ABSTRACT
Multiple novel members of the genus Hepacivirus have recently been discovered in diverse mammalian species. However, to date, their replication mechanisms and zoonotic potential have not been explored in detail. The NS3/4A serine protease of hepatitis C virus (HCV) is critical for cleavage of the viral polyprotein. It also cleaves the cellular innate immune adaptor MAVS, thus decreasing interferon (IFN) production and contributing to HCV persistence in the human host. To investigate the conservation of fundamental aspects of the hepaciviral life cycle, we explored if MAVS cleavage and suppression of innate immune signaling represent a common mechanism employed across different clades of the genus Hepacivirus to enhance viral replication. To estimate the zoonotic potential of these nonhuman hepaciviruses, we assessed if their NS3/4A proteases were capable of cleaving human MAVS. NS3/4A proteases of viruses infecting colobus monkeys, rodents, horses, and cows cleaved the MAVS proteins of their cognate hosts and interfered with the ability of MAVS to induce the IFN-β promoter. All NS3/4A proteases from nonhuman viruses readily cleaved human MAVS. Thus, NS3/4A-dependent cleavage of MAVS is a conserved replication strategy across multiple clades within the genus Hepacivirus. Human MAVS is susceptible to cleavage by these nonhuman viral proteases, indicating that it does not pose a barrier for zoonotic transmission of these viruses to humans.
IMPORTANCE Virus infection is recognized by cellular sensor proteins triggering innate immune signaling and antiviral defenses. While viruses have evolved strategies to thwart these antiviral programs in their cognate host species, these evasion mechanisms are often ineffective in a novel host, thus limiting viral transmission across species. HCV, the best-characterized member of the genus Hepacivirus within the family Flaviviridae, uses its NS3/4A protease to disrupt innate immune signaling by cleaving the cellular adaptor protein MAVS. Recently, a large number of HCV-related viruses have been discovered in various animal species, including wild, livestock, and companion animals. We show that the NS3/4A proteases of these hepaciviruses from different animals and representing various clades of the genus cleave their cognate host MAVS proteins in addition to human MAVS. Therefore, cleavage of MAVS is a common strategy of hepaciviruses, and human MAVS is likely unable to limit replication of these nonhuman viruses upon zoonotic exposure.
INTRODUCTION
Viral zoonotic infections are responsible for numerous emerging infectious diseases (1, 2). Although biological infection barriers (e.g., viral host factor usage and host restriction factors) can limit viral cross-species transmission, the recent epidemics of Ebola virus, Zika virus, and Middle East respiratory syndrome coronavirus (MERS-CoV) in West Africa, Latin America, and the Middle East, respectively, highlight that RNA viruses frequently overcome these barriers, spread to humans, and cause severe disease. Human contacts with wild, livestock, and companion animals, combined with the increasing density of human populations, rising mobility, and climatic changes, facilitate increased human exposure to novel viral pathogens and subsequent spread within human populations. Thus, understanding the biological barriers of viral cross-species transmission has high priority for estimating the risks of viral transmission to humans and enabling development of preventive and/or therapeutic measures. Moreover, detailed information about species-specific viral replication barriers facilitates the development of animal models for human pathogens, for example, hepatitis C virus (HCV).
HCV is a plus-strand RNA virus and a major human pathogen causing chronic liver disease in more than 146 million individuals worldwide (3). It possesses a 9.6-kb genome encoding a polyprotein consisting of 10 viral proteins, including the structural proteins core, E1, and E2, the p7 ion channel protein, and the nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. It is parenterally transmitted among humans primarily through needle sharing, unsafe medical procedures, and use of contaminated medical equipment. Although HCV is generally considered to have a narrow species tropism, naturally infecting only humans (4), chimpanzees are susceptible to experimental infection. HCV was discovered more than 25 years ago (5) and initially recognized as the sole member of the genus Hepacivirus within the family Flaviviridae. Subsequently, numerous additional viruses were discovered and assigned to the genus Hepacivirus, including GB virus (GBV; previously known as GBV-B) (6), with an unknown natural host species, and a large number of HCV-related viruses with different mammalian host species, including wild rodents and bats (7, 8), colobus monkeys, and domesticated animals living in close contact with humans (cattle, dogs, and horses) (9–13). In parallel, several new viruses were discovered and assigned to the related genus Pegivirus within the Flaviviridae. These viruses include simian pegivirus (SPgV; formerly known as GBV-A), which naturally infects New World monkeys, the nonpathogenic human pegivirus virus (HPgV; formerly known as GBV-C), and bat pegivirus (BPgV; formerly known as GBV-D) (6). More recently, a second human pegivirus, tentatively named HPgV-2, was discovered in association with HCV coinfection, and a human hepegivirus that shares features of hepaci- and pegiviruses was found in blood transfusion recipients (7, 14). The pathogenic potential of the last two viruses is currently unknown. Finally, more than 80 different hepaci- and pegiviruses were identified in bats, highlighting the extraordinary diversity of these viruses (8). Nevertheless, the evolutionary origin of HCV in humans remains unknown. Moreover, the potential for cross-species transmission of these novel viruses between animals and to humans is also unknown. In addition, whether key replication and immune evasion mechanisms are conserved across the genus Hepacivirus remains unexplored.
To successfully infect a new host, zoonotic viruses must not only cross the entry barrier but also subvert the innate immune response raised upon sensing of viral danger signals (15, 16). In the case of HCV, the viral NS3/4A serine protease can disrupt innate immune sensing by cleaving human mitochondrial antiviral signaling protein (MAVS) (17) and human TIR domain-containing adaptor-inducing beta interferon (TRIF) (18), two critical adaptor proteins that link recognition of viral double-stranded RNA with interferon (IFN) induction. Intriguingly, MAVS has been under strong positive selection pressure during primate evolution and acquired resistance to HCV protease cleavage in three independent primate species (19). Moreover, hepaciviral GBV and pegiviral SPgV, HPgV, and BPgV proteases also cleave MAVS, indicating that primates have likely been under constant exposure to ancient hepaci- and/or pegiviruses (19). Note that expression of a poorly cleavable, nonhuman MAVS variant limits HCV replication, indicating that resistance of MAVS to hepaciviral protease cleavage is a determinant of cross-species transmission (19, 20). In this regard, it is worth mentioning that the NS3/4A protease of the HCV-related nonprimate hepacivirus (NPHV; formerly known as canine hepacivirus [CHV]) cleaves human MAVS (19, 21, 22), suggesting that, in principle, this virus which circulates among horses may be able to overcome the species barrier imposed by MAVS upon transmission to humans. At the outset of this investigation, it was unclear if hepaciviruses from nonhuman hosts are typically capable of cleaving human MAVS. Moreover, it was uncertain if present-day hepaciviruses cleave their cognate host MAVS proteins. We investigated both these issues in the present study.
MATERIALS AND METHODS
Sequence acquisition and alignment.
Representative hepaciviral genome sequences and their cognate host MAVS sequences were downloaded from GenBank (Tables 1 and 2). In cases where cognate host MAVS sequences were not available (Otomops martiensseni, Hipposideros vittatus, Myodes glareolus, Peromyscus maniculatus, and Rhabdomys pumilio), MAVS sequences from the most closely related species deposited in GenBank were utilized as surrogates (Mus musculus, Eptesicus fuscus, and Myotis brandtii). Nucleotide sequences were trimmed and translated and amino acid sequences were aligned using the ClustalW tool in MEGA5. Amino acid conservation plots for NS3/4A and MAVS were calculated using a 10-amino-acid sliding window in CLC Genomics Workbench v8.1.
TABLE 1.
Representative hepaciviral isolates
| Virus name (abbreviation) | Isolate | Predicted NS3/4A length (no. of amino acids) | Species origin | Accession no. |
|---|---|---|---|---|
| Hepatitis C virus (HCV) | JFH-1 (2A) | 685 | Homo sapiens | AB047639 |
| Nonprimate hepacivirus (NPHV)/canine hepacivirus (CHV) | H10K | 685 | Equus caballus/Canis lupus | KP640276 |
| Bat hepacivirus C (BHV-C) | PDB-452 | 683 | Otomops martiensseni | KC796090 |
| BHV-D | PDB-829 | 684 | Hipposideros vittatus | KC796074 |
| Rodent hepacivirus 1 (RHV-1) | NLR08-365 | 683 | Myodes glareolus | KC411796 |
| RHV | RHV-339 | 679 | Peromyscus maniculatus | KC815310 |
| RHV-3 | SAR-46 | 678 | Rhabdomys pumilio | KC411807 |
| Guereza hepacivirus (GHV) | BWC08 | 686 | Colobus guereza | KC551800 |
| Cattle hepacivirus (Cattle-HV) | GHC25 | 678 | Bos taurus | KP265943 |
TABLE 2.
Mammalian MAVS proteins used in this study
| Species origin of MAVS | Abbreviation | Predicted MAVS length (no. of amino acids) | Accession no. |
|---|---|---|---|
| Homo sapiens (human) | Hu | 540 | NM020746 |
| Macaca mulatta (rhesus macaque) | Rhe | 541 | KC415016 |
| Colobus guereza (colobus monkey) | Gue | 535 | KC415019 |
| Mus musculus (mouse) | Mu | 503 | NM001206385 |
| Canis lupus (dog) | Can | 520 | NM001122609 |
| Bos taurus (cattle) | Bos | 520 | NM001046620 |
| Equus caballus (horse) | Equ | 530 | XM001496561 |
| Eptesicus fuscus (big brown bat) | Fus | 520 | XM005874684 |
| Myotis brandtii (Brandt's bat) | Brn | 525 | XM005874684 |
Molecular phylogenetic analysis.
The evolutionary history of hepaciviral NS3/4A and mammalian MAVS nucleotide sequences was inferred using the maximum likelihood method implemented in MEGA5, based on the data-specific model (23). To assess the significance of clades, the bootstrap approach was employed, whereby 1,000 pseudoreplicate trees were generated. Grouping of sequences was considered significant if the percentage of trees in which the associated taxa clustered together was >70%. For each phylogeny generated, the tree with the highest log likelihood is presented, with significant bootstrap values assigned to the corresponding tree nodes. Trees were generated under a GTR + Γ + I model of substitution: a discrete gamma distribution (Γ) was used to model evolutionary rate differences among sites (4 rate categories), and the proportion of invariant sites (I) was also incorporated into the tree-building process. The trees were drawn to scale, with branch lengths proportional to the number of substitutions per site. All positions containing gaps and missing data were eliminated from the analysis. For hepaciviral NS3/4A, the analysis incorporated nucleotide sequences from nine viral isolates, and a total of 1,950 nucleotide positions were included in the final analysis. For MAVS, the analysis involved nucleotide sequences from nine mammalian species, and a total of 1,416 nucleotide positions were included in the final analysis.
Cell culture.
HEK 293T wild-type (WT), HEK 293T-MAVS−/− (24), and Huh7-Lunet hCD81 (25) cells were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum (FCS) at 37°C and with 5% CO2.
Plasmid construction.
Lentiviral pWPI plasmids expressing either WT or modified NS3/4A or MAVS variants tagged with either a tandem N-terminal hemagglutinin (HA) tag (2×HA tag) (pWPI-HA) or a triple N-terminal FLAG tag (3×FLAG tag) (pWPI-FLAG) were created using either PCR-based cloning strategies or synthesized gene fragments (gBlocks; IDT). To generate protease-inactive mutants, PCR-based site-directed mutagenesis (S139A) was performed on our panel of hepaciviral NS3/4A proteases to abrogate the catalytic activity of the proteases. pWPI vectors expressing either human (Hu) or mouse (Mus musculus; Mu) MAVS were created previously (24). Rhesus macaque (Macaca mulatta; Rhe) and other nonhuman MAVS proteins were generated by PCR, using total cDNA from primary macaque hepatocytes or gBlocks as the PCR template. Each amplicon was then cloned into pWPI-FLAG. PCR-based site-directed mutagenesis (C508R) was also performed to generate a protease-resistant human MAVS. To create red fluorescent protein (RFP)-based reporter constructs, the lentivirus-based plasmid pTRIP-RFP-NLS-Hu MAVS (kindly provided by Charles Rice) was modified by using the Gibson assembly cloning method (NEB) to replace the gene fragment encoding C-terminal Hu MAVS with gBlocks encoding the C-terminal MAVS fragment from each of the indicated nonhuman mammals. More detailed cloning strategies are available upon request. The IFN-β promoter reporter plasmid (pGL3b-IFN-β promoter-firefly luciferase) was kindly provided by Stefan Lienenklaus (Institute for Laboratory Animal Science, Hanover Medical School, Hanover, Germany).
Generation of lentiviral PP.
Lentiviral pseudotype particles (PP) with envelope glycoproteins from vesicular stomatitis virus (VSV-G) were created as previously described (24). In short, HEK 293T WT cells were transfected with the envelope glycoprotein expression construct pcz-VSV-G, the lentiviral Gag-Pol expression construct pCMVΔR.74, and a lentiviral genomic backbone (pWPI or pTRIP) with the desired gene expression by using the polyethylenimine (PEI) transfection method (Sigma-Aldrich). The medium was changed 24 h later, and the pseudoparticles were harvested, filtered, and used directly for gene transduction on the next day.
Transfection and IFN-β promoter reporter assay.
To analyze protein expression, HEK 293T WT or HEK 293T-MAVS−/− cells were transfected with a combination of 100 ng (or as indicated in the figure legends) plasmid DNA expressing the indicated NS3/4A protease plus 100 ng plasmid DNA expressing full-length MAVS or the RFP-based reporter by use of Lipofectamine 2000 (Invitrogen) and then seeded into a poly-l-lysine (Sigma-Aldrich)-coated 24-well plate. The cells were then lysed 48 h later prior to Western blotting. For IFN-β promoter-firefly luciferase reporter assays, 100 ng IFN-β promoter reporter plasmid was transfected into HEK 293T-MAVS−/− cells in a 24-well plate together with plasmid DNAs expressing the indicated protease and MAVS. For protease inhibitor assays, drugs (telaprevir or boceprevir) were given at 24 h posttransfection. At 48 h posttransfection, cells were lysed in 350 μl passive lysis buffer (Promega), and the firefly luciferase activity was measured. Results are shown as values normalized against each indicated MAVS challenged by mock plasmid DNA. Telaprevir and boceprevir were kind gifts from Marc Windisch (Institute Pasteur Korea, Seongnam, South Korea).
Western blotting.
Cell monolayers were lysed in RIPA buffer (0.3 M NaCl, 20 mM Tris-HCl [pH 8], 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1% Triton X-100). Each sample was mixed with 5× denaturing protein sample buffer (200 mM Tris-HCl [pH 8.8], 5 mM EDTA, 0.1% bromophenol blue, 10% sucrose, 3.3% SDS, 2% 2-mercaptoethanol [2-ME]), heated for 5 min at 95°C, loaded onto an SDS-PAGE gel, and resolved by electrophoresis. Subsequently, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare) which was then treated with SuperSignal Western blot enhancer (Thermo) followed by blocking with 5% milk in phosphate-buffered saline containing 0.5% Tween (PBS-T) for 1 h at room temperature (RT). The membrane was then incubated with either anti-HA-tag monoclonal antibody (MAb) (3.3 µg/ml) (16B12; BioLegends), anti-FLAG-tag MAb (2 µg/ml) (M2; Sigma-Aldrich), anti-Hu MAVS MAb (0.6 µg/ml) (E-3; Santa Cruz), anti-Mu MAVS MAb (0.6 µg/ml) (E-6; Santa Cruz), anti-TurboRFP MAb (1 µg/ml) (Evrogen), or anti-β-actin MAb (0.3 µg/ml) (Sigma-Aldrich), followed by incubation with a secondary antibody coupled to horseradish peroxidase (HRP) (Sigma-Aldrich). Bound antibodies were detected with an ECL Plus detection system (GE Healthcare) after mixing with SuperSignal Femto substrate (Thermo) at a 10:1 ratio.
Immunofluorescence and confocal analyses.
Huh7-Lunet hCD81 cells (8 × 104 cells/well) were seeded on coverslips in a 24-well plate followed by overnight transduction with lentiviral pseudoparticles expressing the indicated genes in the presence of 4 μg/ml Polybrene (Sigma-Aldrich). Forty-eight hours later, cells were fixed and permeabilized. Immunostaining was then performed using anti-HA-tag MAb (2 µg/ml) alone or in combination with anti-Hu MAVS MAb (2 µg/ml) or anti-Hu calnexin MAb (2 µg/ml) (Abcam), as indicated, followed by incubation with a secondary antibody coupled to either Alexa 488 (green; Sigma-Aldrich) or Alexa 647 (red; Sigma-Aldrich). Nuclear DNA was stained using DAPI (4′,6-diamidino-2-phenylindole) at a dilution of 1.6 µg/ml. Coverslips were then mounted with Fluoromount-G (Southern Biotech) and processed for confocal analysis using a laser scanning confocal microscope (Olympus). A ×100 lens was used for all pictures. ImageJ software was used to create overlaid pictures.
Statistical methods.
GraphPad Prism 6 software was used for data analysis, using one-way analysis of variance (ANOVA) adjusted with Sidak's multiple-comparison test. P values of <0.05 (*) were considered statistically significant, whereas P values of <0.01 (**), <0.001 (***), and <0.0001 (****) were considered highly significant.
RESULTS
Amino acid diversity, evolutionary relationships, and conserved functional motifs of the predicted hepaciviral NS3/4A and mammalian MAVS proteins.
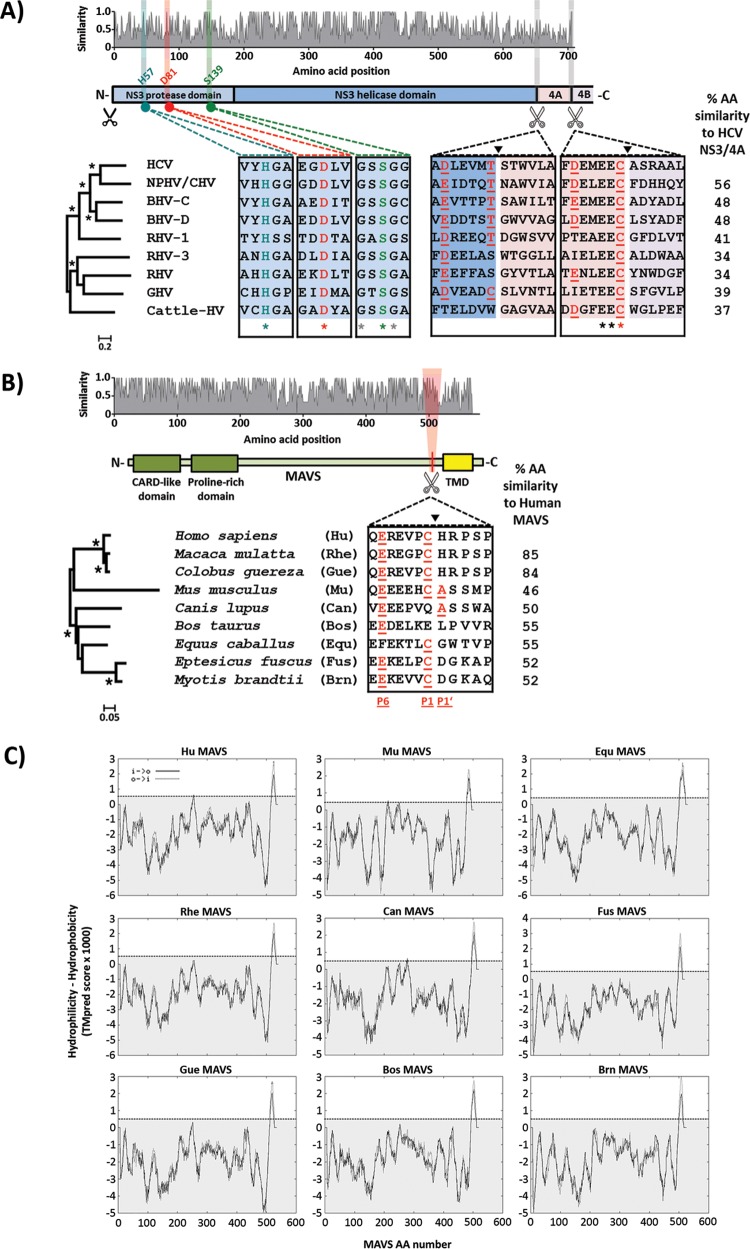
We investigated the properties of hepaciviral NS3/4A proteases by selecting eight isolates from each distinct clade within the genus Hepacivirus, representing viruses isolated from horses/dogs (26), rodents (7, 27), bats (8), nonhuman primates (13), and cattle (9, 11) (Table 1). An amino acid similarity plot (Fig. 1A, top panel) revealed that the helicase domain of NS3 was more conserved than the protease domain and the NS4A transmembrane cofactor. Nevertheless, the catalytic triad of the NS3 protease was completely conserved among all hepaciviruses, suggesting that the encoded proteins possess serine protease activity (Fig. 1A, bottom panels). Interestingly, although the predicted protease cleavage site at the NS3 C terminus was not fully conserved, the conserved EEC sequence clearly marked the NS4A terminus in all hepaciviral isolates analyzed (Fig. 1A, bottom right panel). Phylogenetic analysis of NS3/4A nucleotide sequences revealed a well-supported clade containing HCV, NPHV/CHV, bat hepacivirus C (BHV-C), BHV-D, and rodent hepacivirus 1 (RHV-1), with significant internal branch lengths: the isolates within this cluster are more closely related to HCV (Fig. 1A, bottom left panel). The remainder of the tree was characterized by long internal branches and nonsignificant bootstrap values, suggesting that these isolates are genetically divergent from HCV and from each other. These results support previous reports that NPHV/CHV NS3/4A is the hepaciviral protease most closely related to that of HCV. Indeed, the amino acid similarity to HCV NS3/4A was greatest for NPHV/CHV (56%) and as low as 37% for cattle hepacivirus (cattle-HV) (Fig. 1A).
FIG 1.
Amino acid diversity, evolutionary relationships, and conserved functional motifs of hepaciviral NS3/4A and mammalian MAVS proteins. (A) (Top) Amino acid similarity plot for nine full-length hepaciviral NS3/4A proteins, with relative similarities shown on the y axis and amino acid positions in the encoded proteins presented on the x axis. (Middle) For the purpose of positional referencing, a cartoon of the NS3/4A protein is provided, with the protease and helicase domains of NS3 colored light and dark blue, respectively. The NS4A protein and the N terminus of NS4B are colored pink and gray, respectively. Black scissors positioned below the cartoon represent the NS2 cleavage site, while white scissors represent putative substrates for cleavage by the NS3 protease. (Bottom) Magnifications below the cartoon depict amino acid conservation of the NS3 protease catalytic triad and adjacent residues (three alignment blocks on the left; turquoise for amino acid H57, red for D81, and green for S139) and of putative cleavage substrates for NS3/4A (two alignment blocks on the right; NS3/4A junction and NS4A/4B junction) in the representative hepaciviral proteins. Red underlined amino acid residues show the common consensus cleavage sequence for HCV NS3/4A. A phylogenetic tree depicting the evolutionary relationships of the included hepaciviral NS3/4A sequences is positioned to the left of the alignments, with significant groupings depicted by asterisks. Percent amino acid (AA) similarities of nonhuman hepaciviral NS3/4A proteins and HCV NS3/4A are indicated to the right. (B) (Top) Amino acid similarity plot for nine mammalian MAVS proteins, with relative similarities shown on the y axis and amino acid positions in the encoded proteins presented on the x axis. (Middle) For the purpose of positional referencing, a cartoon of the MAVS protein is presented, with the CARD-like and proline-rich domains colored green and the transmembrane domain (TMD) colored yellow. White scissors positioned below the cartoon represent putative substrate recognition sites for cleavage by hepaciviral NS3/4A proteases. (Bottom) Magnification depicting amino acid conservation at the putative cleavage site and adjacent residues in the representative mammalian MAVS proteins. Red underlined amino acid residues highlight residues that conform to the classical HCV NS3/4A consensus cleavage site. A phylogenetic tree depicting the evolutionary relationships of the included mammalian MAVS sequences is positioned to the left of the magnified alignment, with significant groupings depicted by asterisks. Percent amino acid similarities of mammalian MAVS proteins and human MAVS are located to the right. (C) Transmembrane domain prediction for the nine mammalian MAVS proteins. The hydrophobicity-hydrophilicity analysis was performed using the TMpred server (http://www.ch.embnet.org/software/TMPRED_form.html). The listing gives both inside-to-outside (i → o) and outside-to-inside (o → i) transmembrane helix orientations. A score of >500 (dotted line) predicts a highly hydrophobic transmembrane region.
To explore if MAVS antagonism through NS3/4A protease-dependent cleavage is a conserved strategy among all hepaciviruses, we cloned MAVS variants originating from species harboring the above-mentioned viruses. As NPHV/CHV was initially found in a dog (Canis lupus; Can) and subsequently shown to be circulating among horses (Equus caballus; Equ), the MAVS proteins from both species were included. However, since some viruses included in this study were isolated from poorly characterized species within the very large orders Rodentia and Chiroptera (Table 1), for some of the host species there was no MAVS sequence information deposited in sequence databases. Consequently, we used the sequences for the closest available species as substitutes: mouse (Mus musculus; Mu) MAVS was used as a surrogate for rodents, and big brown bat (Eptesicus fuscus; Fus) and Brandt's bat (Myotis brandtii; Brn) MAVS variants were used as surrogates for bat species (Table 2). Rhesus macaque (Macaca mulatta; Rhe) MAVS was included to provide an example of a MAVS variant that is resistant to hepaciviral protease (HCV NS3/4A) cleavage (19). Comparison of the viral NS3/4A and host species MAVS phylogenies (Fig. 1A and B) revealed a striking lack of congruence between tree topologies: for instance, the colobus monkey (Colobus guereza; Gue), which is closely related to humans, hosts a virus (guereza hepacivirus [GHV]) only distantly related to HCV, whereas NPHV/CHV, the virus most closely related to HCV, infects horses, which are more distantly related to humans than primates are. These data indicate that hepaciviruses did not cospeciate with their cognate hosts from a single ancestral virus and that multiple cross-species transmissions characterize the evolution of the hepaciviral genus. Overall, sequence similarity between MAVS proteins was most pronounced within the CARD domain, and similarities ranged from 85% (rhesus macaque) to 48% (mouse) amino acid conservation for comparisons of full-length proteins to human MAVS. Interestingly, the C-terminal portions of all MAVS proteins investigated comprise a conserved and highly hydrophobic region (Fig. 1B and 2C) which likely serves as a transmembrane domain (TMD) for anchoring in the mitochondrial membrane (28). Using the TMpred program (28), we confirmed the presence of a TMD for all MAVS proteins (Fig. 1C). Cleavage of human MAVS by the HCV NS3/4A protease is dependent on cysteine 508, located within a protein region partially matching the HCV consensus cleavage site (17, 29). Interestingly, the standard HCV NS3/4A protease consensus cleavage site, which includes E/D at position 6 (P6), T/C at P1, and A/S at P1′, is partially conserved among MAVS variants and is only fully matched by the mouse MAVS protein (Fig. 1B) (30–32). Even with less stringent and more accurate cleavage prediction rules, dog, horse, and cattle (Bos taurus; Bos) MAVS proteins are not predicted to be cleaved (32). Thus, MAVS polymorphisms may affect hepacivirus cleavage in a species-specific fashion and potentially contribute to species tropism.
FIG 2.
Expression of predicted hepaciviral NS3/4A proteases and of mammalian MAVS transmembrane domains (TMDs). (A) Diagram illustrating the N-terminally HA-tagged hepaciviral NS3/4A constructs. (B) Detection of HA-NS3 expression in Huh7-Lunet hCD81 cells transduced by lentiviral pseudoparticles (PP) harboring each distinct nonhuman hepaciviral NS3/4A construct. The expression of HA-tagged HCV NS3/4A (1st column) serves as a comparison control, and human calnexin (2nd row) serves as an ER marker. The images shown are representative of two individual experiments. (C) Diagram illustrating the RFP-NLS reporter constructs. The dotted lines indicate the MAVS C-terminal domain from each indicated MAVS species fused to the reporter C terminus. The red arrow and yellow box indicate the predicted cleavage site and hydrophobic TMD of MAVS, respectively, for each indicated species. (D) Expression profiles of reporter constructs harboring the C-terminal domain and TMD from the indicated species (2nd row) in comparison to the expression of endogenous Hu MAVS stained with an N-terminus-specific antibody (1st row) in Huh7-Lunet hCD81 cells. (E) Expression profiles of the indicated reporter constructs (2nd row) in the presence of HCV or nonhuman hepaciviral NS3/4A protease (1st row). Cleavage of the MAVS TMD by the viral NS3/4A proteases causes relocalization of the RFP-NLS reporter to the nuclei. Dotted white lines outline the HA-positive (NS3/4A-expressing) cells. The images shown are representative of two individual experiments.
Expression of representative hepaciviral NS3/4A proteins and mammalian MAVS TMDs.
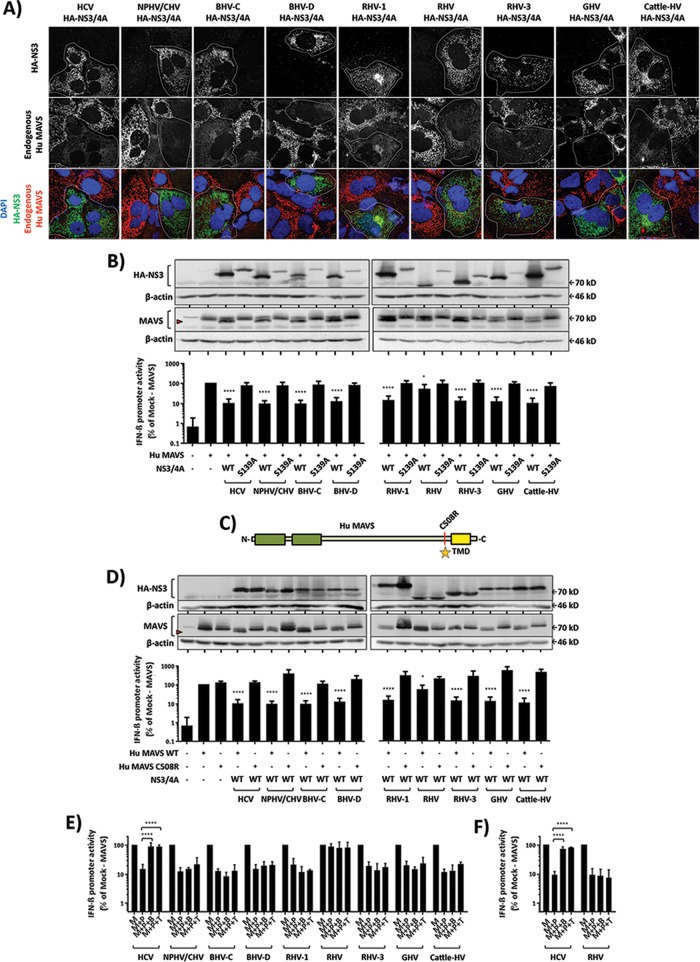
Lentiviral vectors expressing N-terminally HA-tagged NS3/4A proteins (Fig. 2A) from the above-mentioned hepaciviruses were transduced and subsequent expression and subcellular localization monitored in Huh7-Lunet hCD81 cells (Fig. 2B). All hepaciviral proteases expressed displayed cytoplasmic and reticular staining that largely overlapped with the endoplasmic reticulum (ER) marker protein calnexin and was comparable to the staining pattern of the HCV NS3/4A protease (Fig. 2B).
Moreover, we created expression constructs for fusion proteins between a red fluorescent protein carrying a nuclear localization signal (RFP-NLS) and the C-terminal portions of the above-mentioned MAVS variants, as described for human MAVS by Jones et al. (33). In the present study, the RFP-NLS reporter was fused with ±75 C-terminal amino acid residues from each indicated MAVS, including the putative hepaciviral protease recognition site and the predicted TMDs (Fig. 2C). In this context, cleavage of the fusion proteins at the predicted hepaciviral cleavage site will detach the RFP-NLS protein from the MAVS TMD and thus result in nuclear localization of the fluorescent protein. After transduction of Huh7-Lunet hCD81 cells with the expression constructs for the RFP-NLS-MAVS reporter proteins, strong colocalization between endogenous human MAVS and the respective fusion proteins was observed (Fig. 2D), indicating proper expression and subcellular localization at the mitochondrial surface.
Hepaciviral NS3/4A serine proteases cleave MAVS proteins of their cognate (or surrogate) host species.
To monitor cleavage of the MAVS fusion proteins by hepaciviral proteases, Huh7-Lunet hCD81 cells were simultaneously transduced with the indicated NS3/4A proteases and MAVS fusion proteins (Fig. 2E). As expected, cotransduction of these cells with the RFP-NLS human MAVS TMD construct and the HCV NS3/4A protease resulted in cleavage of the MAVS reporter protein, which was clearly evident by the nuclear localization of the RFP-NLS protein (Fig. 2E). In contrast, the RFP-NLS Rhe MAVS TMD construct, which carries a single amino acid difference compared to the human cleavage site (V506G) (19), was not cleaved by the HCV protease, as evidenced by reticular cytoplasmic staining of the RFP-NLS protein in cells coexpressing the HCV protease (Fig. 2E, 2nd column). Interestingly, the colobus monkey, horse, dog, and cattle MAVS C termini were found to be susceptible to cleavage by their cognate hepaciviral NS3/4A proteases. Similarly, the surrogate mouse, big brown bat, and Brandt's bat MAVS C termini were cleaved by the representative rodent and bat hepaciviral NS3/4A proteases (Fig. 2E).
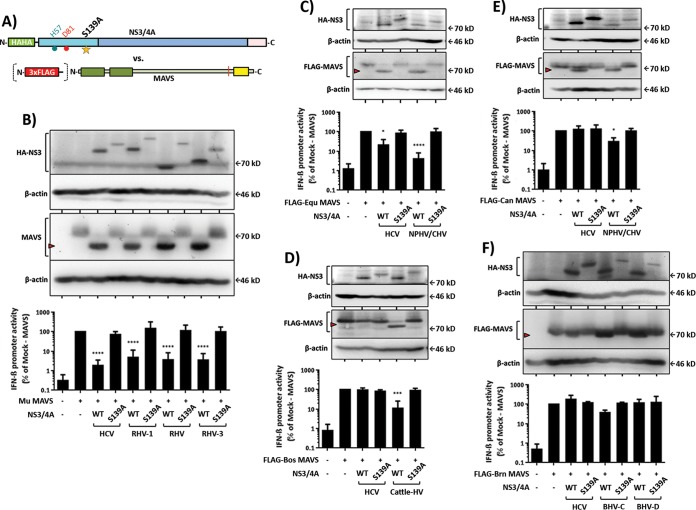
To confirm these observations, we also analyzed cleavage of full-length MAVS proteins by selected hepaciviral proteases (Fig. 3). To ensure the specificity of the cleavage, NS3/4A protease expression constructs carrying an inactivating mutation of the catalytic serine residue (S139A) were used in parallel (Fig. 3A). Since the big brown bat and Brandt's bat MAVS proteins are from bats within the same family, Brandt's bat MAVS was selected for further analysis, and since the predicted colobus monkey MAVS cleavage site matches the human counterpart, this construct was not analyzed further. An N-terminal triple FLAG tag (3×FLAG) was added to MAVS variants with no specific antibody available (horse, dog, cattle, and Brandt's bat MAVS proteins) (Fig. 3A). Subsequently, the indicated wild-type or mutant NS3/4A protease and MAVS expression plasmids were cotransfected into HEK 293T-MAVS−/− cells, which lack endogenous expression of human MAVS (24). To directly monitor the impact of MAVS cleavage on innate immune signaling, a reporter plasmid encoding a firefly luciferase under the control of the IFN-β promoter was cotransfected, and luciferase activity was monitored in the lysates of transfected cells. Interestingly, transient expression of either human or nonhuman MAVS in these HEK 293T-MAVS−/− cells triggered comparable downstream IFN-β promoter activity (as evidenced by similar luciferase activity) (Fig. 3). This indicates that each nonhuman MAVS protein is capable of innate immune signaling in human cells, which likely reflects the high conservation of the CARD-like domain among all MAVS variants. The coexpression of HCV and all rodent hepaciviral NS3/4A proteins resulted in efficient cleavage of and signaling interference with mouse MAVS, as evidenced by the detection of a truncated MAVS species and significantly reduced IFN-β promoter activity (Fig. 3B). Both cleavage and interference with signaling were specific to hepaciviral NS3/4A protease activity, since coexpression of protease variants with an inactive catalytic triad did not reduce MAVS signaling and ablated detection of a truncated MAVS variant. Notably, HCV NS3/4A cleaved and interfered with the signaling of horse MAVS (Fig. 3C), although the predicted cleavage site of this MAVS variant does not fully match the HCV cleavage site rules reported by Rögnvaldsson et al. (32). In contrast, no cleavage and signaling interference were observed in the case of cattle MAVS (Fig. 3E), and only modest cleavage in the absence of overt signaling interference was visible for dog MAVS (Fig. 3D). Both of these MAVS variants are not predicted to be cleaved by the HCV NS3/4A protease according to the Rögnvaldsson rules. Notably, these MAVS variants were susceptible to cleavage and signaling interference by their cognate hepaciviral NS3/4A proteases (NPHV [Fig. 3D] and cattle-HV [Fig. 3E]), which is in full agreement with the results of the RFP-NLS reporter assays depicted in Fig. 2. The Brandt's bat MAVS protein was resistant to the HCV NS3/4A protease (Fig. 3F), and only modest cleavage with weak or no signaling interference was observed for the BHV-C or BHV-D NS3/4A protease, respectively. Notably, the original BHV-C host belongs to the Molossidae family, which is very distantly related to the BHV-D host belonging to the Hipposideridae family. Moreover, the animal host of BHV-C is more closely related to the surrogate Brandt's bat family, Vespertilionidae, than the host of BHV-D, which may explain the relatively inefficient interference with Brandt's bat MAVS and the slightly better interference by the BHV-C NS3/4A protease (34).
FIG 3.
Effects of hepaciviral NS3/4A expression on nonhuman full-length MAVS expression and downstream IFN-β promoter induction. (A) Diagrams illustrating wild-type (WT) and S139A mutant NS3/4A proteases from hepaciviral isolates tested against FLAG-tagged or nontagged full-length MAVS from each indicated species. (B) Effect of NS3/4A from rodent hepacivirus against surrogate mouse (Mu) MAVS. (C and E) Effects of NS3/4A from horse/dog hepacivirus against horse (Equ) or dog (Can) MAVS. (D) Effect of NS3/4A from cattle hepacivirus against cattle (Bos) MAVS. (F) Effect of bat hepacivirus NS3/4A against surrogate Brand's bat (Brn) MAVS. The expression of HCV NS3/4A serves as a comparison control. Western blot (WB) analyses (upper panels; a red arrowhead indicates the cleaved MAVS species) and MAVS-dependent IFN-β promoter reporter assays (lower panels) were performed as described in Materials and Methods. WB images are representative of at least two individual experiments, and all graphical data presented are means ± standard deviations (SD) for at least three independent experiments.
Human MAVS-dependent innate immune signaling is susceptible to interference by nonhuman hepaciviral NS3/4A proteases.
To explore the potential for zoonotic transmission of nonhuman hepaciviruses to humans, we next explored the ability of the nonhuman hepaciviral NS3/4A proteases to cleave human MAVS. To this end, Huh7-Lunet hCD81 cells were transduced with the indicated full-length nonhuman hepaciviral NS3/4A protease expression constructs and subsequently stained for endogenous MAVS expression and ectopic NS3/4A expression (Fig. 4A). Interestingly, in all cells expressing hepaciviral protease, more diffuse or lower endogenous MAVS signals were observed, suggesting that all nonhuman NS3/4A proteases investigated at least partially cleaved human MAVS (Fig. 4A). To confirm this observation and to obtain more quantitative results on human MAVS cleavage and signaling interference by these nonhuman viral proteases, we next coexpressed human MAVS with the indicated proteases in the context of HEK 293T-MAVS−/− cells. Human MAVS was susceptible to cleavage and signaling interference by all hepaciviral NS3/4A proteases, and only the RHV isolate yielded a modest amount of interference (Fig. 4B). MAVS signaling interference could be ablated by addition of boceprevir or telaprevir in the case of HCV NS3/4A protease expression. However, MAVS interference by none of the other hepaciviral proteases was ablated by these protease inhibitors, indicating that these drugs do not interfere with these proteases (Fig. 4E and F). When cysteine 508 of human MAVS, which is critical for cleavage by HCV (17, 29), was mutated to arginine (Fig. 4C), human MAVS was resistant to cleavage by all tested nonhuman hepaciviral NS3/4A proteases (Fig. 4D), indicating that these viral proteases cleave human MAVS at the same site as the HCV protease. Taken together, these results indicate that the NS3/4A serine proteases of nonhuman hepaciviruses examined here not only cleave MAVS proteins of their cognate/surrogate animal hosts but also cleave and interfere with signaling of human MAVS (Fig. 5).
FIG 4.
Nonhuman hepaciviral NS3/4A proteases cleave and inhibit signaling of human MAVS. (A) Localization and expression of endogenous Hu MAVS in Huh7-Lunet hCD81 cells are influenced by the presence of a nonhuman hepaciviral NS3/4A protease. The cells were transduced with lentiviral pseudoparticles (PP) harboring the indicated HA-tagged NS3/4A constructs. The expression of HCV NS3/4A (1st column) serves as a comparison control. Dotted white lines outline HA-positive (NS3/4A-expressing) cells. The confocal immunofluorescence analysis was conducted using HA-tag- and Hu MAVS-specific antibodies. (B) Effects of wild-type (WT) and S139A mutant nonhuman hepaciviral NS3/4A proteases on Hu MAVS expression (upper panels) and downstream MAVS-dependent signaling to the IFN-β promoter (lower panels). HEK 293T MAVS−/− cells were transfected with the indicated expression constructs or mock transfected. Subsequently, protein expression was monitored by Western blotting using HA-, actin-, and Hu MAVS-specific antibodies. IFN-β promoter activity was quantified using luciferase assays. (C) Diagram illustrating wild-type or C508R mutant human MAVS. (D) Effects of wild-type (WT) nonhuman hepaciviral NS3/4A proteases on wild-type or C508R mutant Hu MAVS expression (upper panels) and downstream MAVS-dependent IFN-β promoter activity (lower panels). HCV NS3/4A expression serves as a comparison control. Red arrowheads in WB panels indicate MAVS cleavage products. (E and F) Effects of HCV protease inhibitors (boceprevir [B] or telaprevir [T]) against the indicated hepaciviral proteases (P). Either Hu MAVS (E) or Mu MAVS (F) was used as the protease substrate. The expression of MAVS (M) in the absence of protease served as an untreated control. Each drug was given at a final dose of 2.5 μM. WB analyses and MAVS-dependent IFN-β promoter reporter assays were done as explained in Materials and Methods. WB images are representative of at least two individual experiments, and all graphical data are shown as means ± SD for at least three independent experiments.
FIG 5.
Schematic summary of hepaciviral NS3/4A protease interference with mammalian MAVS function. The phylogenetic trees highlight the relatedness among nucleotide sequences encoding the analyzed proteins. Green box, interference; red box, no interference; *, cognate species; S, surrogate species; white box, untested. Interference was scored when both cleavage of MAVS was detectable and IFN promoter activity was significantly reduced under the chosen experimental conditions.
DISCUSSION
Recently, numerous new members of the genus Hepacivirus were discovered in diverse mammalian species. However, at present, there is very limited information regarding their host range, pathogenic potential, and ability to be transmitted between different animal species and to humans. To address some of these open questions, in this study we explored if these novel viruses cleave the innate immunity signaling adaptor MAVS proteins of their cognate animal hosts and, if so, whether they are also capable of interfering with human MAVS. Previous reports showed that HCV NS3/4A protease efficiently cleaves human MAVS, thus dampening innate immune signaling via RIG-I-dependent double-stranded RNA sensing (17, 35). In contrast, HCV is unable to interfere with rhesus macaque-derived MAVS (19), and since HCV replication in primary macaque hepatocytes is partially controlled by IFN-dependent mechanisms, this indicates that the inability of HCV to overcome rhesus macaque MAVS at least in part limits HCV replication in these cells (20). In contrast, we and others have observed that HCV readily cleaves mouse MAVS (24, 36). Collectively, these results indicate that inefficient MAVS cleavage by HCV represents one of many biological barriers to HCV transmission between hosts and that the HCV protease cleaves and interferes with MAVS proteins in a highly species-specific manner.
To explore if cleavage of MAVS is a common strategy of hepaciviruses, we coexpressed NS3/4A proteases of representative hepaciviruses from each clade together with their cognate or, where necessary due to the lack of deposited sequences, surrogate host-derived MAVS proteins. By monitoring MAVS cleavage via an RFP-NLS relocalization assay (33) or directly through measuring proteolytic cleavage as well as innate immune signaling to the IFN-β promoter, we provide strong evidence that all examined hepaciviruses encode serine proteases that cleave their cognate/surrogate host-derived MAVS proteins. For BHV-C and BHV-D, two bat-derived viruses, we noted relatively inefficient and even absent interference with innate immune signaling, respectively. However, in both cases, a small amount of cleaved bat-derived MAVS was detectable (Fig. 3), suggesting that cleavage does occur, albeit inefficiently. It is possible that bat-tropic hepaciviruses poorly cleave their natural hosts' MAVS proteins. However, we rather believe that the poor bat MAVS cleavage demonstrated here was due to the genetic distance between the cognate host bat species from which the respective viruses were isolated and the surrogate bat species used for our assays. To ultimately clarify this question and also to address subtle differences in cleavage efficiency, future work involving characterization of MAVS proteins and permissive cells from these species are required. Nevertheless, collectively, these results highlight that cleavage of MAVS is a conserved strategy across all currently known clades of the genus Hepacivirus. This conclusion is in agreement with and extends the report by Patel and colleagues, who previously showed that NPHV/CHV and a few members of the genus Pegivirus are capable of cleaving human MAVS (19).
It is worth mentioning that HCV NS3/4A also cleaves TRIF (18). Although the relevance of TRIF cleavage to HCV replication and persistence has been discussed previously (37), it will be interesting to demonstrate whether TRIF cleavage is unique to HCV or also common to other members of this virus genus. Moreover, the HCV NS3/4A protease also cleaves several human host factors that are unrelated to innate immune sensing, including T-cell protein tyrosine phosphatase (TC-PTP) (38), UV damage DNA-binding protein 1 (DDB1) (39), and the membrane-associated peroxidase GPx8 (40). These cleavage events are at least partly important for full replication of HCV in human cells (38, 40). It will be intriguing to find out if the respective nonhuman orthologs of these host factors are also cleaved by the respective nonhuman hepaciviruses and whether this is critical to support replication of these viruses. Unfortunately, infection systems for these novel hepaciviruses are currently not available, making it difficult to explore this question. However, in the case of NPHV, the recently established infectious molecular clone will be instrumental for developing such systems and subsequently exploring this question (22).
If cleavage of these additional host factors is conserved and functionally relevant among other hepaciviruses, it will be important to analyze if these nonhuman viral NS3/4A proteases also cleave the human orthologs in order to fully assess the zoonotic potential of these viruses. In this study, we show that human MAVS is readily susceptible to cleavage by all hepaciviruses examined. These results suggest that unlike the case of rhesus macaque MAVS and HCV, human MAVS would not be able to relay antiviral signaling upon infection of human cells by these HCV-related viruses. Clearly, numerous other host factors codetermine viral species tropism, most notably entry factors (41). Therefore, future work should address if cell entry factor usage is conserved between hepaciviruses. Besides important information regarding the potential zoonotic relevance of these new viruses, these studies certainly will provide important new insights into the infection and replication strategies of this diverse group of viruses.
ACKNOWLEDGMENTS
TWINCORE is a joint venture between the Helmholtz Centre for Infection Research, Braunschweig, Germany, and the Hanover Medical School, Hanover, Germany.
We are very grateful to Charles Rice for the pTRIP-RFP-NLS-Hu MAVS plasmid. We also thank all members of the Institute for Experimental Virology at TWINCORE for helpful comments on and discussions of this work.
REFERENCES
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol 20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Global Burden of Disease Study 2013 Collaborators. 2015. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding Q, von Schaewen M, Ploss A. 2014. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe 16:562–568. doi: 10.1016/j.chom.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapoor A, Simmonds P, Scheel TK, Hjelle B, Cullen JM, Burbelo PD, Chauhan LV, Duraisamy R, Sanchez Leon M, Jain K, Vandegrift KJ, Calisher CH, Rice CM, Lipkin WI. 2013. Identification of rodent homologs of hepatitis C virus and pegiviruses. mBio 4:e00216-13. doi: 10.1128/mBio.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quan PL, Firth C, Conte JM, Williams SH, Zambrana-Torrelio CM, Anthony SJ, Ellison JA, Gilbert AT, Kuzmin IV, Niezgoda M, Osinubi MO, Recuenco S, Markotter W, Breiman RF, Kalemba L, Malekani J, Lindblade KA, Rostal MK, Ojeda-Flores R, Suzan G, Davis LB, Blau DM, Ogunkoya AB, Alvarez Castillo DA, Moran D, Ngam S, Akaibe D, Agwanda B, Briese T, Epstein JH, Daszak P, Rupprecht CE, Holmes EC, Lipkin WI. 2013. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc Natl Acad Sci U S A 110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baechlein C, Fischer N, Grundhoff A, Alawi M, Indenbirken D, Postel A, Baron AL, Offinger J, Becker K, Beineke A, Rehage J, Becher P. 2015. Identification of a novel hepacivirus in domestic cattle from Germany. J Virol 89:7007–7015. doi: 10.1128/JVI.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol 86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman VM, Grundhoff A, Baechlein C, Fischer N, Gmyl A, Wollny R, Dei D, Ritz D, Binger T, Adankwah E, Marfo KS, Annison L, Annan A, Adu-Sarkodie Y, Oppong S, Becher P, Drosten C, Drexler JF. 2015. Highly divergent hepaciviruses from African cattle. J Virol 89:5876–5882. doi: 10.1128/JVI.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. 2011. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A 108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauck M, Sibley SD, Lara J, Purdy MA, Khudyakov Y, Hyeroba D, Tumukunde A, Weny G, Switzer WM, Chapman CA, Hughes AL, Friedrich TC, O'Connor DH, Goldberg TL. 2013. A novel hepacivirus with an unusually long and intrinsically disordered NS5A protein in a wild Old World primate. J Virol 87:8971–8981. doi: 10.1128/JVI.00888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg MG, Lee D, Coller K, Frankel M, Aronsohn A, Cheng K, Forberg K, Marcinkus M, Naccache SN, Dawson G, Brennan C, Jensen DM, Hackett J Jr, Chiu CY. 2015. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog 11:e1005325. doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard CR, Fletcher NF. 2012. Emerging virus diseases: can we ever expect the unexpected? Emerg Microbes Infect 1:e46. doi: 10.1038/emi.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandl JN, Ahmed R, Barreiro LB, Daszak P, Epstein JH, Virgin HW, Feinberg MB. 2015. Reservoir host immune responses to emerging zoonotic viruses. Cell 160:20–35. doi: 10.1016/j.cell.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 18.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M Jr, Lemon SM. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A 102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel MR, Loo YM, Horner SM, Gale M Jr, Malik HS. 2012. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol 10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scull MA, Shi C, de Jong YP, Gerold G, Ries M, von Schaewen M, Donovan BM, Labitt RN, Horwitz JA, Gaska JM, Hrebikova G, Xiao JW, Flatley B, Fung C, Chiriboga L, Walker CM, Evans DT, Rice CM, Ploss A. 2015. Hepatitis C virus infects rhesus macaque hepatocytes and simianized mice. Hepatology 62:57–67. doi: 10.1002/hep.27773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parera M, Martrus G, Franco S, Clotet B, Martinez MA. 2012. Canine hepacivirus NS3 serine protease can cleave the human adaptor proteins MAVS and TRIF. PLoS One 7:e42481. doi: 10.1371/journal.pone.0042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheel TK, Kapoor A, Nishiuchi E, Brock KV, Yu Y, Andrus L, Gu M, Renshaw RW, Dubovi EJ, McDonough SP, Van de Walle GR, Lipkin WI, Divers TJ, Tennant BC, Rice CM. 2015. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A 112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anggakusuma, Frentzen A, Gurlevik E, Yuan Q, Steinmann E, Ott M, Staeheli P, Schmid-Burgk J, Schmidt T, Hornung V, Kuehnel F, Pietschmann T. 2015. Control of hepatitis C virus replication in mouse liver-derived cells by MAVS-dependent production of type I and type III interferons. J Virol 89:3833–3845. doi: 10.1128/JVI.03129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, Pietschmann T. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog 6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaender S, Cavalleri JM, Walter S, Doerrbecker J, Campana B, Brown RJ, Burbelo PD, Postel A, Hahn K, Anggakusuma, Riebesehl N, Baumgartner W, Becher P, Heim MH, Pietschmann T, Feige K, Steinmann E. 2015. Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology 61:447–459. doi: 10.1002/hep.27440. [DOI] [PubMed] [Google Scholar]
- 27.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLoS Pathog 9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A 102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartenschlager R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat 6:165–181. doi: 10.1046/j.1365-2893.1999.00152.x. [DOI] [PubMed] [Google Scholar]
- 31.Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol 67:2832–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rögnvaldsson T, Etchells TA, You LW, Garwicz D, Jarman I, Lisboa PJG. 2009. How to find simple and accurate rules for viral protease cleavage specificities. BMC Bioinformatics 10:149. doi: 10.1186/1471-2105-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pybus OG, Theze J. 2016. Hepacivirus cross-species transmission and the origins of the hepatitis C virus. Curr Opin Virol 16:1–7. doi: 10.1016/j.coviro.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A 103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogt A, Scull MA, Friling T, Horwitz JA, Donovan BM, Dorner M, Gerold G, Labitt RN, Rice CM, Ploss A. 2013. Recapitulation of the hepatitis C virus life-cycle in engineered murine cell lines. Virology 444:1–11. doi: 10.1016/j.virol.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dansako H, Ikeda M, Kato N. 2007. Limited suppression of the interferon-beta production by hepatitis C virus serine protease in cultured human hepatocytes. FEBS J 274:4161–4176. doi: 10.1111/j.1742-4658.2007.05942.x. [DOI] [PubMed] [Google Scholar]
- 38.Brenndorfer ED, Karthe J, Frelin L, Cebula P, Erhardt A, Schulte am Esch J, Hengel H, Bartenschlager R, Sallberg M, Haussinger D, Bode JG. 2009. Nonstructural 3/4A protease of hepatitis C virus activates epithelial growth factor-induced signal transduction by cleavage of the T-cell protein tyrosine phosphatase. Hepatology 49:1810–1820. doi: 10.1002/hep.22857. [DOI] [PubMed] [Google Scholar]
- 39.Kang X, Chen X, He Y, Guo D, Guo L, Zhong J, Shu HB. 2013. DDB1 is a cellular substrate of NS3/4A protease and required for hepatitis C virus replication. Virology 435:385–394. doi: 10.1016/j.virol.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 40.Morikawa K, Gouttenoire J, Hernandez C, Dao Thi VL, Tran HT, Lange CM, Dill MT, Heim MH, Donze O, Penin F, Quadroni M, Moradpour D. 2014. Quantitative proteomics identifies the membrane-associated peroxidase GPx8 as a cellular substrate of the hepatitis C virus NS3-4A protease. Hepatology 59:423–433. doi: 10.1002/hep.26671. [DOI] [PubMed] [Google Scholar]
- 41.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You HN, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]