Abstract
The chemokine CXC ligand 8 (CXCL8)/IL-8 and related agonists recruit and activate polymorphonuclear cells by binding the CXC chemokine receptor 1 (CXCR1) and CXCR2. Here we characterize the unique mode of action of a small-molecule inhibitor (Repertaxin) of CXCR1 and CXCR2. Structural and biochemical data are consistent with a noncompetitive allosteric mode of interaction between CXCR1 and Repertaxin, which, by locking CXCR1 in an inactive conformation, prevents signaling. Repertaxin is an effective inhibitor of polymorphonuclear cell recruitment in vivo and protects organs against reperfusion injury. Targeting the Repertaxin interaction site of CXCR1 represents a general strategy to modulate the activity of chemoattractant receptors.
Leukocyte trafficking into tissue sites of inflammation is directed by chemokines. Chemokines are grouped into four families based on a cysteine motif in the amino terminus of the protein (1, 2). Human CXC ligand 8 (CXCL8)/IL-8 and related molecules are polymorphonuclear cells (PMN) chemoattractants. Two high-affinity human CXCL8 receptors are known, CXC chemokine receptor 1 (CXCR1) and CXC chemokine receptor 2 (CXCR2). Only one corresponding receptor has been identified in the mouse, and this is recognized by ligands that act as neutrophil attractant, although a mouse orthologue of CXCL8 has not been identified. By recruiting and activating PMN, CXCL8 and related rodent molecules have been implicated in a wide range of disease states characterized by PMN infiltration in organs, including reperfusion injury (RI) (3).
G protein-coupled receptors (GPCR) are a prime target for the development of new strategies to control diverse pathologies (4–6). Antichemokine strategies include antibodies, N-terminal modified chemokines, and small-molecule antagonists (7–9). Here we describe a class of GPCR inhibitors that specifically block the inflammatory CXCL8 chemokine receptors CXCR1 and CXCR2 by means of an allosteric noncompetitive mode of interaction and protection against RI.
Materials and Methods
Reagents. Repertaxin (R)(–)-2-(4-isobutylphenyl)propionyl methansulfonamide) salified with l-lysine was dissolved in saline. Chemokines were from PeproTech (London). Chemicals, cell culture reagents, and protease inhibitors were from Sigma.
Migration. Cell migration of human PMN and monocytes and rodent peritoneal PMN were evaluated in a 48-well microchemotaxis chamber with or without Repertaxin. Agonists (1 nM CXCL8, 10 nM N-formyl-l-methionyl-l-leucyl-l-phenylalanine (fMLP), 10 nM CXCL1, 2.5 nM CCL2, 1 nM C5a, 5 nM rat and mouse CXCL1, and 2.5 nM rat and mouse CXCL2) were seeded in the lower compartment. The chemotaxis chamber was incubated for 45 min (human PMN), 1 h (rodent PMN), or 2 h (monocytes). L1.2 migration was evaluated by using 5-μm pore-size Transwell filters (Costar) (10).
CXCL8 Binding. [125I]CXCL8 [0.2–0.02 nM, specific activity 2,200 Ci/mmol (1 Ci = 37 GBq), Amersham Pharmacia] binding on human PMN or CXCR1/L1.2 transfectants was as described in ref. 11. Saturation experiments were performed on CXCR1/L1.2 transfectants. Nonspecific binding was determined by a 200-fold molar excess of unlabeled CXCL8. Scatchard analysis was performed with the ligand program (12).
Molecular Modeling Studies. The starting rhodopsin-based CXCR1 molecular model has been extracted from the GPCR database (www.gpcr.org). (S)-flurbiprofen-bound cyclooxygenase (COX)-1 was from the Protein Data Bank (www.pdb.org, PBD ID code 1EQH). Ligand/receptor interactions were performed by molecular mechanics methods (steepest descent and conjugate gradient algorithms) and molecular dynamics calculations as implemented in the discover (insight ii package Release 2000, Accelrys, San Diego) package by using the consistent valence forcefield. Details can be found in Supporting Text, which is published as supporting information on the PNAS web site.
Mutation Analysis of CXCR1 and Signaling. The human CXCR1 ORF was PCR amplified from a CXCR1/pCEP4 plasmid (kindly provided by P. M. Murphy, National Institutes of Health, Bethesda). Receptor mutants and chimeric receptors, in which the second intracellular loop of CXCR1 (amino acids 133–151) and C5aR (amino acids 132–151) have been swapped, were obtained by a standard two-step PCR by using the high-fidelity Pfx DNA polymerase (Invitrogen). The nucleotide sequence of each construct was confirmed by double-stranded DNA sequencing. G protein activation, Pyk2 tyrosine phosphorylation, and intracellular free calcium concentrations were analyzed as reported in Supporting Text.
Transcriptome Analysis by Using High-Density Oligonucleotide Arrays. PMN (three independent donors) were cultured with 10 nM CXCL8 for 4 h with vehicle or 1 μM Repertaxin. Next, RNA was purified and labeled (13). Gene expression analysis was performed as described in Supporting Text.
Photochemical Crosslinking of CXCR1. PMN membranes were incubated with [14C]R-ketoprofen (0.1 mM final concentration) with vehicle or an excess of Repertaxin (50 mM) and irradiated (10-cm distance) with a handheld UV illuminator. Details are in Supporting Text.
Cecal Ligation and Puncture (CLP). Eight hours after CLP, induced as described in ref. 14, peritoneal cavities were washed with ice-cold PBS and PMN were counted in a hemocytometer. Mice were treated with an optimal dose of Repertaxin (15 mg/kg, s.c.) or vehicle 30 min before CLP and 2 h and 4 h after CLP. A group was treated with dexamethasone (30 mg/kg, i.p.) 30 min before CLP.
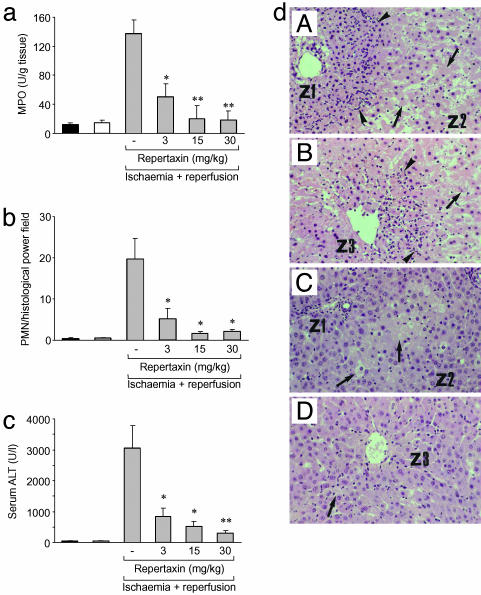
Rat Model of Hepatic Postischaemia RI. Irreversible hepatic damage was induced in the rat by reperfusion of ischaemic liver (1 h of ischaemia and 12 h of reperfusion), as described in ref. 15. RI injury was evaluated by alanin-aminotransferase levels in plasma obtained from posterior cava vein 12 h after reperfusion by using a commercial kit (Sigma). Myeloperoxidase activity and necrosis score were determined as described in refs. 15 and 16. PMN were identified by the naphthol AS-D chloroacetate technique for esterase (17). Red-stained PMN were counted in 20 nonconsecutive, randomly chosen ×400 histological fields. Ischaemic rats were treated with Repertaxin (3, 15, or 30 mg/kg) or vehicle 15 min before reperfusion (i.v.) and 2 h after reperfusion (s.c.).
Results
Identification and Structure Activity Relationship of a CXCL8 Inhibitor. Among nonsteroidal antiinflammatory drugs, phenyl acetic and phenylpropionic acids coordinate a network of polar interactions involving Arg-120, Glu-524, Tyr-355, and Hys90 at the bottom of an hydrophobic channel of COX (18), thus blocking the active site of the enzyme. Because (R)- and (S)-ketoprofen, along with other phenylpropionic acids, inhibit CXCL8-induced chemotaxis of human PMN in a COX-independent manner (11), we postulated the existence of a similar hydrophobic channel in CXCR1 and COX-1. Results, provided in Supporting Text and Fig. 6, which is published as supporting information on the PNAS web site, show the presence in CXCR1 of a putative ketoprofen-binding site in a channel generated by helices 1, 2, 3, 6, and 7 in the outer portion of the CXCR1 transmembrane (TM) domain. This site shows a significant size and shape similarity with the COX-1 channel involved in the binding of the representative nonsteroidal antiinflammatory drug (S)-flurbiprofen.
Starting from the above observation, the synthesis of inhibitors targeting CXCR1 was the goal of a focused Structure Activity Relationship program, provided in Supporting Text and Table 1, which is published as supporting information on the PNAS web site. Repertaxin (Table 1, entry 5) was selected for further investigation.
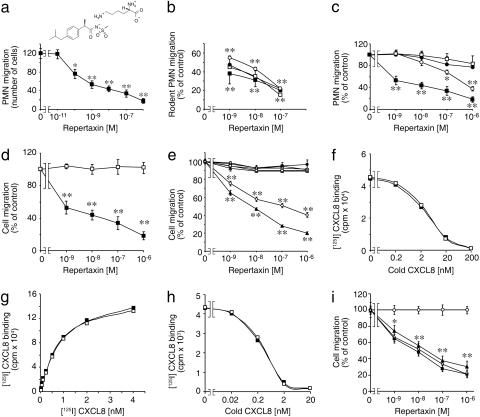
Selectivity of Repertaxin Inhibitory Activity. Repertaxin inhibited human PMN migration induced by CXCL8 (IC50 = 1 nM; Fig. 1a, representative of six independent experiments) and rodent PMN chemotaxis induced by CXCL1 and CXCL2 (Fig. 1b). Repertaxin inhibited the response of human PMN to CXCL1, which interacts with CXCR2 (Fig. 1c, IC50 = 400 nM). The potent inhibition of CXCL8-elicited PMN chemotaxis by Repertaxin is consistent with a dominant role of CXCR1 in mediating CXCL8 chemotaxis of PMN that express both CXCL8 receptors (19). Repertaxin did not inhibit the migration of human PMN to C5a and fMLP (Fig. 1c), monocyte migration induced by CCL2 (Fig. 1d) and spontaneous migration in the absence of chemoattractants (data not shown).
Fig. 1.
Effect of Repertaxin on CXCL8 activity and binding to cellular receptors. Results are absolute numbers (± SD) of migrated cells (a) or percent of control migration (± SD) of one experiment of three. (a) Human PMN migration induced by CXCL8. (b) Rodent PMN migration. Mouse (squares) and rat (circles) PMN migration was induced by mouse or rat CXCL1 (open symbols) or CXCL2 (filled symbols). (c) Human PMN migration induced by different chemoattractants: (CXCL8 (filled squares), CXCL1 (open circles), fMLP (open squares), and C5a (filled circles). (d) Monocyte [CCL2 (open squares)] or PMN [CXCL8 (filled squares)] migration. (e) L1.2 transfectants migration in response to optimal chemokine concentrations: 10 nM CXCL8 for CXCR1 (filled triangles) and CXCR2 (open circles), 10 nM CXCL12 for CXCR4 (filled circles), 10 nM CCL2 for CCR2b (open squares), 3 nM CCL22 for CCR4 (open triangles), 3 nM CCL5 for CCR5 (open diamonds), and 3 nM CCL19 for CCR7 (filled squares). (f) Effects of Repertaxin on CXCL8 binding. PMN were incubated with radiolabeled CXCL8 and increasing amounts of unlabeled CXCL8 in the presence (filled squares) or absence (open squares) of Repertaxin. Data are from one experiment of six. (g) Effects of Repertaxin on CXCL8 saturation curve. CXCR1/L1.2 transfectants were incubated with increasing concentrations of radiolabeled CXCL8 in the presence (filled squares) or absence (open squares) of Repertaxin. Data are from one experiment of three. (h) Effects of Repertaxin on displacement of radiolabeled CXCL8. CXCR1/L1.2 transfectants were incubated with radiolabeled CXCL8 and increasing amounts of unlabeled CXCL8 in the presence (filled squares) or absence (open squares) of Repertaxin. Data are from one experiment of three. (i) Effect of Repertaxin on CXCR1/L1.2 transfectants migration in response to 30 (filled triangles), 10 (filled squares), or 3 (open diamonds) nM CXCL8 and to 10 nM CXCL12 (open circles). *, P < 0.05 versus cell migration in the absence of Repertaxin; **, P < 0.01 versus cell migration in the absence of Repertaxin (Mann–Whitney U test).
Repertaxin inhibited the migration of CXCR1/L1.2 and CXCR2/L1.2 transfectants in response to CXCL8 (Fig. 1e) and of CXCR2/L1.2 transfectants in response to CXCL2 and CXCL7 (data not shown). The IC50 for CXCR1-transfected cells was comparable (1 nM) with that observed for PMN (Fig. 1 c and e). In contrast, the inhibitory activity of Repertaxin on CXCR2-transfected cells was higher (IC50 ≈ 100 nM) than that observed in human PMN, suggesting that the cellular context may influence the susceptibility to Repertaxin. Moreover, Repertaxin did not affect the migration to appropriate ligands of transfectants bearing CC chemokine receptor (CCR)2b, CCR4, CCR5, or CCR7 (Fig. 1e) and the migration to CXCL12 of untransfected L1.2 cells (Fig. 1e).
Surprisingly, despite its potent and selective inhibitory activity, Repertaxin did not compete the binding of radiolabeled CXCL8 (0.2 nM) at a ligand concentration in the range of CXCL8 Kd with CXCR1/2. Repertaxin (1 μM) (Fig. 1f) did not affect CXCL8 receptor numbers (35,518 ± 8,800 and 41,992 ± 12,597 sites per cell in vehicle and Repertaxin groups, respectively; mean ± SD of six different experiments) or affinity (Kd = 6.2 ± 2.3 × 10–10 M and 3.6 ± 1.5 × 10–10 M, respectively). To demonstrate the noncompetitive nature of Repertaxin action by using established methodology (20), CXCL8 binding was evaluated in radiolabeled CXCL8 saturation experiments by using a wide range of agonist concentrations. Repertaxin (1 μM) did not cause dextral displacement of the radioligand saturation curve on CXCR1/L1.2 transfectants (Kd = 5.6 ± 2.1 × 10–10 M and 4.1 ± 2.1 × 10–10 M in vehicle or Repertaxin pretreated cells, respectively; Fig. 1g) exposed to 15 pM–4 nM CXCL8.
Having found that Repertaxin is a noncompetitive antagonist of CXCL8 receptor binding, we examined the allosteric mechanism of action of Repertaxin. Maximal inhibition of radioligand binding by an allosteric antagonist can be observed in displacement experiments with radioligand concentrations much lower that the Kd value (20). Repertaxin (1 μM) did not affect the displacement of 20 pM [125I]CXCL8 by cold CXCL8 on CXCR1/L1.2 transfectants (Fig. 1h) and, in keeping with the binding data, inhibited PMN migration equally well over a wide range of CXCL8 concentrations (3–30 nM) (Fig. 1i). These data support the notion that Repertaxin acts as a noncompetitive allosteric inhibitor of CXCR1. These findings are reminiscent of UCB 35625, which inhibited CCR1 responses with little effect on binding (21).
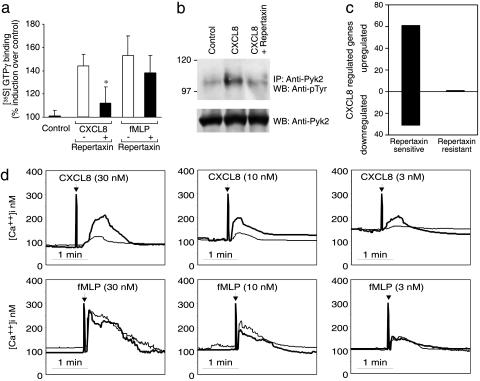
As expected, Repertaxin significantly inhibited CXCL8-induced activation of G protein, Pyk2 tyrosine phosphorylation, increase of intracellular free calcium increase, and transcriptional activation as assessed by Affymetrix technology (Fig. 2). Repertaxin also inhibited CXCL8-induced elastase release and production of reactive oxygen intermediates (data not shown).
Fig. 2.
Effect of Repertaxin on cell signaling activated by CXCL8. (a) G protein activation. Radiolabeled GTP binding was measured in PMN membranes incubated with vehicle (control), CXCL8, or fMLP with or without Repertaxin. Data are percent induction of radiolabeled GTP binding as compared with control cells. Data are mean ± SD of five independent experiments. *, P < 0.05 versus GTP binding without Repertaxin (Student's t test). (b) Pyk2 tyrosine phosphorylation. PMN were treated with CXCL8 with or without Repertaxin. (Upper) Tyrosine phosphorylation was evaluated in the immunoprecipitated Pyk2. (Lower) The same filter was probed with the anti-Pyk2 antibody. Data are from one experiment of three. IP, immunoprecipitate; WB, Western blot. (c) CXCL8-dependent gene regulation in PMN. Data are the number of genes up-regulated or down-regulated by CXCL8 treatment of PMN with or without Repertaxin. (d) Intracellular calcium increase. PMN loaded with FURA-2 were stimulated (arrows) with CXCL8 (3, 10, or 30 nM) or fMLP (3, 10, or 30 nM) with (normal line) or without (bold line) Repertaxin. Data are from one experiment of three.
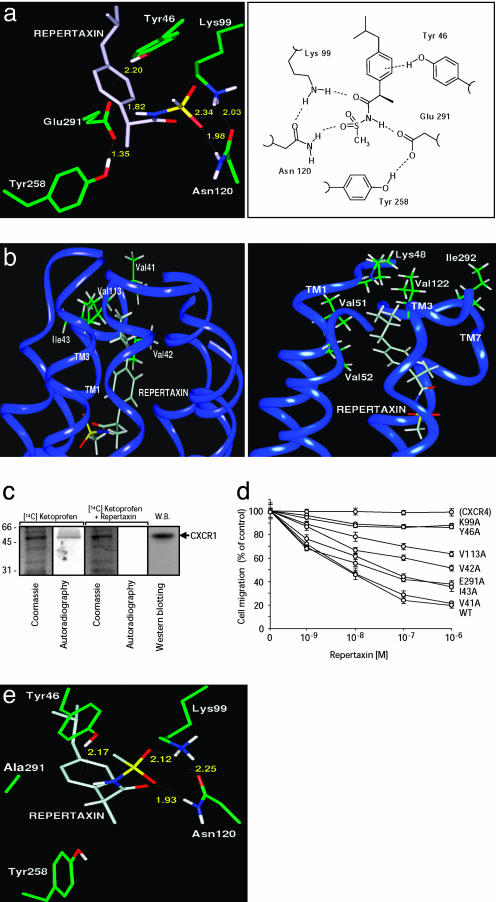
Binding of Repertaxin to CXCR1 and Molecular Modeling of the Interaction. Based on the model described above, molecular dynamics calculations led to a model in which Repertaxin generates strong polar interactions with five residues (Tyr-46 TM1, Lys-99 TM2, Asn-120 TM3, Tyr-258 TM6, and Glu-291 TM7) of CXCR1 (Fig. 3a). An electrostatic interaction directs the phenolic group of Tyr-46 toward the phenyl ring of Repertaxin, whereas a hydrogen bond is established between Repertaxin and the couple Lys-99/Glu-291. Asn-120 is anchored to the sulfonyl moiety of Repertaxin. Furthermore, Repertaxin indirectly promotes the formation of two additional polar interhelical interactions (Lys-99 TM2–Asn-120 TM3 and Tyr-258 TM6–Glu-291 TM7), which are not predicted in CXCR1 in the absence of Repertaxin. Thus, the isobutyl group of Repertaxin comfortably accommodates in an hydrophobic pocket formed by the lipophilic chains of four residues from TM1 and TM3 of CXCR1 (Fig. 3b Left). Val-42 and Val-113 are involved in the recognition of the isobutyl residue of Repertaxin; accordingly, a less favorable interaction with Ile-43 additionally contributes to the stabilization of the structure, whereas Val-41, located close to the hydrophobic cleft, does not directly interact with the isobutyl group.
Fig. 3.
Molecular modeling of Repertaxin interaction with CXCR1/CXCR2. (a) Repertaxin (light gray) in the CXCR1 allosteric site (green) is shown, with polar network in three-dimensional (Left) and two-dimensional (Right) representations. Dashed lines are electrostatic interactions between Repertaxin and CXCR1 residues. Calculated distances (Å) are shown. (b) CXCR1–Repertaxin and CXCR2–Repertaxin structural models viewed from within the plane of the membrane. (Left) Interaction of the isobutyl group of Repertaxin (light gray) with the side chains of five hydrophobic residues (green) of CXCR1 (flat blue ribbon). (Right) Molecular model of Repertaxin and CXCR2 interaction showing the lack of key lipophilic interactions between Repertaxin and the TM1/TM3 hydrophobic pocket of CXCR2. (c) Binding of Repertaxin to CXCR1. Cell membranes were incubated with radiolabeled (R)-ketoprofen with or without an excess of Repertaxin. After photochemical crosslinking, CXCR1 was immunoprecipitated (see Supporting Text) and analyzed by SDS/PAGE and Western blot (W.B.). The arrow marks CXCR1. Data are from one experiment of three. (d) Effect of Repertaxin on the CXCL8-dependent migration of L1.2 transfectants expressing wild-type CXCR1 (WT) on CXCR1 mutants and on L1.2 migration induced by CXCL12 (CXCR4). Results are the percentages of migration without Repertaxin (± SD) of three experiments. (e) Binding of Repertaxin (light gray) to CXCR1 (green) Glu291Ala mutant. Dashed lines are electrostatic interactions between Repertaxin and the CXCR1 mutant. Calculated distances (in Å) are shown.
When Repertaxin and CXCR2 are considered, the isobutyl group of Repertaxin is in a less favorable environment (Fig. 3b Right) because of the lack of specific hydrophobic interactions. These results are in keeping with the observation that Repertaxin is more potent in inhibiting CXCR1 than CXCR2 (see above).
To test the above binding model of Repertaxin with CXCR1, we first carried out experiments to assess whether Repertaxin actually binds to CXCR1. Because (R)-ketoprofen can be photoactivated (22), PMN membranes were incubated with [14C] (R)-ketoprofen and irradiated. The immunoprecipitated CXCR1 was examined by SDS/PAGE. Radiolabeled (R)-ketoprofen binds to CXCR1, and binding of radiolabeled (R)-ketoprofen was displaced by an excess of Repertaxin, thus suggesting that both (R)-ketoprofen and Repertaxin bind to CXCR1 (Fig. 3c).
To put to a test the model of the CXCR1–Repertaxin interactions, the polar amino acids anticipated by the model to be involved in the binding of Repertaxin to CXCR1 were selected for alanine-replacement mutagenesis. The model anticipates that these CXCR1 mutants, expressed in L1.2 cells, still support CXCL8-induced chemotaxis but with reduced sensitiveness to Repertaxin inhibition. Receptor expression levels, CXCL8 binding affinity, and chemotactic migration to CXCL8 of mutated CXCR1 transfectants were similar to wild-type CXCR1. Tyr46Ala or Lys99Ala CXCR1 transfectants completely resisted the action of Repertaxin. The Glu291Ala CXCR1 mutant had partial resistance to Repertaxin (Fig. 3d), in keeping with the finding that the lack of the hydrogen bond between Repertaxin and mutated Glu-291 is replaced by a tight multicentered interaction of Asn-120 and Lys-99 of CXCR1 with the sulfonyl moiety of Repertaxin (Fig. 3e). Next, the hydrophobic residues of CXCR1 interacting with the isobutyl group of Repertaxin have also been mutated to alanine. The partial resistance of Val42Ala and Val113Ala CXCR1 mutants confirms their involvement in Repertaxin binding; the lower resistance exhibited by the Ile43Ala mutant and the lack of resistance shown by Val41Ala confirm the model (Fig. 3d). The finding that CXCL8 binding to mutated CXCR1 is similar to wild-type CXCR1 is in keeping with the conclusion that the interaction sites in CXCR1 with Repertaxin and CXCL8 are distinct.
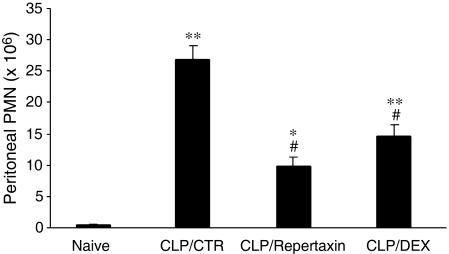
In Vivo Activity and Therapeutic Potential of Repertaxin. Having identified Repertaxin as a new potent CXCL8 inhibitor with a unique mode of action, it was important to assess its therapeutic potential in vivo. PMN recruitment into the peritoneal cavity after CLP was inhibited by Repertaxin treatment by 63%, whereas the inhibition induced by dexamethasone was 45% (Fig. 4).
Fig. 4.
In vivo efficacy of Repertaxin in inhibiting PMN recruitment in CLP. Experimental groups: Naive, animals without CLP; CLP/CTR, animals with CLP and vehicle; CLP/Repertaxin, animal with CLP and Repertaxin (15 mg/kg, s.c.); CLP/DEX, animals with CLP and dexamethasone (30 mg/kg, i.p.). There were five animals per experimental group. Data are PMN in the peritoneal cavity. Data represent the mean ± SE from one experiment of three. *, P < 0.05 versus naive animals; **, P < 0.01 versus naive animals; #, P < 0.01 versus CLP/CTR group (Tukey's test).
To assess the actual therapeutic potential of Repertaxin, we used a rat model of liver postischaemia RI. Repertaxin (15 mg/kg) inhibited PMN recruitment into reperfused livers by 90% as shown by myeloperoxidase content (Fig. 5a) and histopathologic analysis (Fig. 5b). Repertaxin drastically reduced liver damage in terms of alanin-aminotransferase levels and hepatocellular necrosis (Fig. 5 c and d). On the basis of these results, Repertaxin has been evaluated in phase I clinical studies and found safe and well tolerated (data not shown).
Fig. 5.
In vivo efficacy of Repertaxin in inhibiting PMN recruitment and tissue damage in RI. RI was induced by 1-h ischaemia of the liver followed by 12-h reperfusion. Experimental groups (five animals per group): animals without ischaemia and reperfusion (control, black bars); animals with ischaemia but without reperfusion, animals with 1-h ischaemia (white bars); animals with ischaemia plus reperfusion, animals with ischaemia for 1-h followed by 12-h reperfusion (gray bars). Animals with ischaemia and reperfusion were treated either with vehicle or Repertaxin. (a and b) PMN infiltration was assessed by myeloperoxidase (MPO) (a) and cell counts in histopathology examination (b). Data are mean ± SE from one experiment of three. *, P < 0.05; **, P < 0.01 versus ischaemia plus reperfusion vehicle-treated animals (Student's t test). (c) Plasma alanin-aminotransferase (ALT) (five animals per group). Data are mean ± SE from one experiment of three. *, P < 0.05; **, P < 0.01 versus ischaemia plus reperfusion vehicle-treated animals (Student's t test). (d) Histopathological analysis of liver RI. (dA and dC) Periportal areas (Z1 and Z2). (dB and dD) Perivenular areas (Z3). (dA and dB) Control animals. (dC and dD) Repertaxin-treated animals. Arrowheads indicate infiltrating PMN, and arrows indicate necrotic hepatocytes. Microphotographs are representative areas. Data are from one experiment of three.
Discussion
The results reported here show that Repertaxin is a noncompetitive allosteric blocker of the CXCL8 receptors CXCR1 and CXCR2. In this respect, Repertaxin is unique in that we are not aware of previous descriptions of noncompetitive allosteric inhibitors for chemoattractant receptors and, more in general, for peptidergic GPCR (7). This compound is potent (IC50 = 1 nM, CXCR1) and specific inasmuch as other chemoattractant receptors are not affected. Repertaxin was active in vivo as an inhibitor of PMN infiltration and RI.
Mutagenesis studies have implicated a hydrophobic channel defined by helices 1, 2, 3, 6, and 7 in the TM domain as the binding pocket for Repertaxin on CXCR1. Computational docking data of active and inactive analogs of Repertaxin are in keeping with a model in which the engagement of specific interhelical polar interactions accounts for the general inhibitory property of the chemical class, whereas hydrophobic interactions play a crucial role in determining the affinity at the binding site and the potency of the inhibitor. Crucial residues in the Repertaxin binding site are highly conserved in rat and mouse homologue receptors, thus justifying the efficacy of Repertaxin in rodent animal models.
Agonist activation of GPCR induces conformational changes that are, as yet, poorly understood but which seem to involve, at a minimum, rearrangements of membrane helices 6 and 3 (23). Because CXCL8 receptor multimerization and binding to G protein are unaffected by Repertaxin (see Supporting Text and Fig. 7, which is published as supporting information on the PNAS web site), one likely interpretation of the proposed interaction model is that binding of Repertaxin to the CXCR1 TM domain induces a conformational constrain among the TM domains of helices 1, 3, and 6 of CXCR1, thus preventing the rearrangement of helices 6 and 3, which is required for downstream signal transduction. The binding sites of chemokine receptors for small-molecule or peptide-based inhibitors acting as classical receptor antagonists have usually been localized to the extracellular domain acting as classical ligand antagonists (8, 21, 24, 25). A binding pocket for a small-molecule inhibitor of HIV-1 entry has been identified within the TM helices of CCR5 (26), and peptide mimics of the TM helices can disrupt receptor function and the ability of CXCR4 to mediate HIV-1 entry (27). Our data support allosteric sites in the TM domains of GPCR as targets for noncompetitive inhibitors that disrupt receptor function.
RI plays an important role under different pathological conditions, including most states of hypoperfusion, such as limb ischaemia, myocardial infarction, stroke, hypovolemic shock, and transplantation. RI, also referred to as delayed graft function (28), is unavoidable in organ transplantation, occurring during organ retrieval and storage. Organ transplant RI affects ≈20–30% of cadaver-donor kidneys, and its rate may rise as more organs are removed from “marginal” or “extended” donors (29–31). Repertaxin has proven safe and well tolerated in different animal studies and in phase I studies in human volunteers (data not shown). It is therefore a candidate therapeutic agent for the prevention and treatment of RI and, in particular, of delayed graft function.
Repertaxin is a noncompetitive allosteric blocker of the CXCL8 receptors CXCR1 and CXCR2. The mode of interaction of Repertaxin with CXCR1 reported here suggests that allosteric sites in the TM domains of GPCRs could represent valuable targets for noncompetitive inhibitors that disrupt receptor signaling. The significance of the present study may well extend beyond CXCR1 and related chemokine receptors. Indeed, by using the conceptual platform generated by the molecular mechanism of interaction between Repertaxin and CXCR1/2 described here, we have now successfully generated potent and specific inhibitors for other chemoattractant receptors. Therefore, based on the results and model described here, targeting the Repertaxin binding site of CXCR1 represents a general strategy to modulate the activity of chemoattractant GPCR.
Acknowledgments
We thank A. Marchegiani and A. Gismondi for technical assistance. This work was supported by the Ministero dell'Istruzione, dell'Università, e della Ricerca; the Istituto Superiore di Sanità; the Consiglio Nazionale delle Ricerche; and the Italian Association for Cancer Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CLP, cecal ligation puncture; COX, cyclooxygenase; GPCR, G protein-coupled receptors; RI, reperfusion injury; CXCL8, CXC ligand 8; CXCR1/2, CXC receptors 1 and 2; PMN, polymorphonuclear cell; fMLP, N-formyl-l-methionyl-l-leucyl-l-phenylalanine; TM, transmembrane; CCR, CC chemokine receptor.
References
- 1.Mantovani, A. (1999) Immunol. Today 20, 254–257. [DOI] [PubMed] [Google Scholar]
- 2.Rollins, B. J. (1997) Blood 90, 909–928. [PubMed] [Google Scholar]
- 3.Gerard, C. & Rollins, B. J. (2001) Nat. Immunol. 2, 108–115. [DOI] [PubMed] [Google Scholar]
- 4.Howard, O. M., Oppenheim, J. J. & Wang, J. M. (1999) J. Clin. Immunol. 19, 280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy, P. M., Baggiolini, M., Charo, I. F., Hebert, C. A., Horuk, R., Matsushima, K., Miller, L. H., Oppenheim, J. J. & Power, C. A. (2000) Pharmacol. Rev. 52, 145–176. [PubMed] [Google Scholar]
- 6.Proudfoot, A. E. (2002) Nat. Rev. Immunol. 2, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horuk, R. & Ng, H. P. (2000) Med. Res. Rev. 20, 155–168. [DOI] [PubMed] [Google Scholar]
- 8.White, J. R., Lee, J. M., Dede, K., Imburgia, C. S., Jurewicz, A. J., Chan, G., Fornwald, J. A., Dhanak, D., Christmann, L. T., Darcy, M. G., et al. (2000) J. Biol. Chem. 275, 36626–36631. [DOI] [PubMed] [Google Scholar]
- 9.Bizzarri, C., Allegretti, M., Di Bitondo, R., Cervellera, M. N., Colotta, F. & Bertini, R. (2003) Curr. Med. Chem. 2, 67–79. [DOI] [PubMed] [Google Scholar]
- 10.Imai, T., Chantry, D., Raport, C. J., Wood, C. L., Nishimura, M., Godiska, R., Yoshie, O. & Gray, P. W. (1998) J. Biol. Chem. 273, 1764–1768. [DOI] [PubMed] [Google Scholar]
- 11.Bizzarri, C., Pagliei, S., Brandolini, L., Mascagni, P., Caselli, G., Transidico, P., Sozzani, S. & Bertini, R. (2001) Biochem. Pharmacol. 61, 1429–1437. [DOI] [PubMed] [Google Scholar]
- 12.Munson, P. J. & Rodbard, D. (1980) Anal. Biochem. 107, 220–239. [DOI] [PubMed] [Google Scholar]
- 13.Locati, M., Deuschle, U., Massardi, M. L., Martinez, F. O., Sironi, M., Sozzani, S., Bartfai, T. & Mantovani, A. (2002) J. Immunol. 168, 3557–3562. [DOI] [PubMed] [Google Scholar]
- 14.Villa, P., Meazza, C., Sironi, M., Bianchi, M., Ulrich, P., Bothkina, G., Tracey, K. J. & Ghezzi, P. (1997) J. Endotoxin Res. 4, 197–204. [Google Scholar]
- 15.Cutrin, J. C., Boveris, A., Zingaro, B., Corvetti, G. & Poli, G. (2000) Hepatology 31, 622–632. [DOI] [PubMed] [Google Scholar]
- 16.Cavalieri, B., Perrelli, M. G., Aragno, M., Mastrocola, R., Corvetti, G., Durazzo, M., Poli, G. & Cutrin, J. C. (2002) Liver Transplant. 8, 990–999. [DOI] [PubMed] [Google Scholar]
- 17.Moloney, W. C., McPherson, K. & Fliegelman, L. (1960) J. Histochem. Cytochem. 8, 200–207. [DOI] [PubMed] [Google Scholar]
- 18.Selinsky, B. S., Gupta, K., Sharkey, C. T. & Loll, P. J. (2001) Biochemistry 40, 5172–5180. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, M. E., Lapointe, G. R., Feucht, P. H., Hilt, S., Gallegos, C. A., Gordon, C. A., Giedlin, M. A., Mullenbach, G. & Tekamp-Olson, P. (1995) J. Immunol. 155, 1428–1433. [PubMed] [Google Scholar]
- 20.Christopoulos, A. & Kenakin, T. (2002) Pharmacol. Rev. 54, 323–374. [DOI] [PubMed] [Google Scholar]
- 21.Sabroe, I., Peck, M. J., Van Keulen, B. J., Jorritsma, A., Simmons, G., Clapham, P. R., Williams, T. J. & Pease, J. E. (2000) J. Biol. Chem. 275, 25985–25992. [DOI] [PubMed] [Google Scholar]
- 22.Chuang, V. T., Kuniyasu, A., Nakayama, H., Matsushita, Y., Hirono, S. & Otagiri, M. (1999) Biochim. Biophys. Acta. 1434, 18–30. [DOI] [PubMed] [Google Scholar]
- 23.Meng, E. C. & Bourne, H. R. (2001) Trends Pharmacol. Sci. 22, 587–593. [DOI] [PubMed] [Google Scholar]
- 24.Podolin, P. L., Bolognese, B. J., Foley, J. J., Schmidt, D. B., Buckley, P. T., Widdowson, K. L., Jin, Q., White, J. R., Lee, J. M., Goodman, R. B., et al. (2002) J. Immunol. 169, 6435–6444. [DOI] [PubMed] [Google Scholar]
- 25.White, J. R., Lee, J. M., Young, P. R., Hertzberg, R. P., Jurewicz, A. J., Chaikin, M. A., Widdowson, K., Foley, J. J., Martin, L. D., Griswold, D. E. & Sarau, H. M. (1998) J. Biol. Chem. 273, 10095–10098. [DOI] [PubMed] [Google Scholar]
- 26.Dragic, T., Trkola, A., Thompson, D. A., Cormier, E. G., Kajumo, F. A., Maxwell, E., Lin, S. W., Ying, W., Smith, S. O., Sakmar, T. P. & Moore, J. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5639–5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarasova, N. I., Rice, W. G. & Michejda, C. J. (1999) J. Biol. Chem. 274, 34911–34915. [DOI] [PubMed] [Google Scholar]
- 28.Shoskes, A. D., Shahed, A. R. & Kim, S. (2001) Renal Vasc. Dis. Transplant. 28, 721–732. [Google Scholar]
- 29.Ojo, A. O., Wolfe, R. A., Held, P. J., Port, F. K. & Schmouder, R. L. (1997) Transplantation 63, 968–974. [DOI] [PubMed] [Google Scholar]
- 30.Ploeg, R. J., van Bockel, J. H., Langendijk, P. T., Groenewegen, M., van der Woude, F. J., Persijn, G. G., Thorogood, J. & Hermans, J. (1992) Lancet 340, 129–137. [DOI] [PubMed] [Google Scholar]
- 31.Tilney, N. L., Paz, D., Ames, J., Gasser, M., Laskowski, I. & Hancock, W. W. (2001) Transplant. Proc. 33, 843–844. [DOI] [PubMed] [Google Scholar]