ABSTRACT
The human papillomavirus (HPV) capsid protein L2 is essential for viral entry. To gain a deeper understanding of the role of L2, we searched for novel cellular L2-interacting proteins. A yeast two-hybrid analysis uncovered the actin-depolymerizing factor gelsolin, the membrane glycoprotein dysadherin, the centrosomal protein 68 (Cep68), and the cytoskeletal adaptor protein obscurin-like 1 protein (OBSL1) as putative L2 binding molecules. Pseudovirus (PsV) infection assays identified OBSL1 as a host factor required for gene transduction by three oncogenic human papillomavirus types, HPV16, HPV18, and HPV31. In addition, we detected OBSL1 expression in cervical tissue sections and noted the involvement of OBSL1 during gene transduction of primary keratinocytes by HPV16 PsV. Complex formation of HPV16 L2 with OBSL1 was demonstrated in coimmunofluorescence and coimmunoprecipitation studies after overexpression of L2 or after PsV exposure. We observed a strong colocalization of OBSL1 with HPV16 PsV and tetraspanin CD151 at the plasma membrane, suggesting a role for OBSL1 in viral endocytosis. Indeed, viral entry assays exhibited a reduction of viral endocytosis in OBSL1-depleted cells. Our results suggest OBSL1 as a novel L2-interacting protein and endocytosis factor in HPV infection.
IMPORTANCE Human papillomaviruses infect mucosal and cutaneous epithelia, and the high-risk HPV types account for 5% of cancer cases worldwide. As recently discovered, HPV entry occurs by a clathrin-, caveolin-, and dynamin-independent endocytosis via tetraspanin-enriched microdomains. At present, the cellular proteins involved in the underlying mechanism of this type of endocytosis are under investigation. In this study, the cytoskeletal adaptor OBSL1 was discovered as a previously unrecognized interaction partner of the minor capsid protein L2 and was identified as a proviral host factor required for HPV16 endocytosis into target cells. The findings of this study advance the understanding of a so far less well-characterized endocytic pathway that is used by oncogenic HPV subtypes.
INTRODUCTION
Human papillomaviruses (HPVs) are small, nonenveloped DNA viruses that infect dividing basal keratinocytes of skin and mucosa via microlesions of the tissue. HPV is capable of inducing benign epithelial warts on the skin and mucosa, and infection with a high-risk HPV type may cause cervical and other anogenital and oropharyngeal cancers (1, 2). Cervical cancer is the third most common cancer in women worldwide and is associated with HPV infection, more precisely with high-risk HPV types such as HPV16, HPV18, and HPV31 (3). HPV is composed of a viral capsid with the major capsid protein L1, the minor capsid protein L2, and the viral genome. One icosahedral capsid contains 360 copies of L1, which can self-assemble to 72 pentamers, and up to 72 copies of the minor capsid protein L2, located inside the L1 shell (4–6). The capsid proteins L1 and L2 are key players in early events of infection, such as virus binding at the plasma membrane, cell entry, and transport of viral DNA into the nucleus (7–10).
Primary cell binding of the viral capsid is mediated by interaction of major capsid protein L1 with heparan sulfate proteoglycans (HSPGs) or non-HSPG components such as laminin-332 (11–17). After primary binding, both capsid proteins undergo conformational changes initiated by interactions with HSPGs, chaperones, and cellular proteases (18–21). The chaperone cyclophilin B facilitates exposure of the L2 N terminus (22), while furin cleaves the first 12 amino acids of L2 (23–26). Furin cleavage may also occur during virion morphogenesis, as shown for tissue-derived native HPV16, resulting in infection independent of cellular furin (25). The precleaved virus is transferred to tetraspanin-enriched microdomains (TEMs) (27, 28), where L2 directly interacts with the annexin A2 heterotetramer (A2t), a protein localized on the outer and inner leaflet of the plasma membrane and mediating viral endocytosis and infection (29, 30). Interaction occurs between L2 amino acid residues 108 to 126 and A2t subunit S100A10 (29, 30).
Endocytosis of viral particles is mediated by a clathrin-, dynamin-, and caveolin-independent mechanism but requires tetraspanin CD151 and the actin cytoskeleton (10, 27, 28, 31–34). Following internalization, viral particles are found in CD63-positive endosomes recruiting syntenin-1, a CD63-interacting adaptor protein (35). The CD63–syntenin-1 complex was identified as a regulatory component in postendocytic HPV trafficking to multivesicular endosomes (35), where vacuolar ATPase and cyclophilins facilitate capsid disassembly and dissociation of L1 and L2 (36–38). DiGiuseppe and coworkers revealed that L2 amino acid residues 64 to 81 and 163 to 170 and the L2 C-terminal exposure on the cytosolic side of intracellular membranes enable interaction with cytosolic host cell factors (39). Interactions of L2 with actin (40), components of the retrograde transport machinery (37, 41, 42), sorting nexins 17 and 27, TSG101, γ-secretase, and Hsc70, as well as the microtubule network, have been reported (37, 41–48). These interactions result in trafficking to the Golgi network (37, 41, 42, 47), transport toward the nucleus (43, 44), and accumulation at nuclear substructures (49–53). Furthermore, L2 possesses a membrane-destabilizing peptide in its C terminus (54) and a transmembrane domain in its N terminus (55) that are both important for translocation to the cytoplasm. The precise step in viral infection at which L2 becomes accessible on the cytosolic side of the host membrane remains unknown. It may occur after capsid disassembly or earlier in infection.
In this study, we identified the cytoskeletal adaptor protein obscurin-like 1 (OBSL1) as a relevant cellular component during HPV gene transduction and endocytosis. OBSL1 was first discovered and described by Geisler et al. in 2007 as a protein closely related to obscurin (56). Obscurin is a structural protein expressed in myocytes, whereas OBSL1 was found in cells of numerous tissues, indicating that its function is not limited to myocardial muscles. Although OBSL1 has not been characterized well, it is thought to act as a cytoskeletal adaptor protein, for example, in linking the internal cytoskeleton to the plasma membrane (56). Furthermore, it was found to form a protein complex at the Golgi apparatus of mammalian brain neurons (57). Here, we demonstrate that OBSL1 forms a complex with HPV16 L2 and that OBSL1 knockdown inhibits gene transduction of human keratinocytes by pseudoviruses of oncogenic HPV types 16, 18, and 31. Therefore, we propose that OBSL1 plays a general role in infection of keratinocytes by various human papillomaviruses.
MATERIALS AND METHODS
Plasmids and antibodies.
The pcDNA3.1-V5-Obsl1 expression vector was kindly provided by P. E. Clayton (58). Codon-optimized HPV16 L1 and L2 expression plasmids were prepared based on the wild-type origin vector described before (59). FLAG-tagged L2 was cloned into p3xFLAG-CMV-14 using BamHI. For detection of V5-tagged OBSL1 after Western blotting or immunofluorescence staining, either monoclonal antibody (MAb) SV5-Pk1 (Bio-Rad, Hercules, CA) or polyclonal antibody ab9116 (Abcam, Cambridge, UK) was used. HPV16 L1-specific mouse monoclonal antibody 16L1-312F and rabbit polyclonal antibody K75 and 33L1-7 as well as HPV16 L2-specific mouse antibody 33L2-1 have been previously described (L2-1 [60], K75 [61], 312F [62], and L1-7 [63]). Obscurin-like 1-specific polyclonal rabbit antibody HPA036404 and FLAG (M2)- and β-actin (A5441)-specific mouse antibodies were purchased from Sigma-Aldrich, St. Louis, MO. Obscurin-like 1-specific polyclonal goat antibody (E-16) and dysadherin-specific goat antibody (N-20) were obtained from Santa Cruz Biotechnology, Dallas, TX. Cep68-specific polyclonal rabbit antibody (15147-1-AP) was purchased from Proteintech, Rosemont, IL, and gelsolin-specific monoclonal antibody GS-2C4 was purchased from Abcam. Monoclonal mouse antibody against Golgi (Golgin-97) was purchased from Thermo Fisher, Waltham, MA. Horseradish peroxidase (HRP)-coupled secondary antibodies were purchased from Dianova, Hamburg, Germany; secondary antibodies for immunofluorescence detection (Alexa Fluor) as well as the Click-iT EdU imaging kit were purchased from Invitrogen, Carlsbad, CA.
Production of PsVs.
HPV16, -18, and -31 pseudoviruses (PsVs) and HPV16 L1-only PsV were prepared as previously described and published (33, 53, 64). Quantification of the reporter gene plasmid was performed as published before (28, 53, 65).
Yeast two-hybrid screening.
The yeast two-hybrid screening with L2 as bait using a cDNA library (pJG4-5) derived from human serum-starved WI-38 fibroblasts has been described previously (44, 66).
Cell lines and transfection.
The human cervical carcinoma cell line HeLa was purchased from the German Resource Center of Biological Material (DSMZ; Braunschweig, Germany). HaCaT cells (human non-virally transformed keratinocytes) were obtained from Cell Lines Services (CLS), Eppelheim, Germany. Both cell lines were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS; Biochrom AG, Berlin, Germany), 1% Glutamax I (Invitrogen), 1% modified Eagle's medium nonessential amino acids (GE Healthcare, Little Chalfont, UK), and antibiotics (Fresenius Kabi, Bad Homburg vor der Hoehe, Germany). Normal human epidermal keratinocytes (NHEK) were purchased from PromoCell, Heidelberg, Germany, and cultivated according to the manufacturer's instructions in keratinocyte growth medium 2.
siRNA-mediated knockdown and gene transduction assay.
All small interfering RNAs (siRNAs) were purchased from Qiagen, Hilden, Germany. The following gene-specific siRNAs were used: gelsolin, Hs_GSN_3/4/5; dysadherin, Hs_FXYD5_3/5/6; cep68, Hs_CEP68_2/Hs_KIAA0582_2/3; and obsl1, Hs_KIAA0657_4/5/6. Likewise, from Qiagen, AllStar negative-control siRNA was used as a nonsilencing control siRNA. HeLa or HaCaT cells or NHEK were transfected with 30 nM siRNA using Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Subsequent experiments were performed 48 h after siRNA transfection. PsV infection assays were performed as described previously (28). Briefly, HeLa or HaCaT cells or NHEK were seeded into 24-well plates and transfected with siRNA for 48 h. Afterwards, cells were exposed to 100 (HeLa and HaCaT) or 500 (NHEK) luciferase vector-positive PsVs per cell. Twenty-four hours (HeLa and HaCaT) or 48 h (NHEK) postexposure (p.e.), luciferase activity was measured to quantify gene transduction efficiency of HPV PsV and normalized by lactate dehydrogenase (LDH) measurements (CytoTox-ONE homogeneous membrane integrity assay; Promega, Firchburg, WI). Luciferase and LDH activities were measured with a Tristar LB 941 luminometer (Berthold Technologies, Bad Wildbad, Germany). The siRNA knockdown efficiency was quantified by Western blotting and quantitative PCR (qPCR). Western blotting for dysadherin was performed under nonreducing conditions. For qPCR experiments, HeLa or HaCaT cells or NHEK were seeded into 12-well plates and transfected with siRNAs for 48 h as described above. Afterwards, cells were lysed using peqGOLD TriFast (Peqlab, Erlangen, Germany), and total mRNA was isolated using a Direct-zol RNA MiniPrep kit from Zymo Research, Irvine, CA, according to the manufacturer's instructions. The mRNA was then treated with 5 U of RNase-free, recombinant DNase I (Roche Diagnostics, Rotkreuz, Switzerland), and cDNA synthesis was performed by using a qScript cDNA synthesis kit (Quanta Biosciences, Beverly, MA). For real-time quantitative PCR, specific primer sets for OBSL1 were used, while β-actin served as a reference. Primer sequences are available on request. One qPCR mix (20-μl total reaction volume) comprised 5 μl of cDNA template, 1 μl of each forward and reverse primer (10 μM), 10 μl of Fast Start Universal SYBR green Probe Master (Roche Diagnostics, Rotkreuz, Switzerland), and 3 μl of aqua bidest. The qPCR was performed with a 7300 real-time PCR system and Sequence Detection Software 4.0 (Applied Biosystems, Foster City, CA) and analyzed by using the comparative cycle threshold (CT) method.
Immunofluorescence microscopy.
HeLa or HaCaT cells were grown on coverslips. After transfection and/or treatment with about 500 HPV16 PsVs per cell, cells were washed and fixed with methanol at −20°C for 10 min or 30 min according to the 5-ethynyl-2′-deoxyuridine (EdU) staining protocol at different time points after addition of PsVs. For phalloidin costaining, the cells were fixed with 4% paraformaldehyde for 10 min at room temperature and then permeabilized with 0.2% Triton-X in phosphate-buffered saline (PBS) for 2 min at room temperature. Fixed cells were washed three times with PBS and blocked for 30 min with PBS–1% (wt/vol) bovine serum albumin (BSA; AppliChem, Darmstadt, Germany). For L2 staining after PsV exposure, cells were incubated with Click-iT reaction cocktail according to the manufacturer's instructions (without the addition of EdU label for PsV staining) as described previously (22, 53). Afterwards, proteins were stained with the desired antibodies at 37°C for 1 h. After washing with PBS, coverslips were blocked with PBS–1% BSA for 10 min and subsequently incubated with Alexa-conjugated specific secondary antibodies or phalloidin-tetramethyl rhodamine isothiocyanate (TRITC; Sigma-Aldrich, St. Louis, MO) for 1 h at 37°C in the dark. DNA was stained with Hoechst 33342 (Invitrogen, Carlsbad, CA). Coverslips were mounted onto slides using Fluoprep mounting medium (bioMérieux, Marcy-I'Étoile, France). Fluorescence imaging was performed on a Zeiss Axiovert 200 M microscope equipped with a Plan-Apochromat 100× objective lens (1.4 numeric aperture), and Z-stack images were deconvoluted using the software supplied by Zeiss (Axiovision 4.7; Carl Zeiss, Jena, Germany). For confocal microscopy, images were recorded with an LSM880 confocal laser scanning microscope (Carl Zeiss) using a water immersion objective (C-Apochromat 40×/1.2 W). Alexa Fluor 488-labeled OBSL1-V5 was excited with the 488-nm line of an argon laser, and the fluorescence was detected in the wavelength range of 500 to 553 nm. Phalloidin-TRITC that was used for staining of the F-actin structures was excited with the 543-nm line of an HeNe laser, and the fluorescence was detected in the range of 562 to 633 nm. Z-stacks with a distance of 0.5 μm between the slices were acquired and representative slices selected. Image files were assembled into figures using InDesign software (Adobe).
Immunostaining of tissue slices.
Paraffin-embedded cervix tissue slices were stained as described previously (28, 50, 51, 66, 67).
Cell binding assay.
HaCaT cells were grown in 24-well plates and transfected with target-specific siRNA for 48 h. Afterwards, cells were detached with 0.05% trypsin–2.5 mM EDTA, resuspended in DMEM, and transferred into siliconized reaction tubes. Control cells were pretreated with polyethyleneimine (PEI), an inhibitor of HPV infection (Sigma-Aldrich, St. Louis, MO), for 1 h at 4°C as indicated below and described previously (65). Subsequently, all cells were exposed to 200 to 500 HPV16 PsVs for 1 h at 4°C on an overhead rotator. Samples were washed with PBS and collected in SDS sample buffer for Western blotting. Cell-bound HPV16 particles were stained using anti-L1 antibody 312F. β-Actin served as an internal loading control.
Detection of surface-bound particles by flow cytometry and immunofluorescence.
HaCaT cells were seeded in either 12-well plates on coverslips for immunofluorescence or 24-well plates for flow cytometry analysis and transfected with control siRNA or an OBSL1 siRNA pool for 48 h. Afterwards, cells were treated with 200 to 500 HPV16 PsVs for 1 h or 24 h and incubated at 37°C. Cells were washed with fresh medium to remove unbound virus at 1 h postinfection. For immunofluorescence analysis, cells were washed with PBS and fixed with 2% paraformaldehyde. Surface-bound particles were stained with anti-L1 polyclonal antibody K75 and secondary anti-rabbit Alexa Fluor 488 antibody according to the immunofluorescence protocol described above. Cell nuclei were stained with Hoechst 33342. For quantification, the relative amount of surface-bound particles was determined based on the K75-positive pixels relative to the cell nucleus signal (DNA/Hoechst 33342-positive pixels) in 50 randomly selected cells each from three independent experiments. A threshold value was set to exclude background. For flow cytometry analysis, PsV-exposed cells were trypsinized with 0.05% trypsin–2.5 mM EDTA after 1 h and 24 h, as indicated below. Surface-bound particles were stained with anti-L1 polyclonal antibody K75 in PBS–0.5% FCS for 30 min at 4°C and secondary anti-rabbit Alexa Fluor 488 antibody in PBS–0.5% FCS for 20 min at 4°C. Cells were analyzed by flow cytometry (FACScan; Becton, Dickinson, East Rutherford, NJ) and CellQuest3.3 software (Becton, Dickinson).
Measurements of viral particle internalization through protease protection assays.
For the proteinase protection assay as described previously (68), HaCaT cells were transfected with either control siRNA or an OBSL1 siRNA pool for 48 h. Cells were infected with 1,000 to 2,000 HPV16 PsV for 1 h, washed with medium, and incubated for another 24 h. Cells were then washed two times with PBS and incubated with cell culture medium without FCS and supplemented with 20 μg/ml of proteinase K (Sigma-Aldrich, St. Louis, MO) for 15 min at 37°C. Control cells were treated with cell culture medium alone. Digestion was stopped by addition of 2 mM phenylmethylsulfonyl fluoride (PMSF). Subsequently, cells were washed with PBS, lysed in SDS sample buffer, and processed for Western blotting. Undigested L1 protein was stained using anti-L1 antibody 312F and was normalized to L1 input (after 24 h of incubation with HaCaT cells).
Detection of virus capsid disassembly by immunofluorescence.
HaCaT cells were grown on coverslips and transfected with control siRNA or an OBSL1 siRNA pool for 48 h. Cells were treated with 200 to 500 HPV16 PsV for 7 h at 37°C. Subsequently, cells were fixed with methanol and stained with anti-L1-7 MAb and secondary anti-mouse Alexa Fluor 488 antibody according to the immunofluorescence protocol described before (27, 28). Quantification was done as described in the protocol above for detection of surface-bound particles.
Coimmunoprecipitation.
For coimmunoprecipitation after overexpression, HeLa cells were seeded in 60-mm dishes and incubated for 24 h at 37°C. Cells were transfected with the appropriate plasmids for additional 24 h using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Thereafter, cells were lysed using MACS lysis buffer (MACS Miltenyi Biotec, Bergisch Gladbach, Germany), containing dithiothreitol (DTT) and the protease inhibitors aprotinin and leupeptin (10 μg/ml each), and incubated for 20 min at 4°C on an overhead rotator. Lysates were treated 3 times with an ultrasonicator (30% duty cycle; output control, 30%; Branson Sonifier 250; Emerson Industrial Automation, St. Louis, MO) for 20 s and incubated for 20 min at 4°C on an overhead rotator. Lysates were centrifuged for 10 min at 4°C and 13,000 rpm and precleared with 50 μl of protein A/G agarose (Santa Cruz Biotechnology, Dallas, TX). Precleared lysates were incubated with 1 μl of anti-V5 antibody (Bio-Rad) for precipitation of V5-OBSL1 for 1 h at 4°C on a rotating wheel and for 1 h after addition of 50 μl of protein A/G agarose. Agarose was washed with wash buffer containing 500 mM NaCl, 50 mM Tris-HCl (pH 8.0), 1% (vol/vol) NP-40 (Sigma-Aldrich, St. Louis, MO), 0.5% (wt/vol) sodium deoxycholate (Sigma-Aldrich), 0.1% SDS (Carl Roth, Karlsruhe, Germany), and protease inhibitors. The precipitates were boiled in SDS sample buffer and processed for Western blotting. For coimmunoprecipitation after pseudovirus infection, HaCaT cells were seeded in 10-cm dishes and incubated for 24 h at 37°C. Cells were then exposed to HPV16 PsV (L1/L2 and L1 only PsVs) for 4 h, washed on ice with HEPES wash buffer (25 mM HEPES, 150 mM NaCl, 5 mM MgCl2) containing the protease inhibitors aprotinin and leupeptin (10 μg/ml each), lysed with lysis buffer {1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 25 mM HEPES, 150 mM NaCl, 5 mM MgCl2} containing aprotinin and leupeptin (10 μg/ml each), and incubated for 15 min at 4°C on an overhead rotator. Lysates were treated 3 times with an ultrasonicator (30% duty cycle; output control, 30%; Branson Sonifier 250; Emerson Industrial Automation, St. Louis, MO) for 20 s and incubated for 30 min at 4°C on an overhead rotator. M-280 sheep anti-rabbit IgG Dynabeads (Invitrogen, Carlsbad, CA) were preincubated with anti-HPV16 L1 (K75) for 30 min at room temperature and 2 h at room temperature after addition of lysates. Magnetic beads were washed three times with PBS containing 0.02% Tween and aprotinin and leupeptin (10 μg/ml each) and subsequently boiled in SDS sample buffer and processed for Western blotting.
Statistics.
All experiments were reproduced at least three times. Statistical significances (P < 0.05) were calculated with a two-tailed, paired t test using Microsoft Office Excel 2010.
RESULTS
Yeast two-hybrid screening reveals novel putative cellular interaction partners of HPV16 L2.
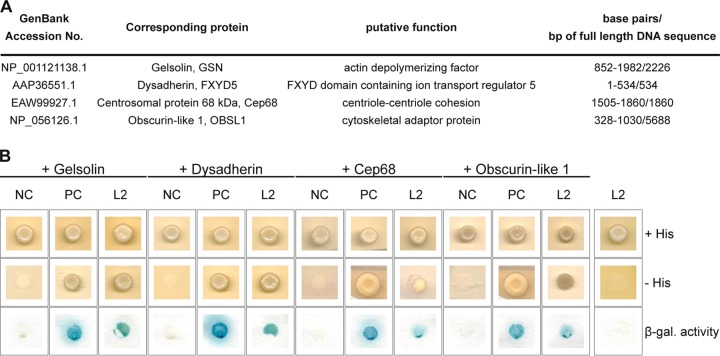
The multifunctional minor capsid protein L2 plays essential role in virus entry and morphogenesis. To gain a deeper insight into the HPV replication cycle, we performed a yeast two-hybrid screen with the L2 protein as previously described (44, 66). Four novel putative cellular interaction partners of L2 were identified in this screen: the actin-depolymerizing factor gelsolin, the membrane glycoprotein dysadherin, the centrosomal protein Cep68, and the cytoskeletal adaptor protein obscurin-like 1 (OBSL1) (Fig. 1).
FIG 1.
Identification of four cellular interaction partners of HPV16 L2. (A) Listed proteins were identified by yeast two-hybrid screen. The GenBank accession no., full name, putative function, and identified base pairs compared to full-length DNA sequence are listed. (B) Yeast strain L40 expressing a negative control (NC; LexA-lamin), a positive control (PC; LexA-Fos2), or the bait LexA-L2 280-473 (L2) fusion construct was transformed with corresponding prey construct B42-HA-GSN (gelsolin), B42-HA-FXYD5 (dysadherin), B42-HA-Cep68 (Cep68), B42-HA-OBSL1 (OBSL1), or empty prey vector pJG4-5 (rightmost column). Transformants were tested for the prototrophic marker histidine (− His) and β-galactosidase (β-gal.) activity reporting bait-prey interaction. Plates were incubated for 4 days at 30°C. Yeast cells were transferred onto nitrocellulose filter and incubated in 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) buffer to detect β-galactosidase activity.
Cep68 and OBSL1 are proviral factors in HPV16 PsV gene transduction.
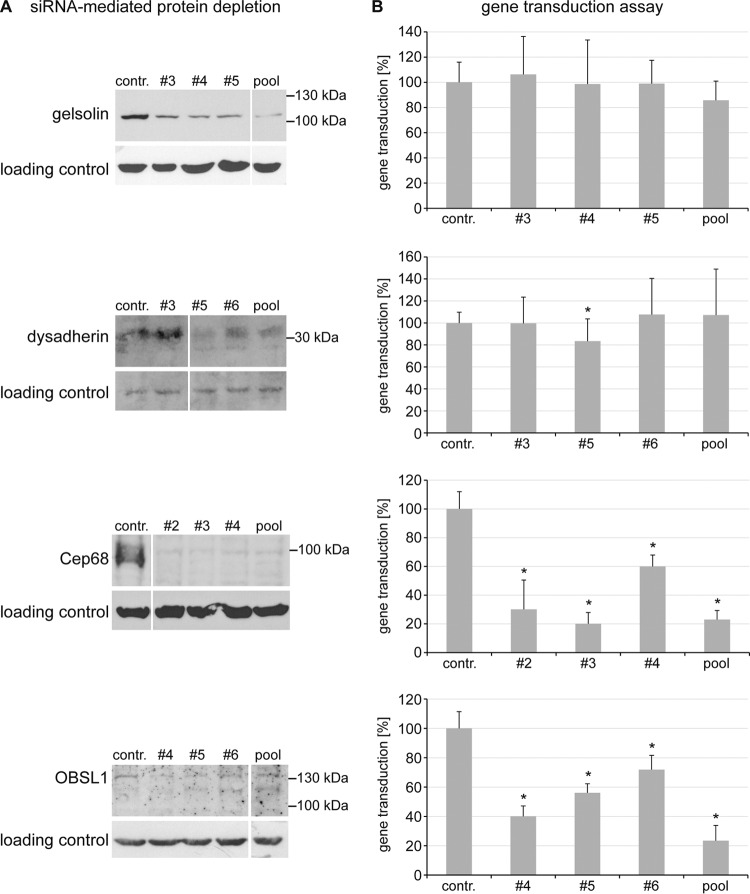
The influence of each of the four putative L2 interaction partners on HPV16 PsV gene transduction was evaluated in siRNA-mediated knockdown experiments with HaCaT cells (Fig. 2) using HPV16 PsV with an encapsidated luciferase expression plasmid. Transfection of the target-specific siRNAs resulted in a depletion of all four proteins (Fig. 2A). However, the knockdown of gelsolin and dysadherin had no or only a minor influence on HPV16 PsV gene transduction, while Cep68- and OBSL1-specific siRNA treatment led to a significant reduction of gene transduction by all tested siRNAs (Fig. 2B). The importance of the cytoskeleton during HPV16 infection has been demonstrated before (69), but a cytoskeletal adaptor with a suggested function as a linker between the cytoskeleton and the plasma membrane has not been described so far. Therefore, OBSL1 was selected for further analysis in this study.
FIG 2.
Influence of the putative cellular L2 interaction partners on HPV16 PsV gene transduction. HaCaT cells were transfected with control siRNA or siRNA targeted against gelsolin, dysadherin, Cep68, or OBSL1 for 48 h and then exposed to HPV16 PsVs for 24 h. (A) Cell lysates of siRNA-mediated protein depletion 48 h after transfection were analyzed by Western blotting with specific antibodies (siRNAs that showed reduction of protein level without off-target effects were selected). (B) Gene transduction efficiency was measured by luciferase activity and was normalized to lactate dehydrogenase (LDH) activity as a control for cell viability. Control siRNA-transfected cells were set to 100% ± SD. *, P < 0.05 compared to the value for the control.
OBSL1 is required for gene transductions by oncogenic HPV types 16, 18, and 31.
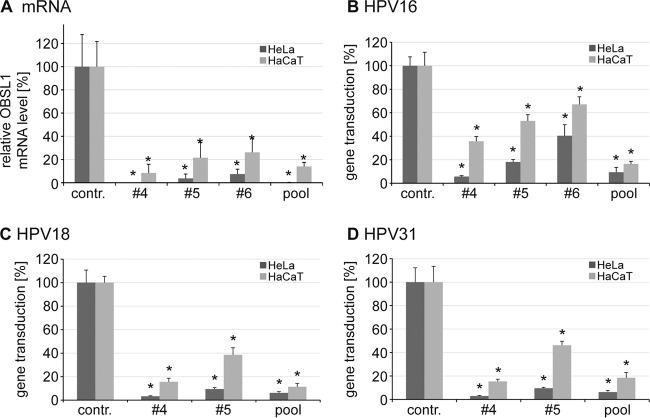
Next, siRNA-mediated OBSL1 mRNA depletion (Fig. 3A) was analyzed. The mRNA level correlated with the reduction of HPV16 gene transduction in HaCaT (Fig. 2 and 3B) and HeLa cells (Fig. 3B). These data suggest a specific function for the cytoskeletal adaptor protein OBSL1 in HPV16 infection. Since HPV16, HPV18, and HPV31 share similar endocytic requirements for entry (33), the influence of OBSL1 depletion was investigated for HPV18 and HPV31 in both cell lines (Fig. 3C and D). Here again, OBSL1 mRNA level knockdown efficiency in HeLa and HaCaT cells correlated with reduced HPV PsV gene transduction. These data suggest that the OBSL1-dependent entry step in infection of keratinocytes with various human papillomaviruses is conserved.
FIG 3.
OBSL1 mRNA level correlates with gene transduction by HPV16, -18, and -31 PsV in HeLa and HaCaT cells. (A) siRNA-mediated OBSL1 depletion on mRNA level in HeLa and HaCaT cells. Experiments were performed as described for Fig. 2. Forty-eight hours after siRNA transfection, total mRNA of siRNA-treated HeLa and HaCaT cells was isolated, reverse transcribed to cDNA, and analyzed by quantitative real-time PCR using gene-specific primers for amplification. Shown are the results of two independent experiments performed in duplicate. (B) HeLa and HaCaT cells were transfected with control siRNA or OBSL1 siRNA for 48 h. Shown is the gene transduction with HPV16 PsV as measured by luciferase activity and normalized to lactate dehydrogenase (LDH) activity as a control for cell viability. Control siRNA-transfected cells were set to 100% ± SD. *, P < 0.05 compared to the value for the control. (C and D) Experiments were performed as described for panel B but with exposure to HPV18 PsV (C) or HPV31 PsV (D).
OBSL1 is expressed in cervical tissue and supports HPV gene transduction of primary keratinocytes.
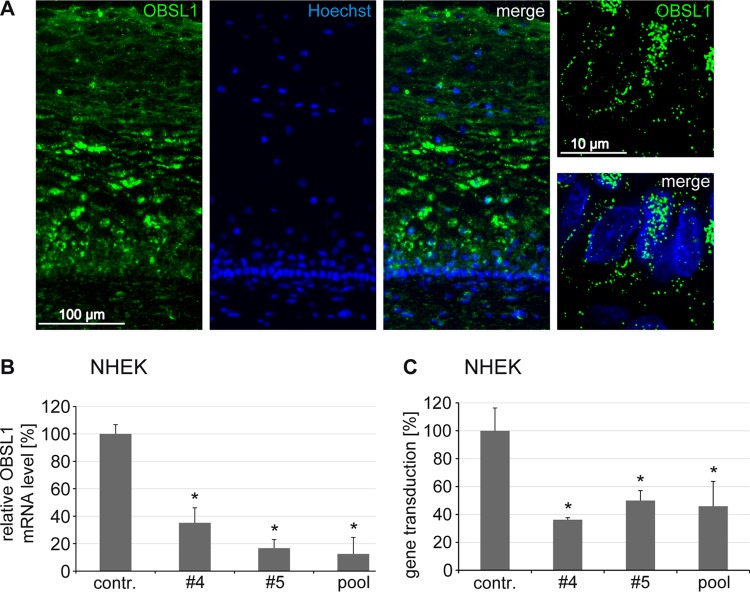
Since HPV16 virions infect keratinocytes of the mucosa (64), we studied the distribution of OBSL1 in cervical tissue sections by immunofluorescence staining. We detected a high expression level of OBSL1 in basal keratinocytes (Fig. 4A). In addition, siRNA-mediated knockdown of OBSL1 in human primary keratinocytes (NHEK) (Fig. 4B) verified its biological relevance in HPV gene transduction (Fig. 4C).
FIG 4.
Obscurin-like 1 is expressed in HPV16 target cells and required for gene transduction of primary keratinocytes. (A) Distribution of OBSL1 in cervical mucosa and expression of OBSL1 in basal cells (right images show higher magnification of basal keratinocytes). Human cervical tissue sections were stained with an OBSL1 antibody (green). Nuclei are shown in blue. (B) Normal human epidermal keratinocytes (NHEK) were transfected with control or OBSL1 siRNA for 48 h. Total mRNA was isolated, reverse transcribed to cDNA, and analyzed by quantitative real-time PCR using gene-specific primers for amplification. (C) NHEK were transfected with control or OBSL1 siRNA for 48 h and then exposed to HPV16 PsV for 24 h. Gene transduction was measured by luciferase activity and normalized by LDH measurements. The control siRNA infection rate was set to 100%. *, P < 0.05 compared to the value for the control.
HPV16 L2 forms a complex with OBSL1.
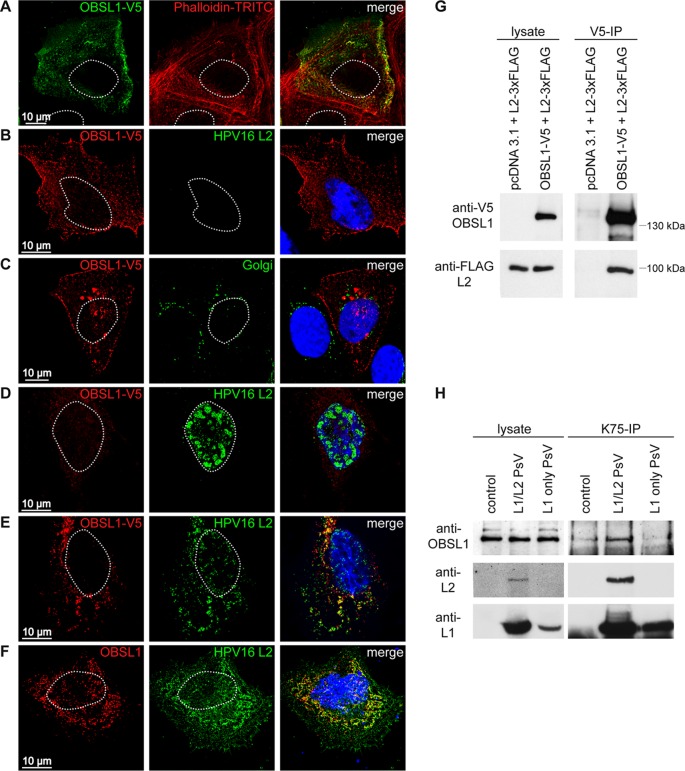
To analyze the OBSL1 localization in keratinocytes and its interaction with HPV16 L2, colocalization and coimmunoprecipitation assays of OBSL1 and L2 were performed (Fig. 5). Confocal and deconvolution fluorescence microscopy showed that OBSL1 is mainly localized in the cell periphery, with partial colocalization with the actin-binding protein phalloidin (Fig. 5A and B), while no colocalization with a Golgi protein was observed (Fig. 5C), which illustrates the role of OBSL1 as a cytoskeletal adaptor protein or a plasma membrane-associated protein. Overexpression of HPV16 L2 led to different subcellular localization patterns: a characteristic punctate pattern in the nucleus (Fig. 5D), diffuse nuclear localization (data not shown), or cytoplasmic localization of L2 (Fig. 5F) due to different expression levels and the diverse localization signals of L2 as described earlier (70–72). In cotransfected cells, the cellular localization patterns of OBSL1 and L2 changed and both proteins were strongly colocalized in the cytoplasm (Fig. 5E). Cytoplasmic L2 also colocalized with endogenous OBSL1 (Fig. 5F). To support these findings, coimmunoprecipitation analyses of OBSL1 and L2 were conducted (Fig. 5G). The precipitation of OBSL1 resulted in a coprecipitation of L2, providing further evidence of OBSL1/L2 complex formation in human keratinocytes. Moreover, coprecipitation of endogenous OBSL1 was also observed after HPV16 L1/L2 PsV infection (Fig. 5H), whereas HPV16 PsV lacking the L2 protein (L1 only) showed a background level of OBSL1 interaction comparable to that with noninfected cells, suggesting complex formation of HPV16 L2 with OBSL1 during virus entry.
FIG 5.
HPV16 L2 forms a complex with OBSL1 in mammalian cells. (A) Confocal fluorescence microscopy shows colocalization of OBSL1-V5 with actin-binding protein phalloidin-TRITC (red). HaCaT cells were transfected with OBSL1-V5 for 24 h. OBSL1 was stained with monoclonal mouse V5 antibody (green). (B to F) Deconvolution microscopy shows OBSL1 distribution pattern. (B) Representative picture of OBSL1-V5 (red). (C) Costaining of OBSL1-V5 and the Golgi apparatus. (D) Representative nuclear HPV16 L2 distribution pattern. (E) Representative colocalization of OBSL1-V5 and L2. OBSL1-V5 and HPV16 L2 were expressed alone or in combination in HeLa cells for 24 h. OBSL1 was stained with a polyclonal rabbit V5 antibody (red), L2 with the monoclonal mouse L2 antibody L2-1 (green). The Golgi vesicles were visualized with the monoclonal mouse antibody Golgin-97 (green), and the DNA was visualized with Hoechst 33342 (blue). (F) Representative colocalization of endogenous OBSL1 and L2 in HaCaT cells. HaCaT cells were transfected with HPV16 L2 for 24 h. OBSL1 was stained using polyclonal rabbit OBSL1 antibody (red), L2 with the monoclonal mouse L2 antibody L2-1 (green), and DNA with Hoechst 33342 (blue). (G) Coimmunoprecipitation studies of L2 and OBSL1. Lysates were prepared from HeLa cells transiently cotransfected with pcDNA3.1 empty vector or OBSL1-V5 and HPV16L2-FLAG as indicated. Protein expression was verified by Western blotting of the lysates with the specific antibodies. Lysates were subjected to immunoprecipitation (IP) with antibodies specific for the V5 tag of OBSL1. The precipitated proteins were detected by Western blotting using anti-V5 or anti-FLAG antibody. (H) Coimmunoprecipitation of HPV16 PsV and OBSL1. Lysates were prepared from mock-, L1/L2 PsV-, or L1-only PsV-exposed HaCaT cells as indicated. Protein input was verified by Western blotting of the lysates with the specific antibodies. Lysates were subjected to immunoprecipitation with antibodies specific for HPV16 L1 (K75). The precipitated proteins were detected by Western blotting using anti-OBSL1 (E-16), anti-HPV16 L2 (L2-1), or anti-HPV16 L1 (312F).
OBSL1 colocalizes with L2, L1 and tetraspanin CD151 during PsV entry.
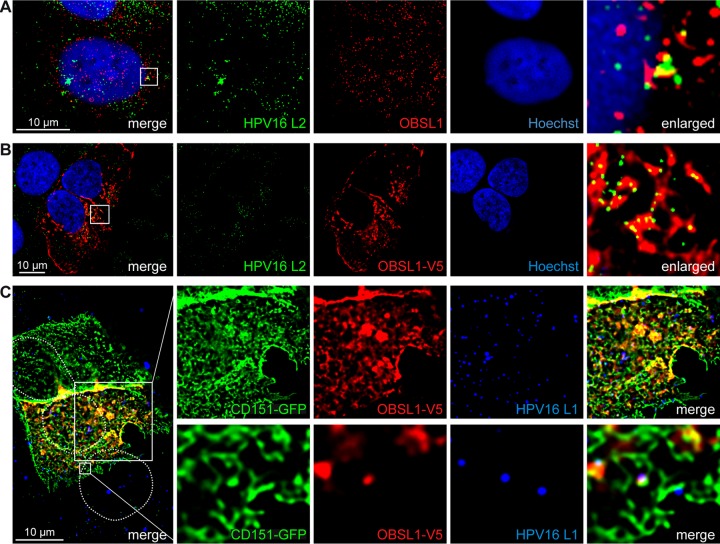
To investigate the role of OBSL1 in HPV infection, we performed coimmunofluorescence studies of endogenous OBSL1 and L2 in HPV16 PsV cell entry. We observed colocalization at the plasma membrane of HaCaT cells 4 h after PsV exposure (Fig. 6A). Due to reduction of OBSL1 antibody reactivity after cell treatment using the L2 staining protocol for PsV-derived L2, we also performed studies in OBSL1-overexpressing cells and found an increase of colocalization with L2 (Fig. 6B). These results imply an involvement of OBSL1 during virus binding or endocytosis. Our earlier studies demonstrated that the virus internalization requires association with the membrane organizer CD151 and integration in tetraspanin-enriched microdomains (27, 28). Therefore, we analyzed the putative association of OBSL1 with CD151 and demonstrated colocalization of both proteins in HaCaT cells (Fig. 6C), suggesting that OBSL1 is a part of the entry receptor complex and involved in HPV endocytosis.
FIG 6.
OBSL1 associates with the HPV16 entry platform. (A and B) HaCaT cells were exposed to HPV16 PsV for 4 h. The cells were fixed with methanol and treated with Click-iT reaction buffer, without the addition of Alexa Fluor label for vDNA staining. Obscurin-like 1 was stained with anti-OBSL1 antibody for staining of endogenous OBSL1 (red) (A) or with anti-V5 antibody after overexpression (B), L2 was stained with monoclonal mouse anti-L2 (L2-1) (green), and nuclear DNA was stained with Hoechst 33342 (blue). (C) HaCaT cells were cotransfected with OBSL1-V5 and CD151-green fluorescent protein (GFP) for 24 h and exposed to HPV16 PsVs for 4 h. OBSL1 was stained with monoclonal mouse V5 antibody (red), L1 was stained with polyclonal rabbit L1 antibody (K75; blue), and nuclei are shown with dotted lines.
OBSL1 is involved in HPV16 PsV endocytosis.
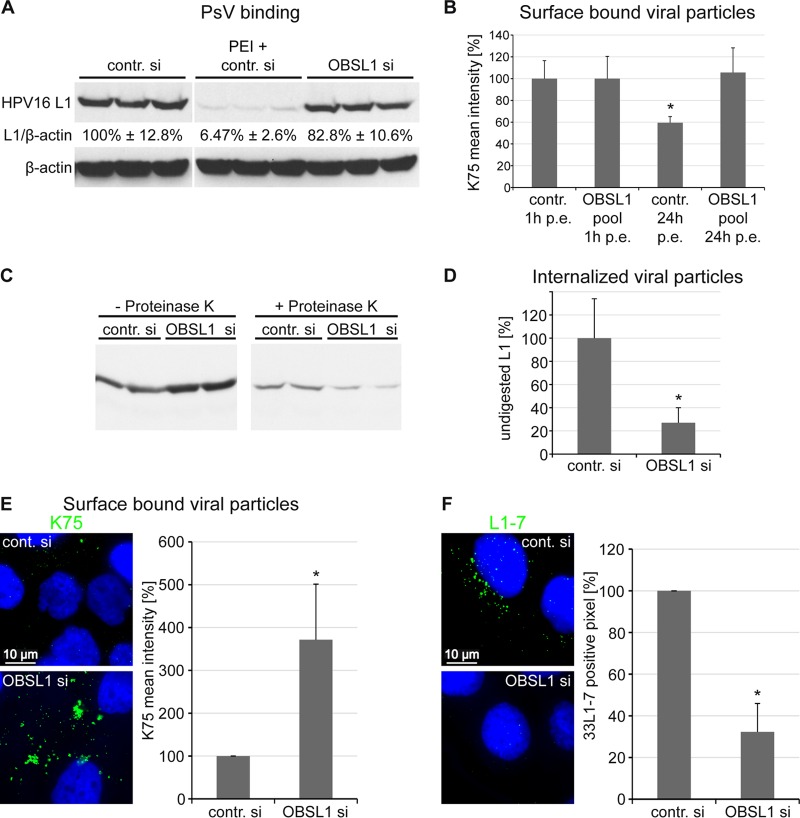
To gain further insights into the function of OBSL1 in HPV entry, we investigated its role during early steps of HPV16 entry. Cells were transfected with either control siRNA or an OBSL1 siRNA pool and incubated with HPV16 PsV (Fig. 7). Surface-bound viral particles were quantified by quantitative Western blot analysis (Fig. 7A), flow cytometry (Fig. 7B), or immunofluorescence analysis (Fig. 7E) using L1-specific antibodies. Quantitative Western blot analysis demonstrated that siRNA-mediated OBSL1 depletion had no or a minor effect on initial HPV16 cell surface binding. As an internal control, HaCaT cells were transfected with control siRNA and treated with polyethyleneimine (PEI), a strong inhibitor for primary HPV16 PsV binding (65). These data were strengthened by results obtained from L1 surface staining by flow cytometry analysis (Fig. 7B, 1 h p.e.), which also showed comparable virus binding in control and OBSL1-depleted cells. After 24 h of incubation with PsV, the amount of surface-bound viral particles decreased to 60% of the initial binding level (Fig. 7B, third bar) in control-treated cells. In contrast, 100% of surface-bound viral particles were still detectable at the cell surface after 24 h of incubation in OBSL1-depleted cells, indicating a lack of HPV16 endocytosis (Fig. 7B, last bar), and were still sensitive for proteinase K digestion, as shown in Fig. 7C and D. Additionally, quantitative immunofluorescence analysis of nonpermeabilized cells revealed a 3-fold-higher level of viral particles on the cell surface of OBSL1-depleted cells than on control cells at 24 h after PsV exposure (Fig. 7E). Taken together, these results demonstrate that OBSL1 is required for virus endocytosis. As an additional control experiment, we analyzed the PsV internalization and capsid disassembly/degradation in endosomes of OBSL1-depleted cells using the disassembly specific anti-L1 MAb 33L1-7 (Fig. 7F). This antibody binds to an epitope revealed only after disassembly in acidified endosomes (27, 38). Control or OBSL1 siRNA-transfected cells were exposed to HPV16 PsV for 7 h. Control cells showed a high degree of 33L1-7 staining, whereas the L1-7 signal was strongly decreased, to 32%, in OBSL1 knockdown cells, further supporting the data showing that OBSL1 is required for HPV16 endocytosis.
FIG 7.
OBSL1 is involved in HPV16 internalization but not in cell surface binding. HaCaT cells were transfected for 48 h with control or OBSL1-specific siRNAs and subsequently exposed to HPV16 PsV. (A) Primary HPV16 cell surface binding is not affected by OBSL1 knockdown. siRNA-transfected cells were incubated with HPV16 PsV for 1 h at 4°C. Cell-bound PsVs were detected in cell lysates by Western blotting using the L1-specific antibody 312F (marker lane was removed). As an internal control, HaCaT cells were transfected with control siRNA and then treated with polyethyleneimine (PEI), an inhibitor of HPV16 binding. Relative band intensities were quantified densitometrically. The L1 expression level normalized to β-actin in control cells was set to 100%. (B) siRNA-transfected cells were exposed to HPV16 PsV for 1 or 24 h as indicated. The amount of surface-bound PsV was measured by flow cytometry using polyclonal L1 antibody K75. The mean fluorescence intensity of cells incubated with HPV16 PsV was adjusted to 100%. (C) siRNA-transfected cells were exposed to HPV16 PsV for 24 h. Noninternalized viral particles were removed by protease digestion (+ Proteinase K), and internalized particles were analyzed by Western blotting using the L1-specific antibody 312F. (D) Relative band intensities were quantified densitometrically: L1 level after protease digestion (+ Proteinase K) normalized to L1 input (− Proteinase K). (E) Cells were treated with siRNAs and HPV16 PsV as for panel C. The amount of surface-bound PsV was measured by quantitative fluorescence microscopy using L1 antibody K75. Representative images are shown on the left. For quantification of surface-bound viral particles, the mean K75 intensity of at least 100 cells was measured. Quantitative analysis was done using ImageJ script. The results of four independent experiments normalized to control siRNA-treated and PsV exposed cells are presented. (F) siRNA-transfected cells were exposed to HPV16 PsV for 7 h. Virus capsid disassembly was analyzed by immunofluorescence using monoclonal antibody L1-7 recognizing an epitope accessible only in internalized and disassembled capsids. Representative images are shown on the left. For quantification of viral capsid disassembly, L1-7-positive pixels of at least 100 cells were analyzed using an ImageJ script. The results of four independent experiments normalized to results for control siRNA-treated and infected cells are given. *, P < 0.05 compared to the value for the control. p.e., postexposure.
DISCUSSION
HPV16 entry and the role of the minor capsid protein L2 therein are not fully understood. Here we present four putative cellular interaction partners of HPV16 L2 identified by a yeast two-hybrid screen: gelsolin, dysadherin, Cep68, and OBSL1. The importance of these proteins in HPV16 infection was studied by siRNA-mediated knockdown experiments in HPV16 PsV gene transduction analyses. We discovered OBSL1 as a novel L2 binding protein involved in the poorly characterized endocytic pathway that is used by the oncogenic HPV types 16, 18, and 31.
Depletion of the putative L2-binding proteins in HaCaT cells demonstrated that the actin-binding protein gelsolin was not required for gene transduction by HPV16 PsV. The interaction between L2 and gelsolin may play a role during the HPV replication cycle, as it was shown that HPV16 E7 interacts with gelsolin during postinfective carcinogenesis, resulting in increased cell survival (73). Besides gelsolin, the membrane glycoprotein dysadherin had no or only a minor influence on HPV16 PsV entry but could still be relevant for HPV16 morphogenesis. In contrast, cellular depletion of Cep68 led to a significant reduction of HPV16 PsV gene transduction. It has been shown that Cep68 affects centrosome cohesion, decorates centrioles, and dissociates from centrosomes during mitosis (74). HPV16 requires cell cycle progression for the complex of L2/viral DNA (vDNA) to enter the host cell nucleus (52, 75). Therefore, the involvement of L2/Cep68 interaction in nuclear entry is conceivable and requires further investigation. More importantly, we found the cytoskeletal adaptor protein OBSL1 as a proviral host cell factor. Cellular depletion of OBSL1 correlated with reduction of HPV16, -18, and -31 PsV gene transductions in HeLa and HaCaT cells. These data demonstrate that this cellular protein plays a general role in infections by various oncogenic human papillomaviruses.
OBSL1 has already been identified in numerous tissues and was described as a membrane-interacting protein with suggested functions similar to those of obscurin, titin, and myomesin. This may allow OBSL1 to function as a cytoskeletal adaptor that links cellular support networks, including the intermediate filament, microfilament, or actin filament system, to membrane-bound structures. The localization of OBSL1 was reminiscent of that of plectin, another cytoskeletal adaptor protein (56). In line with these results, our studies showed the expression of OBSL1 in cervical tissue and plasma membrane localization in keratinocytes. In addition, HPV16 gene transduction of primary keratinocytes was highly regulated by OBSL1, further supporting its biological relevance.
To confirm OBSL1 as a novel binding partner of the minor capsid protein L2, complex formation was demonstrated by coimmunofluorescence and coimmunoprecipitation studies with eukaryotic cells after overexpression of L2 or after PsV exposure. In contrast to reports suggesting OBSL1 localization at the Golgi apparatus (65), no colocalization between the Golgi and OBSL1 was detected in HaCaT cells. Instead, we observed association of OBSL1 with the actin cytoskeleton, the tetraspanin CD151—both required for papillomavirus endocytosis—and colocalization of OBSL1 and L2 at the plasma membrane during HPV16 PsV entry, suggesting a role for OBSL1 in early events of HPV16 entry. We assigned the function of OBSL1 to a postbinding and pre-endocytosis step during viral infection, since the depletion resulted in strong inhibition of viral endocytosis while cell binding was unaffected. These data implicate OBSL1 as part of the entry receptor complex. Interestingly, cyclophilin and annexin A2, two proteins involved in HPV16 entry, have been identified in the OBSL1 interactome (76). Cyclophilin mediates conformational changes of the viral capsid on the cell surface, enabling L2 exposure (22), and annexin A2 is demonstrated to be an L2-specific receptor (29, 30). It will be interesting to demonstrate whether L2 becomes a transmembrane protein on the cell surface as shown for cytosolic localized amino acids of endosomal L2 (39). This translocation may be mediated by the L2-specific receptor annexin A2, since annexins were discovered to bind to the inner and outer leaflet of membranes and able to insert into lipid bilayers (77). The accessibility of L2 on the cytosolic side would allow the interaction with cytosolic OBSL1. Our data indicate that HPV16 forms a complex with OBSL1 at the cell surface in an L2-dependent manner. More specifically, OBSL1 interacts with L2 amino acids 280 to 473 in yeast, suggesting association with the cytoskeletal adaptor protein via the C terminus of L2. This part of L2 exhibits membrane-destabilizing activity and is exposed on the cytoplasmic side of membranes during infection (39, 54). Nevertheless, we cannot exclude a bivalent function for OBSL1. It may also be involved in later steps of infection, as was shown for furin and cyclophilin (22, 23, 36). Additionally, studying the role of OBSL1 during infection with native HPV16 may be of interest since the requirements of HSPG binding and furin cleavage differs between HPV PsV and native viruses (15, 25, 78).
The model of OBSL1 as a part of the entry receptor complex is supported not only by HPV16 PsV coimmunoprecipitation and the requirement of OBSL1 for PsV endocytosis but also by OBSL1/CD151 and OBSL1/actin coimmunofluorescence analyses. We found an association between OBSL1 and the tetraspanin CD151, another protein of the second receptor complex and a mediator of HPV endocytosis (10, 27, 28, 33). Furthermore, the actin cytoskeleton was shown to be required for HPV transport on the cell surface and the virus entry into the cell (32, 69, 79). It was also shown to be regulated by CD151 (80, 81). In this regard, our data suggest that OBSL1 is a membrane-interacting protein and cytoskeletal adaptor supporting the connection between OBSL1, the HPV entry receptor complex, and actin as a precondition for HPV16 internalization.
Combined, our results identified OBSL1 as a novel L2-specific HPV16 interaction partner that is involved in gene transduction of epithelial cells by different oncogenic HPV subtypes and in endocytosis of HPV16 particles. Future research is needed to determine whether this interaction occurs on the plasma membrane in the early steps of infection or whether OBSL1 is associated with viral endocytosis complexes supporting virus internalization without direct L2 contact.
ACKNOWLEDGMENTS
We thank Andrew Woodham, Laura Fast, and Snjezana Mikulicic for their comments on the manuscript, Bob Persiko for proofreading, and Anna Lena Augustin for technical support.
E.W., G.A.S., and L.F. conceived and designed the experiments. E.W., L.H., F.B., M.A.S., G.A.S., I.N., K.K., and L.F. performed the experiments. E.W., L.H., F.B., M.A.S., G.A.S., W.M.K., and L.F. analyzed the data. E.W., M.A.S., W.M.K., and L.F. wrote the main manuscript text. All authors reviewed the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 3.Schiller JT, Lowy DR. 2012. Understanding and learning from the success of prophylactic human papillomavirus vaccines. Nat Rev Microbiol 10:681–692. doi: 10.1038/nrmicro2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. 1991. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J 60:1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck CB, Cheng N, Thompson CD, Lowy DR, Steven AC, Schiller JT, Trus BL. 2008. Arrangement of L2 within the papillomavirus capsid. J Virol 82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck CB, Day PM, Trus BL. 2013. The papillomavirus major capsid protein L1. Virology 445:169–174. doi: 10.1016/j.virol.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Florin L, Sapp M, Spoden GA. 2012. Host-cell factors involved in papillomavirus entry. Med Microbiol Immunol 201:437–448. doi: 10.1007/s00430-012-0270-1. [DOI] [PubMed] [Google Scholar]
- 8.Raff AB, Woodham AW, Raff LM, Skeate JG, Yan L, Da Silva DM, Schelhaas M, Kast WM. 2013. The evolving field of human papillomavirus receptor research: a review of binding and entry. J Virol 87:6062–6072. doi: 10.1128/JVI.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JW, Roden RBS. 2013. L2, the minor capsid protein of papillomavirus. Virology 445:175–186. doi: 10.1016/j.virol.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheffer KD, Berditchevski F, Florin L. 2014. The tetraspanin CD151 in papillomavirus infection. Viruses 6:893–908. doi: 10.3390/v6020893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joyce J, Tung J, Przysiecki C, Cook J, Lehman E, Sands J, Jansen K, Keller P. 1999. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J Biol Chem 274:5810. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- 12.Giroglou T, Florin L, Schafer F, Streeck R, Sapp M. 2001. Human papillomavirus infection requires cell surface heparan sulfate. J Virol 75:1565. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath CAJ, Boulet GAV, Renoux VM, Delvenne PO, Bogers J-PJ. 2010. Mechanisms of cell entry by human papillomaviruses: an overview. Virol J 7:11. doi: 10.1186/1743-422X-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letian T, Tianyu Z. 2010. Cellular receptor binding and entry of human papillomavirus. Virol J 7:2. doi: 10.1186/1743-422X-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz L, Meyers C. 2013. Differential dependence on host cell glycosaminoglycans for infection of epithelial cells by high-risk HPV types. PLoS One 8:e68379. doi: 10.1371/journal.pone.0068379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culp TD, Budgeon LR, Marinkovich MP, Meneguzzi G, Christensen ND. 2006. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J Virol 80:8940–8950. doi: 10.1128/JVI.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards KF, Mukherjee S, Bienkowska-Haba M, Pang J, Sapp M. 2014. Human papillomavirus species-specific interaction with the basement membrane-resident non-heparan sulfate receptor. Viruses 6:4856–4879. doi: 10.3390/v6124856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selinka H-C, Florin L, Patel HD, Freitag K, Schmidtke M, Makarov VA, Sapp M. 2007. Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol 81:10970–10980. doi: 10.1128/JVI.00998-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta J, Bienkowska-Haba M, Ortega ME, Patel HD, Bodevin S, Spillmann D, Bishop B, Sapp M, Chen XS. 2011. Structural basis of oligosaccharide receptor recognition by human papillomavirus. J Biol Chem 286:2617–2624. doi: 10.1074/jbc.M110.160184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerqueira C, Samperio Ventayol P, Vogeley C, Schelhaas M. 2015. Kallikrein-8 proteolytically processes human papillomaviruses in the extracellular space to facilitate entry into host cells. J Virol 89:7038–7052. doi: 10.1128/JVI.00234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards KF, Bienkowska-Haba M, Dasgupta J, Chen XS, Sapp M. 2013. Multiple heparan sulfate binding site engagements are required for the infectious entry of human papillomavirus type 16. J Virol 87:11426–11437. doi: 10.1128/JVI.01721-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bienkowska-Haba M, Patel HD, Sapp M. 2009. Target cell cyclophilins facilitate human papillomavirus type 16 infection. PLoS Pathog 5:e1000524. doi: 10.1371/journal.ppat.1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards RM, Lowy DR, Schiller JT, Day PM. 2006. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A 103:1522–1527. doi: 10.1073/pnas.0508815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Day PM, Lowy DR, Schiller JT. 2008. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol 82:12565–12568. doi: 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz L, Biryukov J, Conway MJ, Meyers C. 2015. Cleavage of the HPV16 minor capsid protein L2 during virion morphogenesis ablates the requirement for cellular furin during de novo infection. Viruses 7:5813–5830. doi: 10.3390/v7112910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bronnimann MP, Calton CM, Chiquette SF, Li S, Lu M, Chapman JA, Bratton KN, Schlegel AM, Campos SK. 2016. Furin cleavage of L2 during papillomavirus infection: minimal dependence on cyclophilins. J Virol 90:6224–6234. doi: 10.1128/JVI.00038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L. 2008. Clathrin- and caveolin-independent entry of human papillomavirus type 16—involvement of tetraspanin-enriched microdomains (TEMs). PLoS One 3:e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheffer KD, Gawlitza A, Spoden GA, Zhang XA, Lambert C, Berditchevski F, Florin L. 2013. Tetraspanin CD151 mediates papillomavirus type 16 endocytosis. J Virol 87:3435–3446. doi: 10.1128/JVI.02906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodham AW, Da Silva DM, Skeate JG, Raff AB, Ambroso MR, Brand HE, Isas JM, Langen R, Kast WM. 2012. The S100A10 subunit of the annexin A2 heterotetramer facilitates L2-mediated human papillomavirus infection. PLoS One 7:e43519. doi: 10.1371/journal.pone.0043519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dziduszko A, Ozbun MA. 2013. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol 87:7502–7515. doi: 10.1128/JVI.00519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fausch SC, Da Silva DM, Kast WM. 2003. Differential uptake and cross-presentation of human papillomavirus virus-like particles by dendritic cells and Langerhans cells. Cancer Res 63:3478–3482. [PubMed] [Google Scholar]
- 32.Schelhaas M, Shah B, Holzer M, Blattmann P, Kühling L, Day PM, Schiller JT, Helenius A. 2012. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raft-independent endocytosis. PLoS Pathog 8:e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spoden G, Kühling L, Cordes N, Frenzel B, Sapp M, Boller K, Florin L, Schelhaas M. 2013. Human papillomavirus types 16, 18, and 31 share similar endocytic requirements for entry. J Virol 87:7765–7773. doi: 10.1128/JVI.00370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Homsi Y, Schloetel J-G, Scheffer KD, Schmidt TH, Destainville N, Florin L, Lang T. 2014. The extracellular δ-domain is essential for the formation of CD81 tetraspanin webs. Biophys J 107:100–113. doi: 10.1016/j.bpj.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gräßel L, Fast LA, Scheffer KD, Boukhallouk F, Spoden GA, Tenzer S, Boller K, Bago R, Rajesh S, Overduin M, Berditchevski F, Florin L. 2016. The CD63-syntenin-1 complex controls post-endocytic trafficking of oncogenic human papillomaviruses. Sci Rep 6:32337. doi: 10.1038/srep32337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienkowska-Haba M, Williams C, Kim SM, Garcea RL, Sapp M. 2012. Cyclophilins facilitate dissociation of the hpv16 capsid protein L1 from the L2/DNA complex following virus entry. J Virol 86:9875–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie M-A, Poffenberger AC, Nelson CDS, Atwood WJ, Dimaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Müller KH, Spoden GA, Scheffer KD, Brunnhöfer R, De Brabander JK, Maier ME, Florin L, Muller CP. 2014. Inhibition of cellular V-ATPase impairs human papillomavirus uncoating and infection. Antimicrob Agents Chemother 58:2905–2911. doi: 10.1128/AAC.02284-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiGiuseppe S, Keiffer TR, Bienkowska-Haba M, Luszczek W, Guion LGM, Müller M, Sapp M. 2015. Topography of the human papillomavirus minor capsid protein L2 during vesicular trafficking of infectious entry. J Virol 89:10442–10452. doi: 10.1128/JVI.01588-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang R, Yutzy W, Viscidi R, Roden R. 2003. Interaction of L2 with β-actin directs intracellular transport of papillomavirus and infection. J Biol Chem 278:12546. doi: 10.1074/jbc.M208691200. [DOI] [PubMed] [Google Scholar]
- 41.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, Dimaio D. 2015. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog 11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Day PM, Thompson CD, Schowalter RM, Lowy DR, Schiller JT. 2013. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J Virol 87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Florin L, Becker KA, Lambert C, Nowak T, Sapp C, Strand D, Streeck RE, Sapp M. 2006. Identification of a dynein interacting domain in the papillomavirus minor capsid protein l2. J Virol 80:6691–6696. doi: 10.1128/JVI.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider MA, Spoden GA, Florin L, Lambert C. 2011. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 45.Bergant Marušič M, Ozbun MA, Campos SK, Myers MP, Banks L. 2012. Human papillomavirus L2 facilitates viral escape from late endosomes via sorting nexin 17. Traffic 13:455–467. doi: 10.1111/j.1600-0854.2011.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broniarczyk J, Bergant M, Goździcka-Józefiak A, Banks L. 2014. Human papillomavirus infection requires the TSG101 component of the ESCRT machinery. Virology 460-461:83–90. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Kazakov T, Popa A, Dimaio D. 2014. Vesicular trafficking of incoming human papillomavirus 16 to the Golgi apparatus and endoplasmic reticulum requires γ-secretase activity. mBio 5:e01777-14. doi: 10.1128/mBio.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pim D, Broniarczyk J, Bergant M, Playford MP, Banks L. 2015. A novel PDZ domain interaction mediates the binding between human papillomavirus 16 L2 and sorting nexin 27 and modulates virion trafficking. J Virol 89:10145–10155. doi: 10.1128/JVI.01499-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day PM, Roden RB, Lowy DR, Schiller JT. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol 72:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Florin L, Schäfer F, Sotlar K, Streeck RE, Sapp M. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 295:97–107. doi: 10.1006/viro.2002.1360. [DOI] [PubMed] [Google Scholar]
- 51.Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Müller M, Streeck RE, Sapp M. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol 78:5546–5553. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aydin I, Weber S, Snijder B, Samperio Ventayol P, Kühbacher A, Becker M, Day PM, Schiller JT, Kann M, Pelkmans L, Helenius A, Schelhaas M. 2014. Large scale RNAi reveals the requirement of nuclear envelope breakdown for nuclear import of human papillomaviruses. PLoS Pathog 10:e1004162. doi: 10.1371/journal.ppat.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bund T, Spoden GA, Koynov K, Hellmann N, Boukhallouk F, Arnold P, Hinderberger D, Florin L. 2014. A L2 SUMO interacting motif is important for PML localization and infection of human papillomavirus type 16. Cell Microbiol 16:1179–1200. doi: 10.1111/cmi.12271. [DOI] [PubMed] [Google Scholar]
- 54.Kämper N, Day PM, Nowak T, Selinka H-C, Florin L, Bolscher J, Hilbig L, Schiller JT, Sapp M. 2006. A membrane-destabilizing peptide in capsid protein L2 is required for egress of papillomavirus genomes from endosomes. J Virol 80:759–768. doi: 10.1128/JVI.80.2.759-768.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bronnimann MP, Chapman JA, Park CK, Campos SK. 2013. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J Virol 87:464–473. doi: 10.1128/JVI.01539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geisler SB, Robinson D, Hauringa M, Raeker MO, Borisov AB, Westfall MV, Russell MW. 2007. Obscurin-like 1, OBSL1, is a novel cytoskeletal protein related to obscurin. Genomics 89:521–531. doi: 10.1016/j.ygeno.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Litterman N, Ikeuchi Y, Gallardo G, O'Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A. 2011. An OBSL1-Cul7 Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol 9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanson D, Murray PG, Sud A, Temtamy SA, Aglan M, Superti-Furga A, Holder SE, Urquhart J, Hilton E, Manson FDC, Scambler P, Black GCM, Clayton PE. 2009. The primordial growth disorder 3-M syndrome connects ubiquitination to the cytoskeletal adaptor OBSL1. Am J Hum Genet 84:801–806. doi: 10.1016/j.ajhg.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leder C, Kleinschmidt JA, Wiethe C, Müller M. 2001. Enhancement of capsid gene expression: preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J Virol 75:9201–9209. doi: 10.1128/JVI.75.19.9201-9209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volpers C, Sapp M, Snijders PJ, Walboomers JM, Streeck RE. 1995. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J Gen Virol 76(Part 11):2661–2667. [DOI] [PubMed] [Google Scholar]
- 61.Rommel O, Dillner J, Fligge C, Bergsdorf C, Wang X, Selinka H-C, Sapp M. 2005. Heparan sulfate proteoglycans interact exclusively with conformationally intact HPV L1 assemblies: basis for a virus-like particle ELISA. J Med Virol 75:114–121. doi: 10.1002/jmv.20245. [DOI] [PubMed] [Google Scholar]
- 62.Knappe M, Bodevin S, Selinka H-C, Spillmann D, Streeck RE, Chen XS, Lindahl U, Sapp M. 2007. Surface-exposed amino acid residues of HPV16 L1 protein mediating interaction with cell surface heparan sulfate. J Biol Chem 282:27913–27922. doi: 10.1074/jbc.M705127200. [DOI] [PubMed] [Google Scholar]
- 63.Sapp M, Kraus U, Volpers C, Snijders PJ, Walboomers JM, Streeck RE. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J Gen Virol 75(Part 12):3375–3383. [DOI] [PubMed] [Google Scholar]
- 64.Buck C, Pastrana D, Lowy D, Schiller J. 2004. Efficient intracellular assembly of papillomaviral vectors. J Virol 78:751. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spoden GA, Besold K, Krauter S, Plachter B, Hanik N, Kilbinger AFM, Lambert C, Florin L. 2012. Polyethylenimine is a strong inhibitor of human papillomavirus and cytomegalovirus infection. Antimicrob Agents Chemother 56:75–82. doi: 10.1128/AAC.05147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider MA, Scheffer KD, Bund T, Boukhallouk F, Lambert C, Cotarelo C, Pflugfelder GO, Florin L, Spoden GA. 2013. The transcription factors TBX2 and TBX3 interact with human papillomavirus 16 (HPV16) L2 and repress the long control region of human papillomaviruses. J Virol 87:4461–4474. doi: 10.1128/JVI.01803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Florin L, Sapp C, Streeck R, Sapp M. 2002. Assembly and translocation of papillomavirus capsid proteins. J Virol 76:10009–10014. doi: 10.1128/JVI.76.19.10009-10014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milne RSB, Nicola AV, Whitbeck JC, Eisenberg RJ, Cohen GH. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J Virol 79:6655–6663. doi: 10.1128/JVI.79.11.6655-6663.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bienkowska-Haba M, Sapp M. 2011. The cytoskeleton in papillomavirus infection. Viruses 3:260–271. doi: 10.3390/v3030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kieback E, Müller M. 2006. Factors influencing subcellular localization of the human papillomavirus L2 minor structural protein. Virology 345:199–208. doi: 10.1016/j.virol.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 71.Lin Z, Yemelyanova AV, Gambhira R, Jagu S, Meyers C, Kirnbauer R, Ronnett BM, Gravitt PE, Roden RBS. 2009. Expression pattern and subcellular localization of human papillomavirus minor capsid protein L2. Am J Pathol 174:136–143. doi: 10.2353/ajpath.2009.080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Becker KA, Florin L, Sapp C, Sapp M. 2003. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology 314:161–167. doi: 10.1016/S0042-6822(03)00447-1. [DOI] [PubMed] [Google Scholar]
- 73.Mileo AM, Abbruzzese C, Vico C, Bellacchio E, Matarrese P, Ascione B, Federico A, Bianca Della S, Mattarocci S, Malorni W, Paggi MG. 2013. The human papillomavirus-16 E7 oncoprotein exerts antiapoptotic effects via its physical interaction with the actin-binding protein gelsolin. Carcinogenesis 34:2424–2433. doi: 10.1093/carcin/bgt192. [DOI] [PubMed] [Google Scholar]
- 74.Graser S, Stierhof Y-D, Nigg EA. 2007. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci 120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 75.Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. 2009. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 5:e1000318. doi: 10.1371/journal.ppat.1000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanson D, Stevens A, Murray PG, Black GCM, Clayton PE. 2014. Identifying biological pathways that underlie primordial short stature using network analysis. J Mol Endocrinol 52:333–344. doi: 10.1530/JME-14-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lizarbe MA, Barrasa JI, Olmo N, Gavilanes F, Turnay J. 2013. Annexin-phospholipid interactions. Functional implications. Int J Mol Sci 14:2652–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Patterson NA, Smith JL, Ozbun MA. 2005. Human papillomavirus type 31b infection of human keratinocytes does not require heparan sulfate. J Virol 79:6838–6847. doi: 10.1128/JVI.79.11.6838-6847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schelhaas M, Ewers H, Rajamäki M-L, Day PM, Schiller JT, Helenius A. 2008. Human papillomavirus type 16 entry: retrograde cell surface transport along actin-rich protrusions. PLoS Pathog 4:e1000148. doi: 10.1371/journal.ppat.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. 2003. CD151 regulates epithelial cell-cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J Cell Biol 163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson JL, Winterwood N, DeMali KA, Stipp CS. 2009. Tetraspanin CD151 regulates RhoA activation and the dynamic stability of carcinoma cell-cell contacts. J Cell Sci 122:2263–2273. doi: 10.1242/jcs.045997. [DOI] [PubMed] [Google Scholar]