23.Imhof, M. O. & McDonnell, D. P. (1996) Mol. Cell. Biol. 16, 2594–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawaz, Z., Lonard, D. M., Smith, C. L., Lev-Lehman, E., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1999) Mol. Cell. Biol. 19, 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Font de Mora, J. & Brown, M. (2000) Mol. Cell. Biol. 20, 5041–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onate, S. A., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (1995) Science 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- 27.Hong, H., Kohli, K., Garabedian, M. J. & Stallcup, M. R. (1997) Mol. Cell. Biol. 17, 2735–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voegel, J. J., Heine, M. J., Zechel, C., Chambon, P. & Gronemeyer, H. (1996) EMBO J. 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 29.Anzick, S. L., Kononen, J., Walker, R. L., Azorsa, D. O., Tanner, M. M., Guan, X. Y., Sauter, G., Kallioniemi, O. P., Trent, J. M. & Meltzer, P. S. (1997) Science 277, 965–968. [DOI] [PubMed] [Google Scholar]
- 30.Torchia, J., Rose, D. W., Inostroza, J., Kamei, Y., Westin, S., Glass, C. K. & Rosenfeld, M. G. (1997) Nature 387, 677–684. [DOI] [PubMed] [Google Scholar]
- 31.Nishihara, E., Yoshida-Komiya, H., Chan, C. S., Liao, L., Davis, R. L., O'Malley, B. W. & Xu, J. (2003) J. Neurosci. 23, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Z., Rose, D. W., Hermanson, O., Liu, F., Herman, T., Wu, W., Szeto, D., Gleiberman, A., Krones, A., Pratt, K., Rosenfeld, R., Glass, C. K. & Rosenfeld, M. G. (2000) Proc. Natl. Acad. Sci. USA 97, 13549–13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, J., Liao, L., Ning, G., Yoshida-Komiya, H., Deng, C. & O'Malley, B. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauritsen, K. J., List, H. J., Reiter, R., Wellstein, A. & Riegel, A. T. (2002) Oncogene 21, 7147–7155. [DOI] [PubMed] [Google Scholar]
- 35.Lonard, D. M., Tsai, S. Y. & O'Malley, B. W. (2004) Mol. Cell. Biol. 24, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willson, T. M., Norris, J. D., Wagner, B. L., Asplin, I., Baer, P., Brown, H. R., Jones, S. A., Henke, B., Sauls, H., Wolfe, S., et al. (1997) Endocrinology 138, 3901–3911. [DOI] [PubMed] [Google Scholar]
- 37.Connor, C. E., Norris, J. D., Broadwater, G., Willson, T. M., Gottardis, M. M., Dewhirst, M. W. & McDonnell, D. P. (2001) Cancer Res. 61, 2917–2922. [PubMed] [Google Scholar]
- 38.Hu, X. & Lazar, M. A. (1999) Nature 402, 93–96. [DOI] [PubMed] [Google Scholar]
- 39.Huibregtse, J. M., Scheffner, M. & Howley, P. M. (1993) Mol. Cell. Biol. 13, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman, B. C. & Yamamoto, K. R. (2002) Science 296, 2232–2235. [DOI] [PubMed] [Google Scholar]
- 41.Kang, Z., Pirskanen, A., Janne, O. A. & Palvimo, J. J. (2002) J. Biol. Chem. 277, 48366–48371. [DOI] [PubMed] [Google Scholar]
- 42.Shiau, A. K., Barstad, D., Loria, P. M., Cheng, L., Kushner, P. J., Agard, D. A. & Greene, G. L. (1998) Cell 95, 927–937. [DOI] [PubMed] [Google Scholar]
- 43.Metivier, R., Stark, A., Flouriot, G., Hubner, M. R., Brand, H., Penot, G., Manu, D., Denger, S., Reid, G., Kos, M., et al. (2002) Mol. Cell 10, 1019–1032. [DOI] [PubMed] [Google Scholar]
- 44.Pike, A. C., Brzozowski, A. M., Walton, J., Hubbard, R. E., Thorsell, A. G., Li, Y. L., Gustafsson, J. A. & Carlquist, M. (2001) Structure (London) 9, 145–153. [DOI] [PubMed] [Google Scholar]
- 45.Fan, M., Bigsby, R. M. & Nephew, K. P. (2003) Mol. Endocrinol. 17, 356–365. [DOI] [PubMed] [Google Scholar]
- 46.Lange, C. A., Shen, T. & Horwitz, K. B. (2000) Proc. Natl. Acad. Sci. USA 97, 1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bautista, S., Valles, H., Walker, R. L., Anzick, S., Zeillinger, R., Meltzer, P. & Theillet, C. (1998) Clin. Cancer Res. 4, 2925–2929. [PubMed] [Google Scholar]
- 48.List, H. J., Reiter, R., Singh, B., Wellstein, A. & Riegel, A. T. (2001) Breast Cancer Res. Treat. 68, 21–28. [DOI] [PubMed] [Google Scholar]
- 49.Osborne, C. K., Bardou, V., Hopp, T. A., Chamness, G. C., Hilsenbeck, S. G., Fuqua, S. A., Wong, J., Allred, D. C., Clark, G. M. & Schiff, R. (2003) J. Natl. Cancer Inst. 95, 353–361. [DOI] [PubMed] [Google Scholar]