ABSTRACT
Primary infection with varicella-zoster virus (VZV), a neurotropic alphaherpesvirus, results in varicella. VZV establishes latency in the sensory ganglia and can reactivate later in life to cause herpes zoster. The relationship between VZV and its host during acute infection in the sensory ganglia is not well understood due to limited access to clinical specimens. Intrabronchial inoculation of rhesus macaques with simian varicella virus (SVV) recapitulates the hallmarks of VZV infection in humans. We leveraged this animal model to characterize the host-pathogen interactions in the ganglia during both acute and latent infection by measuring both viral and host transcriptomes on days postinfection (dpi) 3, 7, 10, 14, and 100. SVV DNA and transcripts were detected in sensory ganglia 3 dpi, before the appearance of rash. CD4 and CD8 T cells were also detected in the sensory ganglia 3 dpi. Moreover, lung-resident T cells isolated from the same animals 3 dpi also harbored SVV DNA and transcripts, suggesting that T cells may be responsible for trafficking SVV to the ganglia. Transcriptome sequencing (RNA-Seq) analysis showed that cessation of viral transcription 7 dpi coincides with a robust antiviral innate immune response in the ganglia. Interestingly, a significant number of genes that play a critical role in nervous system development and function remained downregulated into latency. These studies provide novel insights into host-pathogen interactions in the sensory ganglia during acute varicella and demonstrate that SVV infection results in profound and sustained changes in neuronal gene expression.
IMPORTANCE Many aspects of VZV infection of sensory ganglia remain poorly understood, due to limited access to human specimens and the fact that VZV is strictly a human virus. Infection of rhesus macaques with simian varicella virus (SVV), a homolog of VZV, provides a robust model of the human disease. Using this model, we show that SVV reaches the ganglia early after infection, most likely by T cells, and that the induction of a robust innate immune response correlates with cessation of virus transcription. We also report significant changes in the expression of genes that play an important role in neuronal function. Importantly, these changes persist long after viral replication ceases. Given the homology between SVV and VZV, and the genetic and physiological similarities between rhesus macaques and humans, our results provide novel insight into the interactions between VZV and its human host and explain some of the neurological consequences of VZV infection.
INTRODUCTION
Varicella-zoster virus (VZV) is a neurotropic alphaherpesvirus and the causative agent of varicella (chickenpox) (1). VZV establishes latency in the sensory ganglia and can reactivate later in life to cause herpes zoster (shingles), a painful disease that affects almost 1 million individuals in the United States alone (2). VZV is transmitted through the inhalation of virus-laden saliva droplets or by direct contact with the infectious fluid from vesicles (1). It is believed that VZV replicates in the upper respiratory tract, including the tonsillar lymph nodes, where it infects memory CD4 T cells that disseminate the virus to cutaneous sites where infection of keratinocytes results in varicella exanthem (1).
Two models have been put forth to explain how VZV reaches the ganglia. One model proposes that cell-free virus in varicella lesions infects the nerve terminals and travels to the ganglia via retrograde transport as described for the closely related herpes simplex virus (HSV) (3). A second model proposes that VZV is transported to the ganglia via the hematogenous route within infected T cells (4, 5). The contribution of retrograde axonal transport versus hematogenous spread to VZV dissemination to the ganglia in vivo is poorly understood. Furthermore, whether VZV establishes latency immediately or replicates first in sensory ganglia remains unclear. Experiments using human fetal dorsal root ganglion xenografts directly inoculated with VZV-infected fibroblasts in hu-SCID mice have shown VZV transcription in the ganglia until 14 days postinfection (dpi) (6). However, it is unknown whether viral gene expression is sustained for a similar duration in an immunocompetent host.
Similarly, the host response that develops in the ganglia during primary VZV infection remains poorly understood. Previous studies report the presence of T cells (primarily noncytolytic CD8+ T cells), B cells, macrophages, and natural killer cells within ganglia isolated from individuals who suffered from herpes zoster shortly before they died from other causes (7–9). These studies also showed increased expression of chemokine CXCL10, which binds to CXCR3 to induce migration of memory T cells and natural killer cells (8), as well as increased expression of major histocompatibility complex class I (MHC-I) and MHC-II molecules (9). These observations suggest that both innate and adaptive immunity play a critical role in the resolution of VZV reactivation. However, it is uncertain if similar responses occur in the ganglia during acute infection.
Our incomplete understanding of host-pathogen interactions in the ganglia during acute VZV infection is due to the difficulty in obtaining clinical samples and the strict human host specificity of VZV. Intrabronchial infection of rhesus macaques (RM) with the closely related simian varicella virus (SVV) recapitulates hallmarks of primary VZV infection, including the appearance of varicella, the development of cellular and humoral immunity, and establishment of latency in sensory ganglia (10). Moreover, as described for VZV, SVV can reactivate during episodes of immune suppression and stress (11–13), and this reactivation is accompanied by the upregulation of CXCL10 and T cell infiltration (14). Recent studies using African green monkeys (AGMs) inoculated with SVV-enhanced green fluorescent protein (EGFP) have shown that SVV infects T cells that infiltrate the ganglia during acute infection 9 dpi (15). However, SVV infection of AGMs, in contrast to human VZV infection, results in significant mortality (16), and SVV-GFEP is attenuated compared to the wild type (WT) (15). Moreover, ganglion samples were analyzed only at time points after the appearance of varicella rash (>9 dpi); therefore, the role of T cells versus the axonal route in SVV dissemination to the ganglia remain unclear.
In this study, we leveraged SVV infection of rhesus macaques to interrogate the kinetics by which SVV reaches the ganglia and establishes latency as well as the host response to SVV using ganglia collected from naive animals 3, 7, 10, 14, and 100 days postinfection (dpi). At every time point, we measured viral loads and characterized both viral and host gene expression. We show that SVV DNA and RNA transcripts are detected in the ganglia as early as 3 dpi at the same time as memory T cells. Peripheral T cells from the same animals 3 dpi harbor viral DNA, suggesting a role for these cells in SVV transport to the ganglia. In addition, cessation of viral gene expression in the ganglia coincides with the induction of a robust antiviral innate immune response. Robust changes in expression of genes associated with nervous system development were observed throughout the study, suggesting that SVV infection has a sustained effect on neuronal gene expression long after viral replication ceases.
MATERIALS AND METHODS
Ethics statement.
The study was carried out in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Office of Animal Welfare, and the United States Department of Agriculture (17). All animal work was approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee (IACUC protocol 0779). The ONPRC has been continuously accredited by the American Association for Accreditation of Laboratory Animal Care since 1974 (PHS/OLAW Animal Welfare Assurance A3304-01). Animals were either housed single or paired in caging that allowed for social interactions in a temperature- and humidity-controlled environment. Food and water were available ad libitum, and enrichment was provided daily. All procedures were carried out under ketamine anesthesia in the presence of veterinary staff, and all efforts were made to minimize animal suffering. Animals were euthanized in accordance with the recommendations of the American Veterinary Medical Association guidelines for euthanasia.
Cells and virus.
SVV was propagated in primary rhesus fibroblasts (1° RF) (a generous gift from Scott Wong, Oregon National Primate Center) at 37°C in 175-cm2 flasks with Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). SVV-infected 1° RF were frozen in FetalPlex with 10% dimethyl sulfoxide (DMSO), stored in liquid nitrogen (LN2), and assayed by plaque assay.
Animals and sample collection.
Fifteen colony-bred rhesus macaques (RM; Macaca mulatta) 3 to 5 years of age and of Indian origin were used in these studies. Twelve were inoculated intrabronchially with 4 × 105 PFU of wild-type SVV as previously described (10) and subsequently euthanized at 3 (n = 3), 7 (n = 2), 10 (n = 2), 14 (n = 3), and 100 (n = 2) dpi. An additional 3 animals served as noninfected controls. Animals were housed and handled in accordance with the guidance of the Oregon National Primate Research Center Institutional Animal Care and Use Committee. These animals were previously used to characterize specificity of the anti-SVV T cell response (18). Blood, bronchial alveolar lavage (BAL) fluid, and sensory ganglia were collected from all animals. Blood and BAL fluid were used to measure viral loads and isolate CD20, CD4, and CD8 T cell populations for additional viral load and viral transcription analyses. Trigeminal (TG) and dorsal root ganglia (cervical, thoracic, and lumbar-sacral [DRG-C, DRG-T, and DRG-LS, respectively]) were collected from each animal and were divided to carry out multiple assays described in greater detail below: DNA extraction for viral loads, RNA extraction for viral transcription, digestion and isolation of mononuclear cells, and paraffin embedding for immunohistochemistry (IHC) analysis. DRG ganglia from naive animals were used for transcriptome sequencing (RNA-Seq) analysis. Skin biopsy specimens were collected from the trunk of the animals at 3 dpi.
Isolation of immune cells from ganglia.
Ganglia were digested in 150 U/ml collagenase for 1 h at 37°C with shaking and then homogenized using a cell strainer to generate single-cell suspensions. Cells were pelleted at 2,000 rpm for 5 min and then resuspended in 30% Percoll gradient and spun for 15 min at 2,000 rpm to isolate mononuclear cells. Cells were then stained with antibodies directed against surface markers: CD4 (Tonbo Biosciences, San Diego, CA), CD8β (Beckman Coulter, Brea, CA), CD28 (Tonbo Biosciences), CD95 (BioLegend, San Diego, CA), and CCR7 (BD Pharmingen, San Diego, CA) to delineate naive and memory T cell subsets, as well as CD20 (Southern Biotech, Birmingham, AL), IgD (Southern Biotech, Birmingham, AL), and CD27 (BioLegend) to delineate B cell subsets as previously described (19). Samples were analyzed using the LSRII instrument (Becton, Dickinson and Company, San Jose, CA) and FlowJo software (TreeStar, Ashland, OR). Due to the small amount of lymphocytes in the ganglia, half of the DRG-C, DRG-T, and DRG-LS from the same animal were pooled in order to get a sufficient number of events to quantify the frequency of lymphocytes. The same number of DRG was used for every animal.
Purification of CD4/CD8 T cells.
CD4 and CD8 T cells were isolated from BAL fluid and peripheral blood mononuclear cell (PBMC) samples using magnetic cell sorting (MACS). BAL fluid and PBMCs were first incubated with CD4 microbeads (Miltenyi Biotec, San Diego, CA) for 20 min at 4°C. Cells were then washed in buffer, and the cell suspension was applied to the magnetic column to capture CD4 T cells. The CD4-negative fraction was then stained with CD8-phycoerythrin (PE) (Beckman Coulter) and incubated with the cells for 20 min in the dark at 4°C. Cells were then washed, and anti-PE beads (Miltenyi Biotec) were added and incubated for 15 min in the dark at 4°C. Cells were then washed, and the cell suspension was applied to a magnetic column to isolate CD8 T cells. Finally, the CD4- and CD8-negative population was incubated with CD20 microbeads (Miltenyi Biotec) to isolate B cells. The purity of the fractions was determined using flow cytometry. All samples used had a purity of >90%.
DNA/RNA extraction and quantitative PCR.
Ganglia and skin tissue were digested in proteinase K solution (20 mg/ml) overnight, and DNA was extracted using the Qiagen genomic DNA extraction kit (Qiagen, Valencia, CA). Viral DNA loads were determined exactly as previously described (10) by real-time PCR using primers and probes specific for ORF21 and the ABI StepOne instruments (Applied Biosystems, Foster City, CA). One microgram of DNA was used for ganglion tissue viral loads. For RNA extraction, ganglion tissue in TRIzol was homogenized using a bead beater and zirconia-silica beads followed by extraction using the PureLink RNA minikit (Ambion, Carlsbad, CA).
Measuring anti-SVV T and B cell responses.
The enzyme-linked immunosorbent spot (ELISpot) assay data were derived from a previous study characterizing the specificity of the T cell response to SVV during acute and latent infection (18). The total number of spot-forming cells (SFC) in PBMCs or BAL fluid was determined by adding up the individual responses to each of the 70 open reading frames (ORFs). SVV-specific IgG titer was determined as previously described (10).
Viral gene expression analysis.
Ten nanograms of total RNA from each sample and from the External RNA Controls Consortium (ERCC; internal positive control used for normalization) were randomly primed for reverse transcription and subsequently amplified with 71 custom viral gene-specific and 10 ERCC-targeted primers for 19 cycles of PCR. Barcoded DNA adapters were ligated to amplicons to allow multiplexing libraries for sequencing. Libraries were amplified onto Ion Sphere particles (ISPs) using the OneTouch 2 instrument (Life Technologies, Carlsbad, CA) and sequenced on the Ion Torrent Proton (Life Technologies, Carlsbad, CA) per the manufacturer's protocol for v3 templating and sequencing chemistries. Sequencing reads that were less than 60 bp long were removed from further analysis, and remaining reads were stringently aligned to the viral genome and ERCC sequences using TMAP, an Ion Torrent Proton specific sequence aligner.
Host transcriptome analysis.
RNA was extracted from ganglia using the Ambion PureLink RNA minikit extraction kit (Life Technologies, Carlsbad, CA). RNA library preparation was done using the New England BioLabs (NEB) Next Ultra Direction RNA Prep kit for Illumina (Ipswich, MA). DNA libraries were then multiplexed and sequenced on the Illumina HiSeq2500 (Illumina, San Diego, CA) platform at single ends (100 bp). All data analysis steps were performed with the RNA-Seq workflow module of the systemPiperR package available on Bioconductor (20, 21). Next-generation sequencing (NGS) quality reports were generated with the seeFastq function defined by the same package. RNA-Seq reads were mapped with the splice-junction-aware short-read alignment suite Bowtie2/Tophat2 (22, 23) against the Macaca mulatta genome sequence downloaded from Ensembl (24). The default parameters of Tophat2 optimized for mammalian genomes were used for the alignments. Raw expression values in the form of gene-level read counts were generated with the summarizeOverlaps function (25). Only reads overlapping the exonic regions of genes were counted, while reads mapping to ambiguous regions of exons from overlapping genes were discarded. Given the nonstranded nature of our RNA-Seq libraries, the read counting was performed in a non-strand-specific manner.
Analysis of differentially expressed genes (DEGs) was performed with the general linear modeling (GLM) method from the edgeR package (26, 27). Differentially expressed genes (DEGs) were defined as those with a fold change (FC) of ≥3 and a false-discovery rate (FDR) of ≤0.01. Enrichment analysis of functional annotations was performed to identify significant gene ontology (GO) processes using MetaCore software (GeneGo, Philadelphia, PA).
Gene validation.
RNA was reverse transcribed using random hexamers and SuperScript IV reverse transcriptase (RT) in the SuperScript IV first-strand synthesis system (Invitrogen, Lithuania) to generate cDNA. TaqMan gene expression assays (Thermo Fisher, Waltham, MA) of candidate genes and the housekeeping gene (RPL32) were used with 50 ng of cDNA and carried out in duplicate on the ABI StepOne instrument (Applied Biosystems). mRNA expression levels were calculated relative to our housekeeping gene (RPL32) using threshold cycle (2−ΔΔCT) calculations.
Immunohistochemistry staining.
Four-micrometer ganglion sections were deparaffinized and rehydrated before antigen retrieval. Antigen retrieval was done using a pressure cooker in citrate buffer for 20 min. The sections were then blocked in 10% bovine serum albumin (BSA) and 1% normal goat serum for 1 h followed by avidin and then biotin for 15 min. Tissues were then stained with primary antibodies CD3 (1:200 dilution; Dako M0452) for labeling T lymphocytes, CD68 (1:75 dilution; Dako) for labeling macrophages, CD20 (1:300 dilution; Dako M0755) for labeling B cells, proteoglycan 4 (PRG4) (1:200 dilution; Antibodies-Online), glial fibrillary acidic protein (GFAP) (1:500 dilution; Dako), and complexin 1 (CPLX1) (1:200 dilution; Proteintech Group). Slides were then incubated with the appropriate secondary antibody and treated with 10% peroxidase in methanol for 10 min in the dark. Color development of antigen staining was done with ImmPACT diaminobenzidine (DAB) (Vector Laboratories, Burlingame, CA), and counterstaining was done with hematoxylin QS (Vector Laboratories, Burlingame, CA). Slides were then covered with coverslips using Omnimount (National Diagnostics, Atlanta, GA). Images were taken on the Leica DM5500 B (Leica Biosystems, Buffalo Grove, IL) microscope at ×40 magnification. Isotype IgG antibody was used as a negative control.
IHC quantification for PRG4 was done using ImageJ software. Optical density (OD) values of DAB (brown) and hematoxylin staining (blue) were taken from three different sections of each slide and then averaged. Ganglia from three different infected animals and the three naive controls were used for each staining.
For hematoxylin and eosin (H&E) stains, 4-μm ganglion sections were deparaffinized with Histo-Clear II (National Diagnostics, Atlanta, GA) and rehydrated. Tissue was then stained with hematoxylin for 2 min followed by eosin for 40 s, hydrated back to Histo-Clear, and then covered with coverslips using Omnimount (National Diagnostics, Atlanta, GA).
Statistical analysis.
Statistical analysis and graphing were conducted through the GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA). Significant values were determined using one-way analysis of variance (ANOVA).
RESULTS
SVV DNA is detected before the appearance of the rash.
To determine the kinetics of viral replication in the lung and in the blood, we performed quantitative PCR (qPCR) using primers and probes against open reading frame 21 (ORF21). SVV replication kinetics and viral loads in the bronchial alveolar lavage (BAL) fluid and whole blood (WB) following intrabronchial infection (Fig. 1A) were similar to those in our previous studies, with higher SVV viral loads detected in the BAL fluid than in peripheral blood (10). SVV-specific cellular and humoral immune responses were detected 7, 10, 14, and 100 days postinfection (dpi) (Fig. 1B). Animals euthanized at 10 and 14 dpi showed a characteristic varicella rash (data not shown).
FIG 1.
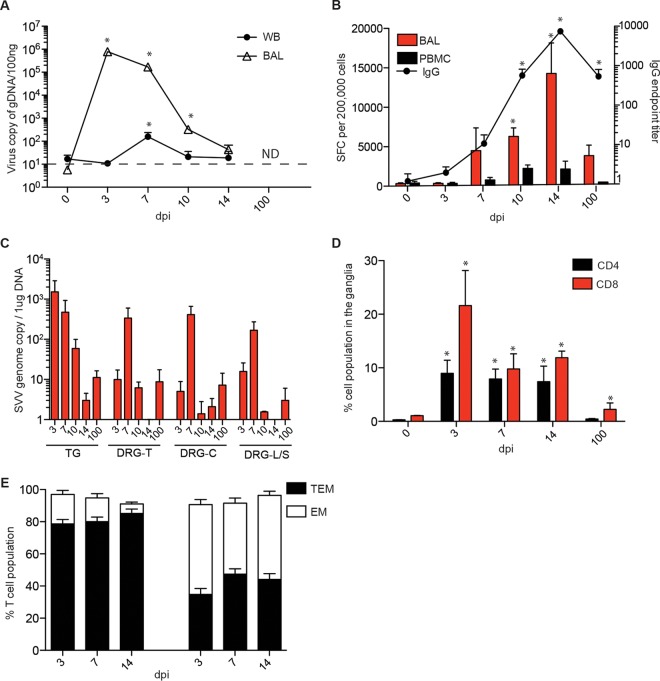
SVV viral DNA and immune infiltrates are detected in sensory ganglia as early as 3 dpi. (A) SVV DNA levels in the BAL fluid and WB were measured by quantitative real-time PCR (100 ng/sample) using primers and probes specific for SVV ORF21 (0 dpi, n = 15; 3 dpi, n = 11; 7 dpi, n = 8; 10 dpi, n = 5; 14 dpi, n = 4; 100 dpi, n = 2) (ND, not detected). Dashed line indicates limit of detection. (B) Frequency of SVV-specific T cells in the BAL fluid was measured using IFN-γ ELISpot assay by enumerating the number of spot-forming cells (SFC) in response to all 69 unique SVV open reading frames as previously described (122) (0 dpi, n = 3; 3 dpi, n = 3; 7 dpi, n = 3; 10 dpi, n = 2; 14 dpi, n = 3; 100 dpi, n = 2). SVV-specific IgG titers were determined using enzyme-linked immunosorbent assay (0 dpi, n = 15; 3 dpi, n = 11; 7 dpi, n = 8; 10 dpi, n = 5; 14 dpi, n = 3; 100 dpi, n = 2). (C) SVV viral DNA was detected in the ganglia using quantitative PCR (1 μg/sample); TG, trigeminal; DRG-T, dorsal root ganglia, thoracic; DRG-C, dorsal root ganglia, cervical; DRG-L/S, dorsal root ganglia, lumbar-sacral (3 dpi, n = 3; 7 dpi, n = 2; 14 dpi, n = 3). No SVV DNA was detected in ganglia from naive animals. (D) DRG-C, -T, and -L/S isolated at the indicated times postinfection were pooled and digested, and mononuclear cells were isolated over a Percoll gradient. Frequencies of CD4 and CD8 T cells were then measured using flow cytometry (0 dpi, n = 2; 3 dpi, n = 3; 7 dpi, n = 2; 14 dpi, n = 3; 100 dpi, n = 2). (E) The percentages of effector memory (EM) or transitional effector memory (TEM) T cells were determined also using flow cytometry (3 dpi, n = 3; 7 dpi, n = 2; 14 dpi, n = 3). *, P < 0.05 compared to day 0.
To determine when SVV reached the ganglia, SVV viral loads were measured in trigeminal ganglia (TG) and dorsal root ganglia (DRG) (cervical, thoracic, and lumbar-sacral) collected from naive animals and from animals euthanized at 3, 7, 10, 14, and 100 dpi. SVV viral DNA was detected in sensory ganglia at 3 dpi, before the appearance of varicella rash. Genome copy numbers were initially higher in the TG than in the DRG (Fig. 1C) but comparable at 7 dpi (Fig. 1C). Since SVV DNA was detected in the ganglia before the appearance of skin lesions and SVV viral DNA was not detected in nonlesioned skin, these data suggest that SVV first reaches the ganglia via means other than retrograde transport from skin lesions.
T cells infiltrate the ganglia during acute SVV infection.
Previous studies have shown a role for T cells in VZV and SVV dissemination (14, 28, 29). To determine whether T cells infiltrate the ganglia early after SVV infection in rhesus macaques, we digested DRG-T, DRG-C, and DRG-LS collected 3, 7, 10, 14, and 100 dpi and analyzed the isolated lymphocytes using flow cytometry and antibodies directed against CD4, CD8, CD28, CD95, and CCR7 in order to delineate naive and memory T cell subsets as well as CD20, CD27, and IgD to delineate naive and memory B cell subsets as recently described (19). Both CD4 and CD8 T cells were detected 3 to 14 dpi (Fig. 1D), and they were highly differentiated into the memory (CD28+/− CD95+ CCR7−) phenotype (Fig. 1E). No T cells were detected in the ganglia isolated from naive animals (day 0) or animals euthanized 100 dpi. No B cells were detected in the ganglia at any time point.
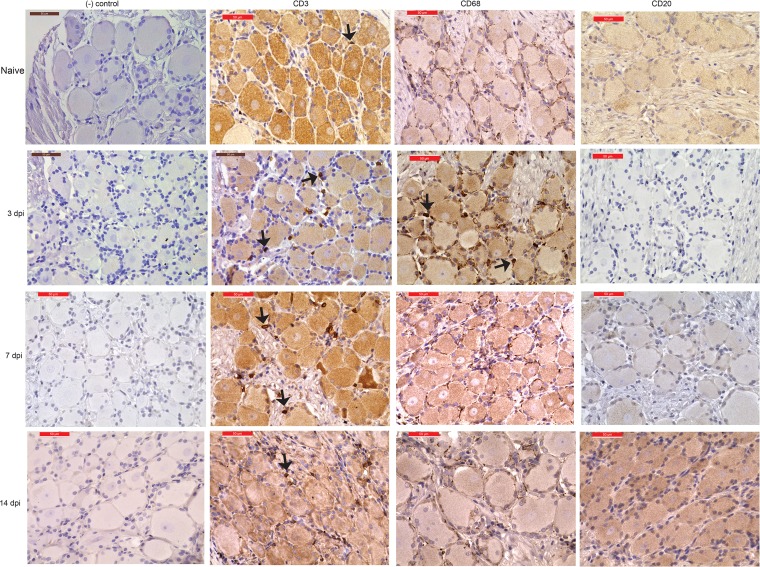
We then carried out immunohistochemistry (IHC) analysis using CD3 antibody to get a better understanding of the spatial distribution of T cells and CD20 antibody to confirm the absence of B cells. Our analysis confirmed the presence of T cells in the ganglia at 3 to 14 dpi (Fig. 2). CD68+ cells were detected on day 3 postinfection (Fig. 2). In line with the flow cytometry data described above, no CD20+ B cells were detected at any time point (Fig. 2).
FIG 2.
Immunohistochemistry staining shows immune infiltration of T cells and macrophages. Representative immunohistochemistry staining for CD3+, CD68+, and CD20+ cells in the dorsal root ganglia (cervical) (DRG-C) from naive animals at 3 and 7 dpi and in the thoracic DRG at 14 dpi. Bar, 50 μm (×40 magnification). Arrows indicate CD3+ and CD68+ cells. Isotype IgG served as our negative control.
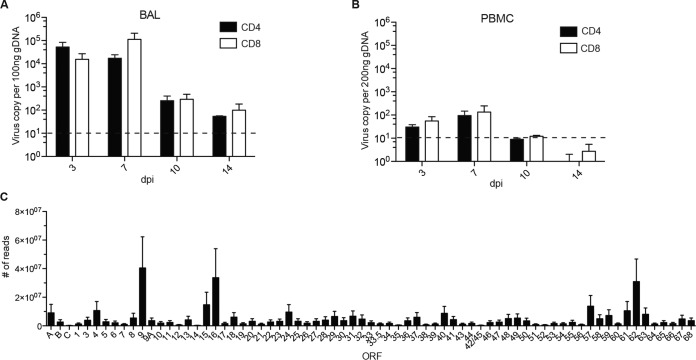
To determine if T cells support SVV replication and dissemination, we isolated CD4 and CD8 T cells from BAL fluid and PBMC samples collected from the same animals from which we collected ganglia 3, 7, 10, and 14 dpi. SVV DNA was detected 3 to 14 dpi in CD4 and CD8 T cells from both BAL fluid and PBMCs (Fig. 3A and B). No viral DNA was detected in T cells obtained from naive animals at 100 dpi or from B cells detected at any time point. To determine whether T cells can support SVV replication, viral transcripts were measured using Ion AmpliSeq technology. Due to the limited amount of RNA and because the viral loads were comparable, we pooled RNA from BAL fluid CD4 and CD8 T cells isolated 3 and 7 dpi. Transcripts from immediate early, early, and late genes were detected in both CD4 and CD8 T cells, albeit at a lower frequency in CD8 T cells (Fig. 3C and data not shown). The most abundant transcripts were ORF9 (tegument protein), ORF16 (viral DNA polymerase subunit), and ORF62 (viral transactivator) (30) (Fig. 3C).
FIG 3.
SVV trafficking and replication are supported in T cells of the BAL fluid. (A and B) SVV viral loads in purified CD4 and CD8 T cells from BAL fluid (A) and PBMC (B) were measured by qRT-PCR with primers and probes for ORF21. Dashed line indicates limit of detection. (C) SVV viral transcripts in BAL fluid T cells (n = 5 [n = 3, CD4; n = 2, CD8]) isolated 3 and 7 dpi using Ion AmpliSeq technology.
SVV transcripts are detected during acute infection.
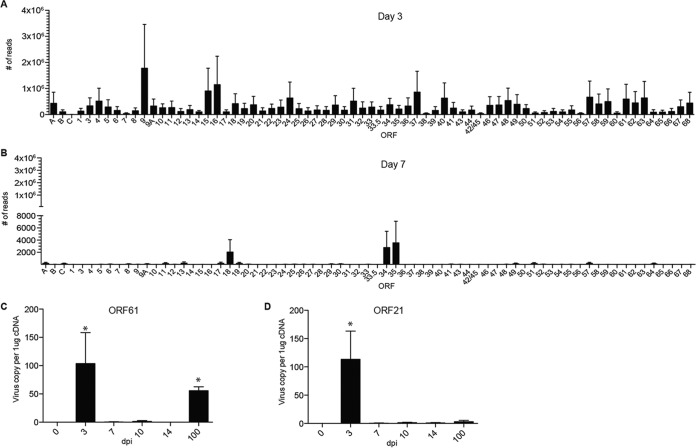
We next determined when SVV establishes latency in the ganglia by measuring the abundance of SVV viral transcripts using Ion Torrent AmpliSeq technology (Fig. 4). Viral transcripts associated with almost all SVV ORFs were detected 3 dpi in the ganglia (Fig. 4A). As previously described for infected BSC-1 and Vero cells (31), transcripts of ORF9 (tegument protein) were the most abundant at 3 dpi. In addition, transcripts of ORF16, ORF15 (membrane protein), ORF 37 (glycoprotein H), and ORF62/ORF63 (viral transactivators) were also abundant (30). In contrast, only transcripts from ORF18 (viral ribonucleotide reductase), ORF34 (viral DNA packaging protein), and ORF35 (a nuclear matrix protein required for infectivity of skin and T cells [32]) were detected at 7 dpi (Fig. 4B). No viral transcripts were detected at 10 and 14 dpi.
FIG 4.
SVV replicates in the ganglia before establishing latency at 7 dpi. (A and B) Abundances of SVV viral gene transcripts at day 3 (A) (n = 3) and day 7 (B) (n = 3) postinfection in the ganglia were measured using Ion AmpliSeq technology. (C and D) Viral transcripts of ORF61 (A) and ORF21 (B) (control transcript) were measured using qRT-PCR (0 dpi, n = 3; 3 dpi, n = 3; 7 dpi, n = 2; 10 dpi, n = 2; 14 dpi, n = 3; 100 dpi, n = 2). *, P < 0.05 compared to day 0.
These data suggest that SVV begins to establish latency 7 dpi. Previous studies from our lab have shown that ORF61 transcripts were the most abundant latency-associated transcripts during SVV latency (>70 dpi) (33). Interestingly, ORF61 transcripts were not detected by AmpliSeq on days 7, 10, and 14 postinfection. Since our previous studies looked at later time points (>70 dpi) (34), we hypothesized that ORF61 transcription may first be turned off and then restart after 14 dpi. To test this hypothesis, we measured ORF61 and ORF21 transcripts using reverse transcriptase quantitative PCR (qRT-PCR) using the same RNA samples, including samples from 100 dpi. ORF21 served as our control transcript. In line with our AmpliSeq data, ORF61 and ORF21 transcripts were detected at 3 dpi but not at 7, 10, and 14 dpi (Fig. 4C and D). More importantly, ORF61 but not ORF21 transcripts were detected at 100 dpi, in agreement with our previous data (33) (Fig. 4C).
SVV infection results in large and sustained changes in gene expression in the ganglia.
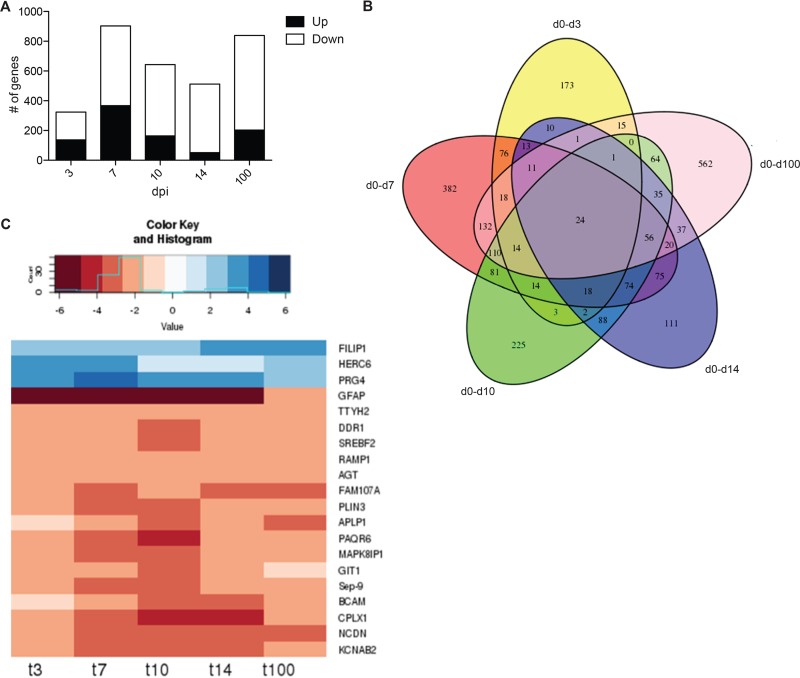
To characterize the host response in the ganglia during acute SVV infection, we used RNA-Seq to determine the transcriptome of sensory ganglia isolated from naive animals (n = 3) and SVV-infected animals 3 (n = 3), 7 (n = 2), 10 (n = 2), 14 (n = 3), and 100 (n = 2) dpi. Types of ganglia and average viral loads of samples used for RNA-Seq are shown in Table 1. Given the limited number of animals and available ganglia harboring SVV DNA, it was not possible to use the same type of ganglia at every time point. To address the issue of our small sample size, we used a cutoff fold change (FC) of ≥3 and a false-discovery rate (FDR) corrected P value of ≤0.01 compared to naive animals (instead of the traditional cutoff FC of ≥2 and FDR corrected P value of ≤0.05) to identify differentially expressed genes (DEGs). SVV infection resulted in large changes in gene expression at all time points examined (Table 1; Fig. 5A; also see Data Sets S1 to S5 in the supplemental material). We confirmed changes in the expression of 8 genes by qRT-PCR and 3 genes by IHC analysis (Fig. 6 and 7). Functional enrichment of the DEGs was carried out using Metacore software, which requires the use of human gene identifiers for analysis. On average, 83% of rhesus DEGs were successfully mapped to human homologs.
TABLE 1.
Number of DEGs found in ganglia after SVV infection
| dpi | Samplesa | Avg viral load ± SEMb | No. of DEGs |
|||
|---|---|---|---|---|---|---|
| Total | With human homologsc | Upregulated | Downregulated | |||
| 3 | DRG-L/S, DRG-C, DRG-T | 28 ± 6 | 393 | 324 | 137 | 187 |
| 7 | DRG-T, DRG-C | 26 ± 6 | 1,118 | 904 | 368 | 536 |
| 10 | DRG-C, TG | 17 ± 5 | 809 | 643 | 164 | 479 |
| 14 | TG, DRG-C, TG | 17 ± 9 | 576 | 512 | 51 | 461 |
| 100 | TG, TG | 39 ± 3 | 1100 | 839 | 203 | 636 |
Types of ganglia used for RNA-Seq analysis.
Average SVV genome copy number of all ganglia used in the RNA-Seq experiment per 1 μg DNA.
Number of rhesus macaque DEGs with human homologs used for enrichment analysis.
FIG 5.
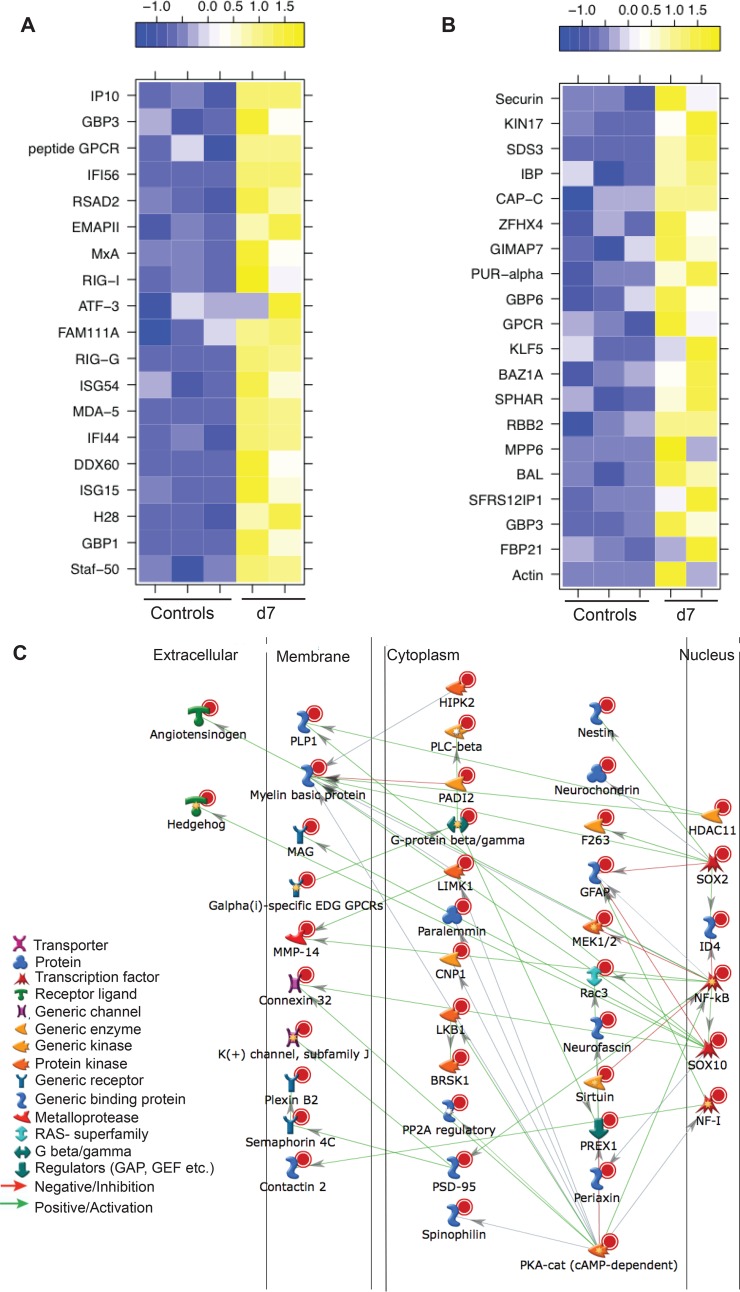
SVV infection results in robust changes in gene expression. (A) Number of upregulated and downregulated differentially expressed genes (DEGs) detected at days 3, 7, 10, 14, and 100 postinfection. (B) Five-way Venn diagram of the DEGs detected at each time point with human homologs. (C) Heat map analysis of the 20 characterized DEGs common at all time points. Numbers at bottom are days postinfection.
FIG 6.
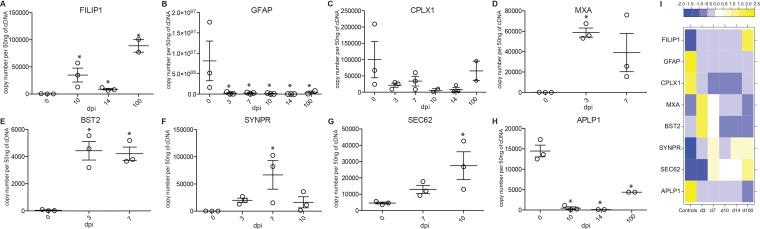
Gene validation. (A to H) TaqMan assays were done on FILIP1, upregulated at 10, 14, and 100 dpi (A); GFAP, downregulated at 3, 7, 10, 14, and 100 dpi (B); CPLX1, downregulated at 3, 7, 10, 14, and 100 dpi (C); MXA, upregulated at 3 and 7 dpi (D); BST2, upregulated at 3 and 7 dpi (E); SYNPR, upregulated at 7 dpi (F); SEC62, upregulated at 10 dpi (G); and APLP1, downregulated at 10, 14, and 100 dpi (H) (one additional ganglion tissue from the same animal was used at 7 dpi and 10 dpi). *, P < 0.05 compared to day 0. (I) Heat map of genes confirmed by TaqMan in panels A to H showing the median normalized transcript number for each day postinfection.
FIG 7.
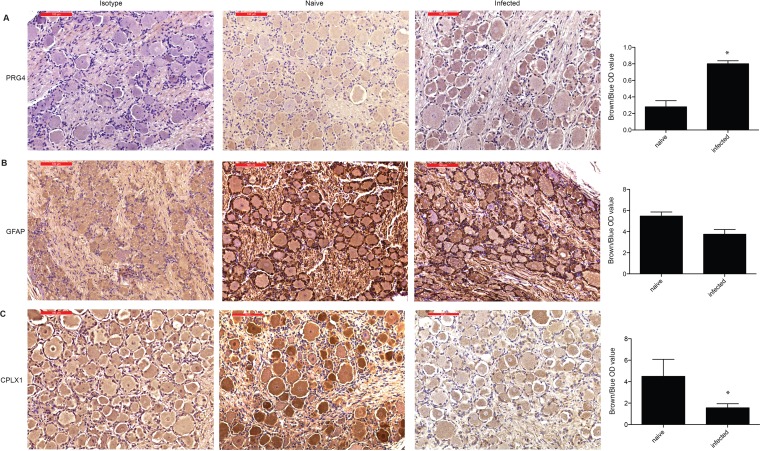
Immunohistochemistry gene validation. IHC staining and IHC quantification of PRG4 (A), GFAP (B), and CPLX1 (C) on isotype controls and naive and infected ganglion tissue (n = 3, for naive and infected tissue used for quantification). Representative images are all from DRG-L/S isolated 10 dpi. Quantification data on infected tissue were from 3 dpi (DRG-T), 7 dpi (DRG-T), and 10 dpi (DRG-L/S). Unpaired t test; *, P < 0.05 compared to naive tissue.
A total of 24 (20 characterized; 3 upregulated and 17 downregulated) genes were differentially expressed at all time points examined (Fig. 5B and C). The three genes that remained upregulated throughout infection were proteoglycan 4 (PRG4), filamin A-interacting protein 1 (FILIP1), and HECT/RCC1-like domain-containing protein 6 (HERC6) (Fig. 5C). PRG4 (confirmed by IHC staining [Fig. 7A]) is a lubricating glycoprotein that can induce Schwann cell proliferation (35). FILIP1 (confirmed by qRT-PCR [Fig. 6A]) interacts with filamin A to control neuron migration out of the ventricular zone (36), and it interferes with myosin 2b binding to F-actin in glutamatergic neurons, affecting the neural spine structure (37). The current function of HERC6 is unknown; however, HERC5, with which it shares 50% nucleotide sequence identity, plays a role in innate antiviral responses (38).
Glial fibrillary acidic protein (GFAP), an intermediate filament protein that plays a critical role in maintaining astrocyte function (39), was the most downregulated common gene throughout infection (Fig. 5C) (confirmed by IHC and qRT-PCR [Fig. 6B and 7B]). Other genes that remained downregulated during infection were neurochondrin (NCDN), involved in neurite outgrowth (40); complexin 1 (CPLX1), expressed by neurons and regulating synaptic vesicle exocytosis (41) (confirmed by IHC and qRT-PCR [Fig. 6C and 7C]); and progestin and adipoQ receptor 6 (PAQR6) a G-coupled membrane progesterone receptor that exhibits neuroprotective properties (42) (Fig. 5C).
DEGs on day 3 play a role in axon transport, immunity, and neuronal development.
Of the 137 upregulated DEGs in the ganglia 3 dpi, several mapped to gene ontology (GO) terms associated with filament movement (Table 2). The 2 most significantly upregulated genes were subunits C and T of the troponin complex (TNNC, FC of 272, and TNNT, FC of 226), which plays a role in intra-axonal movements in rat dorsal root ganglia (43). Additional genes that were highly upregulated include myosin essential light chain (MYL3, FC of 44), which plays a role in synaptic plasticity (44), and tetranectin (CLEC3B, FC of 24), a glycoprotein that regulates proteolytic processes in neurons and dendrites (45). In addition, a number of innate immune genes were upregulated at 3 dpi (Fig. 8A), most notably several interferon (IFN)-induced genes that play a critical role in the antiviral immune response (46), such as MXA (FC of 7, confirmed by qRT-PCR [Fig. 6D]) (47); chemokine ligand 1 (CCL1, FC of 6), secreted by activated T cells to recruit monocytes (48); and bone marrow stromal cell antigen 2 (BST2, FC of 5), a restriction factor that interferes with the release of enveloped viruses, including herpes simplex virus 2 (HSV-2) (confirmed by qRT-PCR [Fig. 6E]) (49).
TABLE 2.
Ten most statistically significant GO terms to which DEGs enriched at each time point
| GO process | No. of genes | −log P value |
|---|---|---|
| Day 3 | ||
| Upregulated | ||
| Muscle filament sliding | 13 | 14.53 |
| Actin-myosin filament sliding | 13 | 14.53 |
| Muscle contraction | 22 | 14.5 |
| Actin-mediated cell contraction | 13 | 13.38 |
| Muscle system process | 22 | 12.79 |
| Actin filament-based movement | 13 | 11.69 |
| Striated muscle contraction | 12 | 8.44 |
| Heart process | 10 | 7.64 |
| Regulation of ATP activity | 8 | 6.07 |
| Regulation of muscle contraction | 12 | 5.98 |
| Downregulated | ||
| Localization | 75 | 3.21 |
| Regulation of synaptic plasticity | 11 | 3.21 |
| Transport | 64 | 3.21 |
| Establishment of localization | 65 | 3.21 |
| Anion transport | 18 | 2.96 |
| Ion transport | 31 | 2.92 |
| Developmental process | 82 | 2.85 |
| Single-organism developmental process | 81 | 2.74 |
| Single-organism transport | 52 | 2.47 |
| Endocytosis | 17 | 2.46 |
| Day 7 | ||
| Upregulated | ||
| Response to virus | 23 | 5.188 |
| Defense response to virus | 17 | 5.188 |
| Nucleobase-containing compound metabolic process | 125 | 5.09 |
| Nucleic acid metabolic process | 107 | 5.05 |
| Smooth muscle metabolic process | 11 | 4.77 |
| Skeletal myofibril assembly | 6 | 4.76 |
| Cellular nitrogen compound metabolic process | 130 | 4.76 |
| Heterocycle metabolic process | 125 | 4.47 |
| RNA metabolic process | 92 | 4.41 |
| Cellular aromatic compound metabolic process | 125 | 4.41 |
| Downregulated | ||
| Nervous system development | 155 | 20.63 |
| Neurogenesis | 111 | 14.38 |
| System development | 206 | 12.66 |
| Generation of neurons | 102 | 12.16 |
| Anatomical structure development | 219 | 10.38 |
| Regulation of signaling | 149 | 10.38 |
| Ensheathment of neurons | 23 | 10.38 |
| Axon ensheathment | 23 | 10.38 |
| Regulation of cell communication | 149 | 10.38 |
| Multicellular organismal development | 216 | 9.61 |
| Day 10 downregulated | ||
| Nervous system development | 120 | 10.49 |
| Regulation of cellular component organization | 101 | 10.49 |
| Regulation of signaling | 134 | 9.38 |
| Regulation of cell communication | 133 | 9.16 |
| System development | 176 | 9.16 |
| Negative regulation of cellular process | 156 | 8.27 |
| Neurogenesis | 87 | 8.15 |
| Multicellular organismal development | 190 | 7.96 |
| Negative regulation of cellular component organization | 43 | 7.96 |
| Negative regulation of biological process | 163 | 7.32 |
| Day 14 downregulated | ||
| Regulation of signaling | 132 | 9.84 |
| Regulation of cell communication | 132 | 9.84 |
| Regulation of signal transduction | 114 | 7.32 |
| Nervous system development | 106 | 7.08 |
| Phosphate-containing compound metabolic process | 107 | 6.48 |
| Phosphorylation | 63 | 6.42 |
| Phosphorus metabolic process | 108 | 6.42 |
| Intracellular signal transduction | 79 | 5.97 |
| Epidermal growth factor receptor signaling pathway | 22 | 5.81 |
| ERBB signaling pathway | 22 | 5.79 |
| Day 100 | ||
| Upregulated | ||
| Cellular component organization or biogenesis | 82 | 8.4 |
| Cellular component organization | 79 | 7.73 |
| Organelle organization | 53 | 7.44 |
| Cell division | 22 | 6.36 |
| Cell cycle | 32 | 5.97 |
| Microtubule-based process | 18 | 5.94 |
| RNA splicing | 14 | 5.53 |
| Nucleic acid metabolic process | 63 | 5.4 |
| RNA metabolic process | 55 | 5.02 |
| Mitotic nuclear division | 13 | 4.92 |
| Downregulated | ||
| Nervous system development | 194 | 30.15 |
| Localization | 293 | 26.92 |
| Regulation of cellular component organization | 150 | 22.31 |
| Establishment of localization | 246 | 22.13 |
| Neurogenesis | 138 | 21.55 |
| Single-organism metabolic process | 281 | 21.16 |
| Transport | 239 | 21.05 |
| Protein localization | 135 | 20.14 |
| Cellular localization | 157 | 20 |
| System development | 259 | 19.85 |
FIG 8.
DEGs at 3 dpi play a role in axon transport, immunity, and neuronal development. (A) Heat map of the immune genes upregulated 3 dpi. (B) Heat map of the downregulated DEGs that mapped to the GO processes “regulation of synaptic plasticity,” “ion transport,” “anion transport,” and “endocytosis.”
Almost half of the 187 downregulated DEGs at 3 dpi mapped to GO terms associated with transport and localization (Table 2). The most downregulated genes among these DEGs were myelin-associated oligodendrocyte basic protein (MOBP, FC of 41, involved in myelin formation [50]), ATP-binding cassette subfamily A member 2 (ABCA2, FC of 13, regulating transmembrane lipid transport in neural tissues [51]), myelin-associated glycoprotein (MAG, FC of 10, important for myelination and maintaining the axon-Schwann cell arrangement [52]), and sodium channel voltage-gated type VII alpha subunit (SCN7A, FC of 7, regulating sodium homeostasis in neural cells [53]).
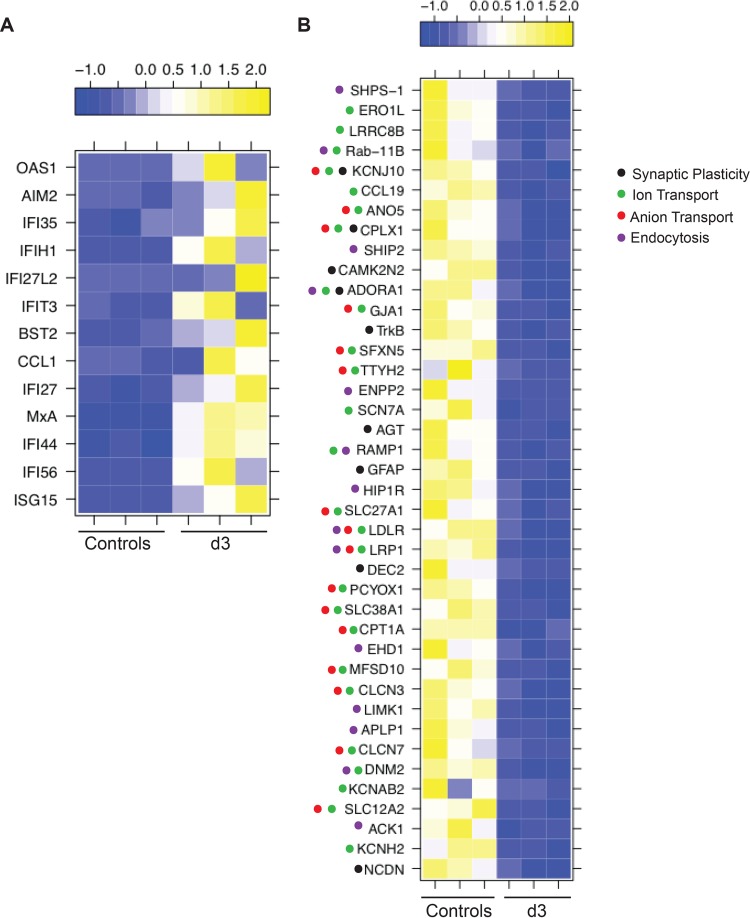
The most downregulated gene among the 9 DEGs that mapped to the GO term “regulation of synaptic plasticity” (Fig. 8B) was GFAP (FC of 43), one of the 17 genes that remained downregulated throughout infection. Other notable DEGs that mapped to this GO term are calcium/calmodulin independent protein kinase II inhibitor 2 (CAMK2N2, FC of 6, regulating neuronal synaptic plasticity through phosphorylation of glutamate receptors [54]), helix-loop-helix member e41 (BHLHE41, FC of 6, a transcriptional repressor suppressed by axonal injury [55]), synaptoporin (SYNPR, FC of 4, synaptic vesicle component [56], confirmed by qRT-PCR [Fig. 6F]), and angiotensinogen (AGT, FC of 4, regulating salt uptake by astrocytes and neurons [57]). DEGs that mapped to both GO terms “ion transport” and “anion transport” (Fig. 8B) include fatty acid transport protein (SLC27A1, FC of 5) (58); solute carrier family 38 member 1 (SLC38A1, FC of 4, a glutamine transporter in the central nervous system [CNS] [59]); complexin 1 (CPLX1, FC of 4, a regulator of synaptic vesicle exocytosis), and sideroflexin 5 (SFXN5, FC of 4, involved in iron transport in the brain [60]). DEGs that mapped to “endocytosis” (Fig. 8B) are known to regulate phagocytosis (e.g., DNM2, FC of 7, and signal-regulatory protein alpha [SHPS-1], FC of 5) and chemotaxis (e.g., ectonucleotide pyrophosphatase/phosphodiesterase 2 [ENPP2, FC of 6]) by macrophages (61–63).
DEGs at 7 dpi play a role in antiviral immunity and neuron development.
“Response to virus” and “defense response to virus” were the most significant GO terms to which DEGs upregulated at 7 dpi mapped (Table 2 and Fig. 9A). Five of these genes were also upregulated at 3 dpi: ISG15, IFI44, MxA, RIG-G, and MDA-5. The 5 most upregulated genes at 7 dpi were ISG15 (FC of 16), IFI56 (FC of 12), IFI44-like (IF44L, FC of 9), guanylate binding proteins 1 and 3 (GBP1 and GBP3, respectively; FC of 8), and IFI44 (FC of 7). These genes are induced by a type I interferon (IFN) response and block viral replication (46). Other immune genes that were upregulated but did not map to these GO terms were C-X-C motif chemokine 10 (CXCL10, FC of 7), a chemokine that binds to CXCR3 to induce monocyte, NK, and activated T cell migration to sites of infection (64); chemokine ligand 8 (CCL8, FC of 6), which is primarily produced by monocytes and may be involved in neurodegeneration (65); tumor necrosis factor ligand superfamily member 10 (TNSF10, FC of 4), AIM2 (FC of 4), and IFI27 (FC of 4), which induce apoptosis of virus-infected cells (66–68); and monocyte-to-macrophage differentiation protein (MMD, FC of 3) (69). Upregulated DEGs that mapped to the remaining 8 GO terms were associated with nucleic acid metabolism (Fig. 9B) and play a role in splicing, e.g., splicing factor arginine/serine rich 12 (SFRS12, FC of 6) and formin binding protein 21 (FBP21, FC of 9) (70, 71), or antiviral defense, e.g., guanylate binding proteins 3 and 6 (GBP3 and GBP6, respectively; FCs of 3 and 5), which repress viral polymerases (72).
FIG 9.
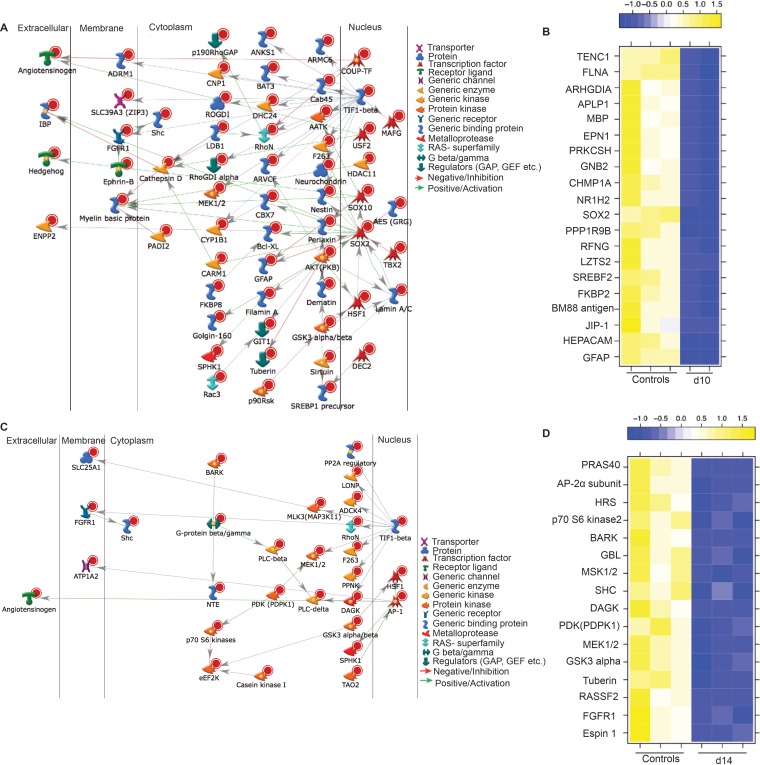
DEGs at 7 dpi play a role in antiviral immunity and neuronal development. (A) Heat map of the upregulated DEGs that mapped to the GO process “response to virus.” (B) The 20 most upregulated DEGs that mapped to GO processes involved with nucleic acid metabolism. (C) Network image showing direct interaction between the downregulated DEGs that mapped to the GO process “nervous system development.”
DEGs downregulated at 7 dpi mapped to GO terms associated with neuron development and function (Table 2). Network analysis of 155 DEGs that mapped to the GO term “nervous system development” showed that 41 are regulated by transcription factors important for nervous system function: nuclear factor I/X (NF1, FC of 4), sex-determining region Y-box 2 and 10 (SOX 2 and SOX10, respectively; FC of 3), and nuclear factor kappa-light-chain-enhancer of activated B cells (NFkb, FC of 4) (73–75) (Fig. 9C). The most downregulated gene in this group was GFAP (FC of 41), while other DEGs play an important role in myelination, such as MAG (FC of 38), MOBP (FC of 24), plasmolipin (PLLP, FC of 23), claudin 11 (CLDN11, FC of 14), PAQR6 (FC of 13), myelin basic protein (MBP, FC of 12), fatty acid 2-hydroxylase (FA2H, FC of 10), and gap junction protein beta 1 (GJP1, FC of 8) (52, 76–79). Other highly downregulated genes include ABCA2 (FC of 16), which was also significantly downregulated at 3 dpi, and the lipid-anchored membrane paralemmin (PALM, FC of 8), which regulates neuron cell shape and motility (80). Some of the most downregulated DEGs that mapped to these GO terms were also detected at 3 dpi, notably GFAP, MAG, MOBP, and CLDN11. Other downregulated DEGs play a role in the maintenance of the blood-brain barrier, such as matrix-remodeling associated 8 (Mxra8; FC of 10) and vasohibin (VASH1, FC of 9) (81, 82). Interestingly, T cell differentiation protein (MAL, FC of 10), which is involved in T cell signal transduction, was also downregulated (83).
DEGS at 10 dpi play a role in neuronal development.
The 5 most upregulated DEGs at 10 dpi were proteoglycan 4 (PRG4, FC of 15), coiled-coil domain-containing 80 (CCDC8, FC of 8), small nucleolar RNA c/d box 112 (SNORD112, FC of 8), S-phase response (SPHAR, FC of 8), actin (FC of 7), and translocation protein 1 (SEC62, FC of 3) (confirmed by qRT-PCR) (Fig. 6G). PRG4 is one of the genes that remained upregulated throughout acute infection, while the other genes play a role in DNA replication (CCDC80 [84] and SPHAR [85]), translation (SNORD112 [86]), axon elongation (actin [87]), and protein transport (SEC62 [88]).
Among the 479 DEGs downregulated at 10 dpi, 120 DEGs mapped to “nervous system development” and “neurogenesis” (Table 2). In addition to GFAP, which was the most downregulated gene (FC of 75), other notable DEGs mapping to these GO terms include periaxin (PRX, FC of 20, important for the ensheathment of neurons and axons [89]), CPLX1 (FC of 16, regulating synaptic vesicle exocytosis), and proprotein convertase subtilisin/kexin type 1 inhibitor (ProSAAS, FC of 17, neural tissue inhibitor of prohormone convertase 1, which is involved in insulin synthesis [90]).
Of 176 DEGs that mapped to the GO term “system development,” 60 directly interact with each other through transcription factors SOX2 (FC of 8), SOX10 (FC of 6), and transcription intermediate factor 1 beta (TIF1-beta, FC of 6) (Fig. 10A). Interestingly, TIF1-beta, which associates with the KRAB domain of zinc finger proteins, has been reported to play a critical role in the latency of another herpesvirus, human cytomegalovirus (91). Several of the DEGs that enriched to the GO term “regulation of cellular processes” (Fig. 10B) play an important role in neuron survival, such as hepatic and glial cell adhesion molecule (HEPACAM, FC of 11, an axon protein that plays a role in neuron motility [92]), mitogen-activated protein (MAP) kinase 8 interacting protein 1 (JIP1, FC of 10, part of the MAP kinase signal transduction pathway in neurons [93]), FK056-binding protein 8 (FKBP8, FC of 9, inhibiting programmed cell death in neurons [94]), and BM88 antigen (FC of 10, involved in neuron differentiation [95]).
FIG 10.
DEGs at 10 dpi and 14 dpi are associated with neuronal development. (A) Network image of DEGs downregulated at 10 dpi that mapped to the GO process “multicellular organismal development” and directly interacted. (B) Heat map of the 20 most downregulated genes at 10 dpi mapping to the GO process “negative regulation of biological process.” (C) Network image of DEGs downregulated at 14 dpi that mapped to the GO process “phosphorus metabolic process” and directly interacted. (D) Heat map of DEGs downregulated at 14 dpi mapping to the GO process “ERBB signaling pathway.”
DEGs downregulated at 14 dpi play a role in neuron signaling and development.
The most upregulated DEGs at 14 dpi are PRG4 (FC of 10) and FILIP1 (FC of 7), important for myelination and neuron migration; fibroblast growth factor 7 (FGF7, FC of 7), which regulates balance between excitatory and inhibitory synapse signaling in neurons (96); and NIMA-related kinase 1 (NEK1, FC of 5), which plays a role in axon development (97). The 509 DEGs downregulated at 14 dpi enriched to GO terms associated with signaling and nervous system development (Table 2). Thirty of the DEGs enriching to the GO term “phosphorus metabolic process” have direct interactions (Fig. 10C) and are regulated by transcription factors AP-1 (FC of 3), heat shock factor protein 1 (HSF1, FC of 4), and binding protein TIF1-beta (FC of 5). Casein kinase 1 gamma 2 (CSNK1G2, FC of 11) was among the most downregulated genes in these GO terms and is critical in the regulation of glutamatergic synaptic transmission in the brain (98) as well as neuronal gene expression through its interaction with eukaryotic elongation factor 2 kinase (eEF2K, FC of 3) (99). Other interesting DEGs that were associated with nervous system development include secreted frizzled-related protein 2 (SFRP2, FC of 35, important in neuron development [100]), C-C motif chemokine ligand 19 (CCL19, FC of 29, important for lymphocyte recirculation and homing to lymphoid tissue [101]), and aquaporin 4 (AQP4, FC of 13, a water channel expressed in astrocytes [102]). The most downregulated DEGs that mapped to the GO term “nervous system development” include GFAP (FC of 59), SFRP2 (FC of 35), MOBP (FC of 26), CPLX1 (FC of 18), and patatin-like phospholipase domain-containing 2 (PNPLA2, FC of 14). PNPLA2 plays a role in the interactions between neurons and glial cells (103).
In addition, we also saw several downregulated DEGs known to regulate apoptotic processes in neurons, notably NRBP2 (FC of 11) (104), proline-rich Akt substrate 40 (PRAS40, FC of 8) (105), mitogen-activated protein kinase 11 (MAP3K11, FC of 5) (106), homeodomain interacting protein kinase 2 (HIPK2, FC of 6) (107), and mannosylinositol phosphorylceramide (BCL21, FC of 3) (108). Finally, 16 downregulated DEGs mapped to the GO terms “ERBB (Erythroblastic Leukemia Viral Oncogene Homolog) signaling pathway” (Fig. 10D). Insufficient ERBB signaling is associated with the development of neurodegenerative diseases (109, 110). Some of the most significantly downregulated genes found in this GO term include diacylglycerol kinase1 (DAGK1, FC of 6, playing a role in myelination and neuronal plasticity [111]), Epsin1 (EPN1, FC of 6, contributing to clathrin-coated pit formation and endocytosis [112]), and RAS association domain-containing protein 2 (RASSF2, FC of 5, a tumor suppressor in gliomas [113]).
SVV infection results in a sustained downregulation of genes important for nervous system development.
Upregulated DEGs at 100 dpi played a role in cell cycle progression (Table 2). The 20 most upregulated DEGs enriched to cellular component organization were as follows: intraflagellar transport 74 (IFT74, FC of 8), important for survival and differentiation of sensory neurons (114); thyroid hormone receptor interactor 11 (TRIP11, FC of 7), important for maintenance of Golgi structure and protein transport (115); and chromodomain helicase DNA binding protein 9 (PRIC320, FC of 6), which controls the expression of genes involved in energy homeostasis and the cell cycle (116) (Fig. 11A). Interestingly, MBP (FC of 4), which was significantly downregulated during acute infection, is upregulated at latency (Fig. 11A). DEGs that mapped to the GO term “nucleic acid metabolism” are involved in the regulation of gene expression, such as bromodomain adjacent to zinc finger domain protein 2B (BAZ2B, FC of 6), zinc finger homeobox 4 (ZFHX4, FC of 4), zinc finger protein 292 (ZNF292, FC of 4), polymerase RNA III polypeptide G (POLR3G, FC of 4), and TWIST neighbor protein (TWISTNB, FC of 4) (117) (Table 2).
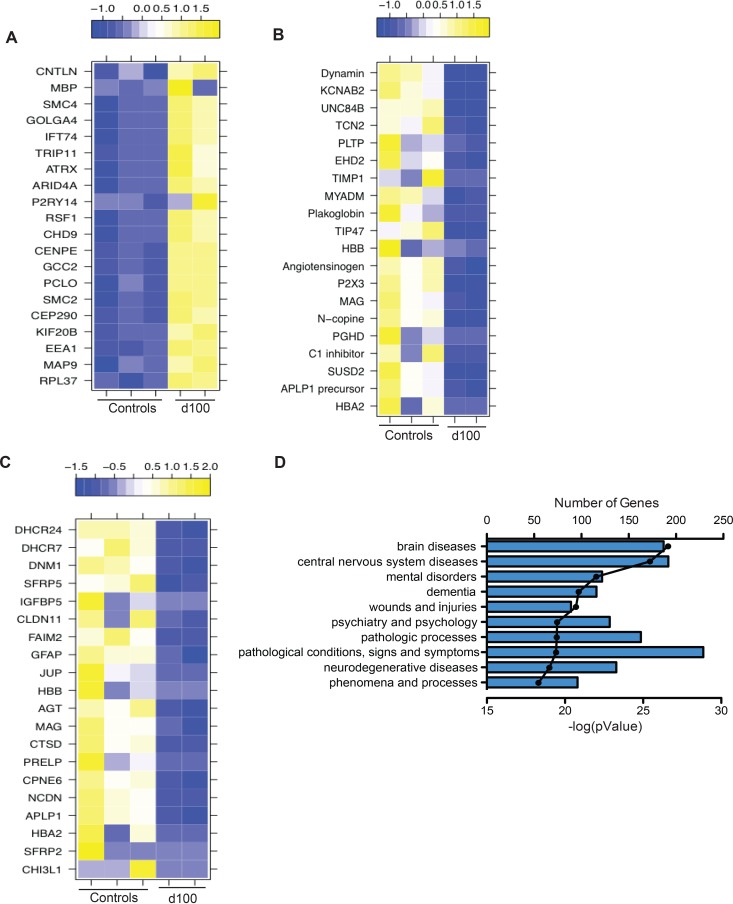
FIG 11.
DEGS in latently infected ganglion tissue (100 dpi) play a role in the development of the nervous system and are associated with neurological diseases. (A) Heat map of the 20 most upregulated genes that mapped to the GO process “cellular component organization or biogenesis.” (B and C) Heat map of the 20 most downregulated genes that mapped to the GO processes “localization” (B) and “system development” (C). (D) The 10 most statistically significant diseases (by biomarkers) to which downregulated DEGs mapped.
Interestingly, 636 DEGs remained downregulated at 100 dpi, with the majority (456 DEGs) enriching to GO terms associated with the nervous system and cellular localization (Table 2). The most significantly downregulated DEGs in these GO terms play a role in immunity (myeloid differentiation marker gene, MYADM, FC of 6 [118] and serpin peptidase inhibitor, SERPIN, FC of 9, an inhibitor of the complement classical pathway [119]), pain (purinergic receptor, P2X3, FC of 7) (120), and cell division (sushi domain-containing 2, SUSD2, FC of 9) (121) (Fig. 11B). Several of the 20 most downregulated genes enriched to “nervous system development” and play a role in astrocyte function, e.g., chitinase 3-like protein 1 (CHI3L, FC of 17) and GFAP (FC of 6) (122); inflammation, e.g., hemoglobulin alpha 2 (HBA2, FC of 13) (123); neurite growth, e.g., amyloid beta (A4) precursor-like protein 1 (APLP1, FC of 10) (confirmed by qRT-PCR [Fig. 6H]) and neurochondrin (NCDN, FC of 8) (124, 125); synaptic plasticity, e.g., copine 6 (CPNE6, FC of 7) (126); and myelination, e.g., MAG (FC of 7) and CLDN11 (FC of 6) (Fig. 11C). Interestingly, a secondary functional enrichment analysis for diseases by biomarkers using the downregulated genes at 100 dpi showed enrichment for processes involved with neurodegeneration and mental health disease (Fig. 11D).
DISCUSSION
In this study, we leveraged a rhesus macaque model of intrabronchial SVV infection to gain a better insight into host-virus interactions during acute and latent VZV infection in the ganglia. We analyzed viral loads, viral transcriptome, and host transcriptome in sensory ganglia collected from naive animals and on days 3, 7, 10, 14, and 100 post-SVV infection. Our analysis shows that SVV DNA is detected in the trigeminal ganglia as early as 3 dpi before the appearance of the rash (typically seen 7 to 10 dpi). These results are in agreement with previous studies that reported the presence of SVV DNA in the ganglia before the appearance of rash in intratracheally infected AGMs, the detection of VZV DNA in the ganglia of immunocompromised children who died from varicella before the appearance of the rash (4, 6), and the ability of the Oka vaccine strain to establish latency in sensory ganglia in the absence of varicella rash (127). Given that SVV rapidly reaches the ganglia before the appearance of vesicular rash and that we cannot detect SVV DNA in nonlesioned skin, we believe that SVV is most likely transported to the ganglia via the hematogenous route, which has been previously suggested for VZV (128). DNA viral loads in the DRG were higher at 7 dpi than at 3 dpi, suggesting additional seeding via axonal transport (rash is observed 7 dpi) and/or as a result of viral replication detected 3 dpi in the ganglia.
In addition, our data suggest that T cells play a role in varicella virus dissemination since memory T cells are detected in the ganglia 3 dpi at the same time as viral DNA. Our data also suggest that this is a selective recruiting of T cells since no B cells were detected at any time point. It is highly unlikely that these T cells are SVV specific given that SVV-specific T cell responses are not detected in the blood or BAL fluid until 7 dpi in this animal model (10, 18, 19). Similarly, VZV-specific T cells are also not detected in the blood until 3 days postrash (approximately 14 days after exposure) in children during primary infection (129). Rather, we hypothesize that these T cells are transporting SVV to the ganglia. This hypothesis is in line with data from previous studies that have suggested an initial role for T cells in VZV dissemination. In vitro experiments showed an increased propensity of VZV to infect tonsillar memory CD4 T cells (130, 131), and intravenous injection of VZV-infected CD4 T cells and not fibroblasts results in a rash on human skin explant in SCID-hu mice (28). In this study, we examined ganglia at earlier time points than in previous similar studies (15, 29) and were able to show T cell infiltration as early as 3 dpi and before the onset of varicella. Although we were not able to determine whether T cells isolated 3 dpi from the ganglia were infected with SVV, we were able to show that BAL fluid CD4 and CD8 T cells isolated 3 dpi from the same animals harbor SVV DNA and viral transcripts. Collectively, these data suggest that T cells play a critical role in SVV dissemination and may potentially support viral replication.
Additionally, given the intrabronchial challenge route that we used, SVV may be reaching the ganglia at this early time point postinfection by traveling directly from the lung (site of infection) to the ganglia through the vagus nerve as observed in mice infected intranasally with influenza virus (132) and intraperitoneally with HSV (133). We also report for the first time that SVV transcripts are detected 3 dpi in the ganglia, before viral gene expression. Since we did not measure infectious virus, these transcripts could be the result of abortive replication either in neurons or in the surrounding T cells. This issue will have to be clarified in future studies using ganglionic explants. SVV transcription in the ganglia ceased 7 dpi, at which point we observed low-level gene expression from ORF18, ORF34, and ORF35. This was followed by a quiescent period before reexpression of SVV ORF61 (viral transactivator) at 100 dpi, which suggests additional regulatory mechanisms controlling the transcription of ORF61. This hypothesis is supported by the fact that ORF61 transcripts detected during latency are antisense whereas those detected during acute/lytic infection are sense (34). Similarly to our observations, HSV-1 replication was also seen only at 4 dpi in the ganglia of mice inoculated through the snout or the cornea (134). Our results differ from previous studies that showed that VZV replication of directly inoculated fetal xenografts in SCID mice is sustained until 14 dpi (6). This discrepancy could be due to the lack of adaptive immunity in the SCID mouse, the immature nature of fetal DRG, and/or the direct inoculation of the virus into xenografts, which bypasses host restriction factors.
Viral gene expression at 3 dpi was accompanied by several significant changes in host gene expression. The most highly upregulated genes were involved in the troponin complex. It is believed that the troponin complex may control intra-axonal movement in neurons and that SVV might be using this mechanism for neuronal spread as previously described for HSV (43). Elevated troponin levels in the blood are also often seen in patients during an acute stroke (135, 136). Interestingly, both primary VZV infection and reactivation have been associated with an increased risk of stroke after infection (137, 138). In addition, genes associated with synaptic plasticity and ion transport were downregulated 3 dpi, indicative of potentially impaired neuronal signal transduction.
Several IFN-stimulated genes (ISGs) were also highly upregulated 3 dpi (OAS1, ISG15, and MXA). This innate immune response is likely mediated by the satellite glial cells (SGCs), which share phenotypic and functional characteristics with dendritic cells and macrophages (139). Importantly, SGCs express Toll-like receptor 9 (TLR-9) (140), an intracellular sensor of double-stranded DNA viruses such as SVV and VZV (141). Therefore, recognition of SVV viral DNA by TLR-9 may result in the induction of a type I interferon response. Type I interferons play a critical role in the host defense against VZV since treatment of immunocompromised patients with alpha interferon (IFN-α) within the first 72 h after the appearance of the rash reduced the number of lesions in children with cancer (142). Moreover, IFN-α plays an important role in controlling VZV spread in the skin of the SCID mouse model (28). Interestingly, we did not detect alpha/beta interferon transcripts. This may be due to the small number of infected cells and the ability of SVV to suppress interferon expression within those infected cells (143, 144), which could have diluted the strength of the IFN signal, especially since we were using total ganglion RNA. IFN-stimulated genes could potentially be upregulated within infiltrating innate immune cells. Although we detected increased expression of CD68 in ganglia at 3 dpi, it should be noted that CD68 can also be expressed by SGCs (139). Unfortunately, due to the limited number of cells recovered after ganglion digestion, we were unable to quantify other immune cells that may have infiltrated the ganglia during acute infection using flow cytometry. Expression of ISGs and other innate immune genes remained increased at 7 dpi and coincided with the establishment of latency.
As previously reported for ganglion explants experimentally infected with VZV (6) and ganglia collected from acutely infected AGMs (14), we saw an upregulation of CXCL10 in the ganglia at 7 dpi. CXCL10 is a chemokine that can be produced by neurons, astrocytes, and oligodendrocytes (145) and binds the receptor CXCR3 on memory T cells and NK cells to induce their migration to the site of infection (146). Indeed, the NK-specific transcript, natural killer cell triggering receptor (NKTR), was upregulated 7 and 10 dpi (FC of 3 and 4, respectively). NK cells have also been shown to infiltrate the ganglia in patients who have had herpes zoster a few months prior to death (7). Increased expression of CXCL10 may contribute to the infiltration by antigen-specific T cells in the ganglia after 7 dpi. However, DEGs associated with T cell effector function (such as IFN-γ, granzyme B, or CD69) were not detected, possibly due to the low frequency of SVV-specific T cells in the ganglia. These data differ from those in a recent study that reported an increase in granzyme B 9 and 21 dpi in the ganglia of SVV-infected AGMs (29). This discrepancy could be due to the fact that in contrast to that in rhesus macaques, SVV infection in AGMs is often fatal (16). Expression of genes important for macrophage-mediated phagocytosis (DNM2, SHIPS, and ENPP2) was reduced 3 and 7 dpi. The reduced expression of genes involved in endocytosis may be due to increased expression of ISGs, several of which inhibit viral uptake through endocytosis as a strategy to stop viral spread (147, 148).
Many of the downregulated DEGs at 7, 10, 14, and 100 dpi mapped to the GO terms associated with nervous system development. Interestingly, these DEGs remained downregulated long after virus replication had ended, suggesting that SVV infection of the ganglia results in long-term changes in the ganglion transcriptome. Several of these genes were involved in myelination (MOBP, MAG, MBP, and PRG4), which suggested that SVV may lead to demyelination, as has been described for HSV-1 and -2 (149, 150). Genes downregulated 14 dpi were involved in neuronal apoptosis, which may be another strategy that SVV utilizes to establish latency in the ganglia. VZV ORF63 has indeed been shown to inhibit apoptosis in neurons (151). We also report a downregulation of genes involved in ERBB signaling, a critical pathway for DRG development (152), at 14 dpi. As previously reported for VZV-infected human astrocytes (153), expression of GFAP, which plays a critical role in astrocyte function, remained downregulated in sensory ganglia of SVV-infected macaques until 100 dpi.
A recent study examined gene expression changes in keratinocyte cell lines infected in vitro with VZV using RNA-Seq (154); however, this study reported only changes in epidermal genes (cytokeratins and desmosomal genes) and did not discuss changes in the innate immune response to VZV. An earlier study using microarray studies showed altered gene expression in T cells infected with VZV in vitro and in skin xenografts harvested from VZV-infected SCID-hu mice (155). Only 5 DEGs in the skin xenografts were detected in our study. These include RGS (G-coupled protein signaling, 3 and 7 dpi), RPS (ribosomal protein, 7 dpi), GAS (apoptosis, 3 and 10 dpi), GADD (growth arrest and DNA damage, 100 dpi), and CHI3L (angiogenesis, 100 dpi). Together with the recent study using keratinocytes (154), these observations strongly suggest that skin cells generate a different response than neuronal cells do to varicella virus infection. In contrast, and in line with the fact that T cells infiltrate the ganglia after SVV infection, 31 DEGs were detected in VZV-infected T cells and SVV-infected ganglia. These include genes such as TRIP11 (protein transport, upregulated 7 dpi), S100A11 (cell cycle, downregulated 3 dpi), BAZ2B (transcriptional regulation, upregulated 7 and 10 dpi), and APOB (lipid transport, upregulated 3 dpi).
In summary, this study is the first to use next-generation sequencing of ganglia collected during both acute and latent infection following experimental inoculation of a robust nonhuman primate model of VZV infection to study both host and viral transcriptomes. Although a small number of animals were used in these studies, the outbred nature of rhesus macaques coupled with the stringent bioinformatics approaches provides novel insight into host-pathogen interactions during acute varicella and latent infection. Our data show that SVV reaches the ganglia very quickly after infection, most likely via hematogenous spread within infected T cells. SVV transcription is detected briefly before establishment of latency. Cessation of viral transcription correlates with a robust upregulation of IFN-stimulated genes and antiviral innate immune response. Importantly, SVV infection results in significant changes in the expression of genes involved in neurogenesis and nervous system development that persist long after cessation of viral replication. It is possible that some of these gene changes could in part be mediated by the robust immune response that develops during acute infection. Given the similarities between SVV and VZV, these data enhance our current understanding of the neurological consequences of VZV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank the veterinarians and the husbandry staff at the Oregon National Primate Research Center for their expert care of the animals and Miranda Fischer, Reed Norris, and Flora Engelmann for assisting in sample collection and tissue processing. We also thank Vincent Funari and Lindsay Spurka at the Cedars-Sinai Genomics core for running our AmpliSeq data.
This work was funded by the National Institutes of Health (NIH) (RO1AG037042-06).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01272-16.
REFERENCES
- 1.Zerboni L, Sen N, Oliver SL, Arvin AM. 2014. Molecular mechanisms of varicella zoster virus pathogenesis. Nat Rev 12:197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fashner J, Bell AL. 2011. Herpes zoster and postherpetic neuralgia: prevention and management. Am Fam Physician 83:1432–1437. [PubMed] [Google Scholar]
- 3.Bearer EL, Breakefield XO, Schuback D, Reese TS, LaVail JH. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc Natl Acad Sci U S A 97:8146–8150. doi: 10.1073/pnas.97.14.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahalingam R, Wellish M, Soike K, White T, Kleinschmidt-DeMasters BK, Gilden DH. 2001. Simian varicella virus infects ganglia before rash in experimentally infected monkeys. Virology 279:339–342. doi: 10.1006/viro.2000.0700. [DOI] [PubMed] [Google Scholar]
- 5.Taha Y, Scott FT, Parker SP, Syndercombe Court D, Quinlivan ML, Breuer J. 2006. Reactivation of 2 genetically distinct varicella-zoster viruses in the same individual. Clin Infect Dis 43:1301–1303. doi: 10.1086/508539. [DOI] [PubMed] [Google Scholar]
- 6.Zerboni L, Ku CC, Jones CD, Zehnder JL, Arvin AM. 2005. Varicella-zoster virus infection of human dorsal root ganglia in vivo. Proc Natl Acad Sci U S A 102:6490–6495. doi: 10.1073/pnas.0501045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowrishankar K, Steain M, Cunningham AL, Rodriguez M, Blumbergs P, Slobedman B, Abendroth A. 2010. Characterization of the host immune response in human ganglia after herpes zoster. J Virol 84:8861–8870. doi: 10.1128/JVI.01020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steain M, Gowrishankar K, Rodriguez M, Slobedman B, Abendroth A. 2011. Upregulation of CXCL10 in human dorsal root ganglia during experimental and natural varicella-zoster virus infection. J Virol 85:626–631. doi: 10.1128/JVI.01816-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steain M, Sutherland JP, Rodriguez M, Cunningham AL, Slobedman B, Abendroth A. 2014. Analysis of T cell responses during active varicella-zoster virus reactivation in human ganglia. J Virol 88:2704–2716. doi: 10.1128/JVI.03445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messaoudi I, Barron A, Wellish M, Engelmann F, Legasse A, Planer S, Gilden D, Nikolich-Zugich J, Mahalingam R. 2009. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog 5:e1000657. doi: 10.1371/journal.ppat.1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahalingam R, Traina-Dorge V, Wellish M, Deharo E, Singletary ML, Ribka EP, Sanford R, Gilden D. 2010. Latent simian varicella virus reactivates in monkeys treated with tacrolimus with or without exposure to irradiation. J Neurovirol 16:342–354. doi: 10.3109/13550284.2010.513031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahalingam R, Traina-Dorge V, Wellish M, Lorino R, Sanford R, Ribka EP, Alleman SJ, Brazeau E, Gilden DH. 2007. Simian varicella virus reactivation in cynomolgus monkeys. Virology 368:50–59. doi: 10.1016/j.virol.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Kolappaswamy K, Mahalingam R, Traina-Dorge V, Shipley ST, Gilden DH, Kleinschmidt-Demasters BK, McLeod CG Jr, Hungerford LL, DeTolla LJ. 2007. Disseminated simian varicella virus infection in an irradiated rhesus macaque (Macaca mulatta). J Virol 81:411–415. doi: 10.1128/JVI.01825-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouwendijk WJ, Abendroth A, Traina-Dorge V, Getu S, Steain M, Wellish M, Andeweg AC, Osterhaus AD, Gilden D, Verjans GM, Mahalingam R. 2013. T-cell infiltration correlates with CXCL10 expression in ganglia of cynomolgus macaques with reactivated simian varicella virus. J Virol 87:2979–2982. doi: 10.1128/JVI.03181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouwendijk WJ, Mahalingam R, de Swart RL, Haagmans BL, van Amerongen G, Getu S, Gilden D, Osterhaus AD, Verjans GM. 2013. T-cell tropism of simian varicella virus during primary infection. PLoS Pathog 9:e1003368. doi: 10.1371/journal.ppat.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soike KF. 1992. Simian varicella virus infection in African and Asian monkeys. The potential for development of antivirals for animal diseases. Ann N Y Acad Sci 653:323–333. [DOI] [PubMed] [Google Scholar]
- 17.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 18.Haberthur K, Kraft A, Arnold N, Park B, Meyer C, Asquith M, Dewane J, Messaoudi I. 2013. Genome-wide analysis of T cell responses during acute and latent simian varicella virus infections in rhesus macaques. J Virol 87:11751–11761. doi: 10.1128/JVI.01809-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haberthur K, Meyer C, Arnold N, Engelmann F, Jeske DR, Messaoudi I. 2014. Intrabronchial infection of rhesus macaques with simian varicella virus results in a robust immune response in the lungs. J Virol 88:12777–12792. doi: 10.1128/JVI.01814-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girke T. 2015. systemPipeR: NGS workflow and report generation environment. University of California—Riverside, Riverside, CA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kahari AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SM, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P. 2015. Ensembl 2015. Nucleic Acids Res 43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 28.Ku C-C, Zerboni L, Ito H, Graham BS, Wallace M, Arvin AM. 2004. Varicella-zoster virus transfer to skin by T cells and modulation of viral replication by epidermal cell interferon-alpha. J Exp Med 200:917–925. doi: 10.1084/jem.20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouwendijk WJ, Getu S, Mahalingam R, Gilden D, Osterhaus AD, Verjans GM. 2015. Characterization of the immune response in ganglia after primary simian varicella virus infection. J Neurovirol 22:376–388. doi: 10.1007/s13365-015-0408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohen JI. 2010. The varicella-zoster virus genome. Curr Top Microbiol Immunol 342:1–14. doi: 10.1007/82_2010_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohrs RJ, Hurley MP, Gilden DH. 2003. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J Virol 77:11718–11732. doi: 10.1128/JVI.77.21.11718-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito H, Sommer MH, Zerboni L, Baiker A, Sato B, Liang R, Hay J, Ruyechan W, Arvin AM. 2005. Role of the varicella-zoster virus gene product encoded by open reading frame 35 in viral replication in vitro and in differentiated human skin and T cells in vivo. J Virol 79:4819–4827. doi: 10.1128/JVI.79.8.4819-4827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer C, Kerns A, Barron A, Kreklywich C, Streblow DN, Messaoudi I. 2011. Simian varicella virus gene expression during acute and latent infection of rhesus macaques. J Neurovirol 17:600–612. doi: 10.1007/s13365-011-0057-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou Y, Davis KA, Traina-Dorge V, Gray WL. 2007. Simian varicella virus expresses a latency-associated transcript that is antisense to open reading frame 61 (ICP0) mRNA in neural ganglia of latently infected monkeys. J Virol 81:8149–8156. doi: 10.1128/JVI.00407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratner N, Bunge RP, Glaser L. 1985. A neuronal cell surface heparan sulfate proteoglycan is required for dorsal root ganglion neuron stimulation of Schwann cell proliferation. J Cell Biol 101:744–754. doi: 10.1083/jcb.101.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato M, Nagano T. 2005. Involvement of filamin A and filamin A-interacting protein (FILIP) in controlling the start and cell shape of radially migrating cortical neurons. Anat Sci Int 80:19–29. doi: 10.1111/j.1447-073x.2005.00101.x. [DOI] [PubMed] [Google Scholar]
- 37.Yagi H, Nagano T, Xie MJ, Ikeda H, Kuroda K, Komada M, Iguchi T, Tariqur RM, Morikubo S, Noguchi K, Murase K, Okabe M, Sato M. 2014. Filamin A-interacting protein (FILIP) is a region-specific modulator of myosin 2b and controls spine morphology and NMDA receptor accumulation. Sci Rep 4:6353. doi: 10.1038/srep06353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, Zhu LH, Wang C. 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol 30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petzold A. 2015. Glial fibrillary acidic protein is a body fluid biomarker for glial pathology in human disease. Brain Res 1600:17–31. doi: 10.1016/j.brainres.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Dateki M, Horii T, Kasuya Y, Mochizuki R, Nagao Y, Ishida J, Sugiyama F, Tanimoto K, Yagami K, Imai H, Fukamizu A. 2005. Neurochondrin negatively regulates CaMKII phosphorylation, and nervous system-specific gene disruption results in epileptic seizure. J Biol Chem 280:20503–20508. doi: 10.1074/jbc.M414033200. [DOI] [PubMed] [Google Scholar]
- 41.Kielar C, Sawiak SJ, Navarro Negredo P, Tse DH, Morton AJ. 2012. Tensor-based morphometry and stereology reveal brain pathology in the complexin1 knockout mouse. PLoS One 7:e32636. doi: 10.1371/journal.pone.0032636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas P, Pang Y. 2012. Membrane progesterone receptors: evidence for neuroprotective, neurosteroid signaling and neuroendocrine functions in neuronal cells. Neuroendocrinology 96:162–171. doi: 10.1159/000339822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roisen FJ, Wilson FJ, Yorke G, Inczedy-Marcsek M, Hirabayashi T. 1983. Immunohistochemical localization of troponin-C in cultured neurons. J Muscle Res Cell Motil 4:163–175. doi: 10.1007/BF00712028. [DOI] [PubMed] [Google Scholar]
- 44.Kneussel M, Wagner W. 2013. Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nat Rev Neurosci 14:233–247. doi: 10.1038/nrn3445. [DOI] [PubMed] [Google Scholar]
- 45.Stoevring B, Jaliashvili I, Thougaard AV, Ensinger C, Hogdall CK, Rasmussen LS, Sellebjerg F, Christiansen M. 2006. Tetranectin in cerebrospinal fluid of patients with multiple sclerosis. Scand J Clin Lab Invest 66:577–583. doi: 10.1080/00365510600863929. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B, Jiang Z. 2013. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci 9:200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoshino A, Kawamura YI, Yasuhara M, Toyama-Sorimachi N, Yamamoto K, Matsukawa A, Lira SA, Dohi T. 2007. Inhibition of CCL1-CCR8 interaction prevents aggregation of macrophages and development of peritoneal adhesions. J Immunol 178:5296–5304. doi: 10.4049/jimmunol.178.8.5296. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Luo S, He S, Zhang M, Wang P, Li C, Huang W, Hu B, Griffin GE, Shattock RJ, Hu Q. 2015. Tetherin restricts HSV-2 release and is counteracted by multiple viral glycoproteins. Virology 475:96–109. doi: 10.1016/j.virol.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Montague P, McCallion AS, Davies RW, Griffiths IR. 2006. Myelin-associated oligodendrocytic basic protein: a family of abundant CNS myelin proteins in search of a function. Dev Neurosci 28:479–487. doi: 10.1159/000095110. [DOI] [PubMed] [Google Scholar]
- 51.Schmitz G, Kaminski WE. 2002. ABCA2: a candidate regulator of neural transmembrane lipid transport. Cell Mol Life Sci 59:1285–1295. doi: 10.1007/s00018-002-8508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carenini S, Montag D, Schachner M, Martini R. 1999. Subtle roles of neural cell adhesion molecule and myelin-associated glycoprotein during Schwann cell spiralling in P0-deficient mice. Glia 27:203–212. [PubMed] [Google Scholar]
- 53.Watanabe E, Hiyama TY, Shimizu H, Kodama R, Hayashi N, Miyata S, Yanagawa Y, Obata K, Noda M. 2006. Sodium-level-sensitive sodium channel Na(x) is expressed in glial laminate processes in the sensory circumventricular organs. Am J Physiol Regul Integr Comp Physiol 290:R568–R576. [DOI] [PubMed] [Google Scholar]
- 54.Ashpole NM, Song W, Brustovetsky T, Engleman EA, Brustovetsky N, Cummins TR, Hudmon A. 2012. Calcium/calmodulin-dependent protein kinase II (CaMKII) inhibition induces neurotoxicity via dysregulation of glutamate/calcium signaling and hyperexcitability. J Biol Chem 287:8495–8506. doi: 10.1074/jbc.M111.323915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabos P, Kabosova A, Neuman T. 2002. Neuronal injury affects expression of helix-loop-helix transcription factors. Neuroreport 13:2385–2388. doi: 10.1097/00001756-200212200-00002. [DOI] [PubMed] [Google Scholar]
- 56.Sun T, Xiao HS, Zhou PB, Lu YJ, Bao L, Zhang X. 2006. Differential expression of synaptoporin and synaptophysin in primary sensory neurons and up-regulation of synaptoporin after peripheral nerve injury. Neuroscience 141:1233–1245. doi: 10.1016/j.neuroscience.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Morimoto S, Cassell MD, Sigmund CD. 2002. Neuron-specific expression of human angiotensinogen in brain causes increased salt appetite. Physiol Genomics 9:113–120. doi: 10.1152/physiolgenomics.00007.2002. [DOI] [PubMed] [Google Scholar]
- 58.Gimeno RE. 2007. Fatty acid transport proteins. Curr Opin Lipidol 18:271–276. doi: 10.1097/MOL.0b013e3281338558. [DOI] [PubMed] [Google Scholar]
- 59.Ogura M, Takarada T, Nakamichi N, Kawagoe H, Sako A, Nakazato R, Yoneda Y. 2011. Exacerbated vulnerability to oxidative stress in astrocytic C6 glioma cells with stable overexpression of the glutamine transporter slc38a1. Neurochem Int 58:504–511. doi: 10.1016/j.neuint.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Lockhart PJ, Holtom B, Lincoln S, Hussey J, Zimprich A, Gasser T, Wszolek ZK, Hardy J, Farrer MJ. 2002. The human sideroflexin 5 (SFXN5) gene: sequence, expression analysis and exclusion as a candidate for PARK3. Gene 285:229–237. doi: 10.1016/S0378-1119(02)00402-X. [DOI] [PubMed] [Google Scholar]
- 61.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. 1999. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med 190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H. 2002. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol 158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamao T, Noguchi T, Takeuchi O, Nishiyama U, Morita H, Hagiwara T, Akahori H, Kato T, Inagaki K, Okazawa H, Hayashi Y, Matozaki T, Takeda K, Akira S, Kasuga M. 2002. Negative regulation of platelet clearance and of the macrophage phagocytic response by the transmembrane glycoprotein SHPS-1. J Biol Chem 277:39833–39839. doi: 10.1074/jbc.M203287200. [DOI] [PubMed] [Google Scholar]
- 64.Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. 2002. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol 168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- 65.Villa C, Venturelli E, Fenoglio C, Clerici F, Marcone A, Benussi L, Ghidoni R, Gallone S, Cortini F, Scalabrini D, Serpente M, Binetti G, Cappa S, Mariani C, Rainero I, Bresolin N, Scarpini E, Galimberti D. 2009. CCL8/MCP-2 association analysis in patients with Alzheimer's disease and frontotemporal lobar degeneration. J Neurol 256:1379–1381. doi: 10.1007/s00415-009-5138-y. [DOI] [PubMed] [Google Scholar]
- 66.Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA, Goodwin RG. 1995. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 67.Warke RV, Becerra A, Zawadzka A, Schmidt DJ, Martin KJ, Giaya K, Dinsmore JH, Woda M, Hendricks G, Levine T, Rothman AL, Bosch I. 2008. Efficient dengue virus (DENV) infection of human muscle satellite cells upregulates type I interferon response genes and differentially modulates MHC I expression on bystander and DENV-infected cells. J Gen Virol 89:1605–1615. doi: 10.1099/vir.0.2008/000968-0. [DOI] [PubMed] [Google Scholar]
- 68.Li S, Xie Y, Zhang W, Gao J, Wang M, Zheng G, Yin X, Xia H, Tao X. 2015. Interferon alpha-inducible protein 27 promotes epithelial-mesenchymal transition and induces ovarian tumorigenicity and stemness. J Surg Res 193:255–264. doi: 10.1016/j.jss.2014.06.055. [DOI] [PubMed] [Google Scholar]
- 69.Li W, He F. 2014. Monocyte to macrophage differentiation-associated (MMD) targeted by miR-140-5p regulates tumor growth in non-small cell lung cancer. Biochem Biophys Res Commun 450:844–850. doi: 10.1016/j.bbrc.2014.06.075. [DOI] [PubMed] [Google Scholar]
- 70.Barnard DC, Patton JG. 2000. Identification and characterization of a novel serine-arginine-rich splicing regulatory protein. Mol Cell Biol 20:3049–3057. doi: 10.1128/MCB.20.9.3049-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang X, Beullens M, Zhang J, Zhou Y, Nicolaescu E, Lesage B, Hu Q, Wu J, Bollen M, Shi Y. 2009. Structure and function of the two tandem WW domains of the pre-mRNA splicing factor FBP21 (formin-binding protein 21). J Biol Chem 284:25375–25387. doi: 10.1074/jbc.M109.024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordmann A, Wixler L, Boergeling Y, Wixler V, Ludwig S. 2012. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J 26:1290–1300. doi: 10.1096/fj.11-189886. [DOI] [PubMed] [Google Scholar]
- 73.Wilczynska KM, Singh SK, Adams B, Bryan L, Rao RR, Valerie K, Wright S, Griswold-Prenner I, Kordula T. 2009. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem Cells 27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiefer JC. 2007. Back to basics: Sox genes. Dev Dyn 236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- 75.Kaltschmidt B, Kaltschmidt C. 2009. NF-kappaB in the nervous system. Cold Spring Harb Perspect Biol 1:a001271. doi: 10.1101/cshperspect.a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barton DE, Arquint M, Roder J, Dunn R, Francke U. 1987. The myelin-associated glycoprotein gene: mapping to human chromosome 19 and mouse chromosome 7 and expression in quivering mice. Genomics 1:107–112. doi: 10.1016/0888-7543(87)90002-4. [DOI] [PubMed] [Google Scholar]
- 77.Hamacher M, Pippirs U, Kohler A, Muller HW, Bosse F. 2001. Plasmolipin: genomic structure, chromosomal localization, protein expression pattern, and putative association with Bardet-Biedl syndrome. Mamm Genome 12:933–937. doi: 10.1007/s00335-001-3035-5. [DOI] [PubMed] [Google Scholar]
- 78.Kruer MC, Paisan-Ruiz C, Boddaert N, Yoon MY, Hama H, Gregory A, Malandrini A, Woltjer RL, Munnich A, Gobin S, Polster BJ, Palmeri S, Edvardson S, Hardy J, Houlden H, Hayflick SJ. 2010. Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann Neurol 68:611–618. doi: 10.1002/ana.22122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mandich P, Grandis M, Geroldi A, Acquaviva M, Varese A, Gulli R, Ciotti P, Bellone E. 2008. Gap junction beta 1 (GJB1) gene mutations in Italian patients with X-linked Charcot-Marie-Tooth disease. J Hum Genet 53:529–533. doi: 10.1007/s10038-008-0280-4. [DOI] [PubMed] [Google Scholar]
- 80.Kutzleb C, Petrasch-Parwez E, Kilimann MW. 2007. Cellular and subcellular localization of paralemmin-1, a protein involved in cell shape control, in the rat brain, adrenal gland and kidney. Histochem Cell Biol 127:13–30. [DOI] [PubMed] [Google Scholar]
- 81.Yonezawa T, Ohtsuka A, Yoshitaka T, Hirano S, Nomoto H, Yamamoto K, Ninomiya Y. 2003. Limitrin, a novel immunoglobulin superfamily protein localized to glia limitans formed by astrocyte endfeet. Glia 44:190–204. doi: 10.1002/glia.10279. [DOI] [PubMed] [Google Scholar]
- 82.Watanabe K, Hasegawa Y, Yamashita H, Shimizu K, Ding Y, Abe M, Ohta H, Imagawa K, Hojo K, Maki H, Sonoda H, Sato Y. 2004. Vasohibin as an endothelium-derived negative feedback regulator of angiogenesis. J Clin Invest 114:898–907. doi: 10.1172/JCI200421152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Copie-Bergman C, Gaulard P, Maouche-Chretien L, Briere J, Haioun C, Alonso MA, Romeo PH, Leroy K. 1999. The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood 94:3567–3575. [PubMed] [Google Scholar]
- 84.Ferraro A, Schepis F, Leone V, Federico A, Borbone E, Pallante P, Berlingieri MT, Chiappetta G, Monaco M, Palmieri D, Chiariotti L, Santoro M, Fusco A. 2013. Tumor suppressor role of the CL2/DRO1/CCDC80 gene in thyroid carcinogenesis. J Clin Endocrinol Metab 98:2834–2843. doi: 10.1210/jc.2012-2926. [DOI] [PubMed] [Google Scholar]
- 85.Digweed M, Gunthert U, Schneider R, Seyschab H, Friedl R, Sperling K. 1995. Irreversible repression of DNA synthesis in Fanconi anemia cells is alleviated by the product of a novel cyclin-related gene. Mol Cell Biol 15:305–314. doi: 10.1128/MCB.15.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. 2015. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Ed Engl 54:451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- 87.Letourneau PC. 2009. Actin in axons: stable scaffolds and dynamic filaments. Results Probl Cell Differ 48:65–90. doi: 10.1007/400_2009_3. [DOI] [PubMed] [Google Scholar]
- 88.Muller L, de Escauriaza MD, Lajoie P, Theis M, Jung M, Muller A, Burgard C, Greiner M, Snapp EL, Dudek J, Zimmermann R. 2010. Evolutionary gain of function for the ER membrane protein Sec62 from yeast to humans. Mol Biol Cell 21:691–703. doi: 10.1091/mbc.E09-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takashima H, Boerkoel CF, De Jonghe P, Ceuterick C, Martin JJ, Voit T, Schroder JM, Williams A, Brophy PJ, Timmerman V, Lupski JR. 2002. Periaxin mutations cause a broad spectrum of demyelinating neuropathies. Ann Neurol 51:709–715. doi: 10.1002/ana.10213. [DOI] [PubMed] [Google Scholar]
- 90.Fricker LD, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J. 2000. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci 20:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, Trono D. 2015. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. eLife 4:e06068. doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lopez-Hernandez T, Sirisi S, Capdevila-Nortes X, Montolio M, Fernandez-Duenas V, Scheper GC, van der Knaap MS, Casquero P, Ciruela F, Ferrer I, Nunes V, Estevez R. 2011. Molecular mechanisms of MLC1 and GLIALCAM mutations in megalencephalic leukoencephalopathy with subcortical cysts. Hum Mol Genet 20:3266–3277. doi: 10.1093/hmg/ddr238. [DOI] [PubMed] [Google Scholar]
- 93.Whitmarsh AJ, Kuan CY, Kennedy NJ, Kelkar N, Haydar TF, Mordes JP, Appel M, Rossini AA, Jones SN, Flavell RA, Rakic P, Davis RJ. 2001. Requirement of the JIP1 scaffold protein for stress-induced JNK activation. Genes Dev 15:2421–2432. doi: 10.1101/gad.922801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zak M, Bress A, Pfister M, Blin N. 2011. Temporal expression pattern of Fkbp8 in rodent cochlea. Cell Physiol Biochem 28:1023–1030. doi: 10.1159/000335789. [DOI] [PubMed] [Google Scholar]
- 95.Mamalaki A, Boutou E, Hurel C, Patsavoudi E, Tzartos S, Matsas R. 1995. The BM88 antigen, a novel neuron-specific molecule, enhances the differentiation of mouse neuroblastoma cells. J Biol Chem 270:14201–14208. doi: 10.1074/jbc.270.23.14201. [DOI] [PubMed] [Google Scholar]
- 96.Terauchi A, Timmons KM, Kikuma K, Pechmann Y, Kneussel M, Umemori H. 2015. Selective synaptic targeting of the excitatory and inhibitory presynaptic organizers FGF22 and FGF7. J Cell Sci 128:281–292. doi: 10.1242/jcs.158337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Surpili MJ, Delben TM, Kobarg J. 2003. Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry 42:15369–15376. doi: 10.1021/bi034575v. [DOI] [PubMed] [Google Scholar]
- 98.Chergui K, Svenningsson P, Greengard P. 2005. Physiological role for casein kinase 1 in glutamatergic synaptic transmission. J Neurosci 25:6601–6609. doi: 10.1523/JNEUROSCI.1082-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Monteggia LM, Gideons E, Kavalali ET. 2013. The role of eukaryotic elongation factor 2 kinase in rapid antidepressant action of ketamine. Biol Psychiatry 73:1199–1203. doi: 10.1016/j.biopsych.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kele J, Andersson ER, Villaescusa JC, Cajanek L, Parish CL, Bonilla S, Toledo EM, Bryja V, Rubin JS, Shimono A, Arenas E. 2012. SFRP1 and SFRP2 dose-dependently regulate midbrain dopamine neuron development in vivo and in embryonic stem cells. Stem Cells 30:865–875. doi: 10.1002/stem.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marsland BJ, Battig P, Bauer M, Ruedl C, Lassing U, Beerli RR, Dietmeier K, Ivanova L, Pfister T, Vogt L, Nakano H, Nembrini C, Saudan P, Kopf M, Bachmann MF. 2005. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity 22:493–505. doi: 10.1016/j.immuni.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 102.Satoh J, Tabunoki H, Yamamura T, Arima K, Konno H. 2007. Human astrocytes express aquaporin-1 and aquaporin-4 in vitro and in vivo. Neuropathology 27:245–256. doi: 10.1111/j.1440-1789.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- 103.Saarela J, Jung G, Hermann M, Nimpf J, Schneider WJ. 2008. The patatin-like lipase family in Gallus gallus. BMC Genomics 9:281. doi: 10.1186/1471-2164-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Larsson J, Forsberg M, Brannvall K, Zhang XQ, Enarsson M, Hedborg F, Forsberg-Nilsson K. 2008. Nuclear receptor binding protein 2 is induced during neural progenitor differentiation and affects cell survival. Mol Cell Neurosci 39:32–39. doi: 10.1016/j.mcn.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Shang YC, Chong ZZ, Wang S, Maiese K. 2012. Wnt1 inducible signaling pathway protein 1 (WISP1) targets PRAS40 to govern beta-amyloid apoptotic injury of microglia. Curr Neurovasc Res 9:239–249. doi: 10.2174/156720212803530618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sugino T, Nozaki K, Hashimoto N. 2000. Activation of mitogen-activated protein kinases in gerbil hippocampus with ischemic tolerance induced by 3-nitropropionic acid. Neurosci Lett 278:101–104. doi: 10.1016/S0304-3940(99)00906-4. [DOI] [PubMed] [Google Scholar]
- 107.Doxakis E, Huang EJ, Davies AM. 2004. Homeodomain-interacting protein kinase-2 regulates apoptosis in developing sensory and sympathetic neurons. Curr Biol 14:1761–1765. doi: 10.1016/j.cub.2004.09.050. [DOI] [PubMed] [Google Scholar]
- 108.Harder JM, Ding Q, Fernandes KA, Cherry JD, Gan L, Libby RT. 2012. BCL2L1 (BCL-X) promotes survival of adult and developing retinal ganglion cells. Mol Cell Neurosci 51:53–59. doi: 10.1016/j.mcn.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gondi CS, Dinh DH, Klopfenstein JD, Gujrati M, Rao JS. 2009. MMP-2 downregulation mediates differential regulation of cell death via ErbB-2 in glioma xenografts. Int J Oncol 35:257–263. [PMC free article] [PubMed] [Google Scholar]
- 110.Erlich S, Shohami E, Pinkas-Kramarski R. 2000. Closed head injury induces up-regulation of ErbB-4 receptor at the site of injury. Mol Cell Neurosci 16:597–608. doi: 10.1006/mcne.2000.0894. [DOI] [PubMed] [Google Scholar]
- 111.Tu-Sekine B, Raben DM. 2011. Regulation and roles of neuronal diacylglycerol kinases: a lipid perspective. Crit Rev Biochem Mol Biol 46:353–364. doi: 10.3109/10409238.2011.577761. [DOI] [PubMed] [Google Scholar]
- 112.Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM. 2006. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 7:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- 113.Perez-Janices N, Blanco-Luquin I, Tunon MT, Barba-Ramos E, Ibanez B, Zazpe-Cenoz I, Martinez-Aguillo M, Hernandez B, Martinez-Lopez E, Fernandez AF, Mercado MR, Cabada T, Escors D, Megias D, Guerrero-Setas D. 2015. EPB41L3, TSP-1 and RASSF2 as new clinically relevant prognostic biomarkers in diffuse gliomas. Oncotarget 6:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsujikawa M, Malicki J. 2004. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 42:703–716. doi: 10.1016/S0896-6273(04)00268-5. [DOI] [PubMed] [Google Scholar]
- 115.Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM. 1999. GMAP-210, a cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol 145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. 2006. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochem Biophys Res Commun 343:535–543. doi: 10.1016/j.bbrc.2006.02.160. [DOI] [PubMed] [Google Scholar]
- 117.Camarena V, Cao L, Abad C, Abrams A, Toledo Y, Araki K, Araki M, Walz K, Young JI. 2014. Disruption of Mbd5 in mice causes neuronal functional deficits and neurobehavioral abnormalities consistent with 2q23.1 microdeletion syndrome. EMBO Mol Med 6:1003–1015. doi: 10.15252/emmm.201404044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pettersson M, Dannaeus K, Nilsson K, Jonsson JI. 2000. Isolation of MYADM, a novel hematopoietic-associated marker gene expressed in multipotent progenitor cells and up-regulated during myeloid differentiation. J Leukoc Biol 67:423–431. [DOI] [PubMed] [Google Scholar]
- 119.Davis AE III, Cai S, Liu D. 2004. The biological role of the C1 inhibitor in regulation of vascular permeability and modulation of inflammation. Adv Immunol 82:331–363. doi: 10.1016/S0065-2776(04)82008-X. [DOI] [PubMed] [Google Scholar]
- 120.North RA. 2004. P2X3 receptors and peripheral pain mechanisms. J Physiol 554:301–308. doi: 10.1113/jphysiol.2003.048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Watson AP, Evans RL, Egland KA. 2013. Multiple functions of sushi domain containing 2 (SUSD2) in breast tumorigenesis. Mol Cancer Res 11:74–85. doi: 10.1158/1541-7786.MCR-12-0501-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kzhyshkowska J, Gratchev A, Goerdt S. 2007. Human chitinases and chitinase-like proteins as indicators for inflammation and cancer. Biomark Insights 2:128–146. [PMC free article] [PubMed] [Google Scholar]
- 123.Richter F, Meurers BH, Zhu C, Medvedeva VP, Chesselet MF. 2009. Neurons express hemoglobin alpha- and beta-chains in rat and human brains. J Comp Neurol 515:538–547. doi: 10.1002/cne.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhou F, van Laar T, Huang H, Zhang L. 2011. APP and APLP1 are degraded through autophagy in response to proteasome inhibition in neuronal cells. Protein Cell 2:377–383. doi: 10.1007/s13238-011-1047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hermann JS, Skroblin P, Bertinetti D, Hanold LE, von der Heide EK, Wagener EM, Zenn HM, Klussmann E, Kennedy EJ, Herberg FW. 2015. Neurochondrin is an atypical RIIalpha-specific A-kinase anchoring protein. Biochim Biophys Acta 1854:1667–1675. doi: 10.1016/j.bbapap.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nakayama T, Yaoi T, Yasui M, Kuwajima G. 1998. N-copine: a novel two C2-domain-containing protein with neuronal activity-regulated expression. FEBS Lett 428:80–84. doi: 10.1016/S0014-5793(98)00497-9. [DOI] [PubMed] [Google Scholar]
- 127.Gershon AA, Chen J, Davis L, Krinsky C, Cowles R, Reichard R, Gershon M. 2012. Latency of varicella zoster virus in dorsal root, cranial, and enteric ganglia in vaccinated children. Trans Am Clin Climatol Assoc 123:17–33. [PMC free article] [PubMed] [Google Scholar]
- 128.Arvin AM, Moffat JF, Sommer M, Oliver S, Che X, Vleck S, Zerboni L, Ku CC. 2010. Varicella-zoster virus T cell tropism and the pathogenesis of skin infection. Curr Top Microbiol Immunol 342:189–209. doi: 10.1007/82_2010_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arvin AM, Koropchak CM, Williams BR, Grumet FC, Foung SK. 1986. Early immune response in healthy and immunocompromised subjects with primary varicella-zoster virus infection. J Infect Dis 154:422–429. doi: 10.1093/infdis/154.3.422. [DOI] [PubMed] [Google Scholar]
- 130.Ku CC, Padilla JA, Grose C, Butcher EC, Arvin AM. 2002. Tropism of varicella-zoster virus for human tonsillar CD4(+) T lymphocytes that express activation, memory, and skin homing markers. J Virol 76:11425–11433. doi: 10.1128/JVI.76.22.11425-11433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Moffat JF, Stein MD, Kaneshima H, Arvin AM. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J Virol 69:5236–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Matsuda K, Park CH, Sunden Y, Kimura T, Ochiai K, Kida H, Umemura T. 2004. The vagus nerve is one route of transneural invasion for intranasally inoculated influenza a virus in mice. Vet Pathol 41:101–107. doi: 10.1354/vp.41-2-101. [DOI] [PubMed] [Google Scholar]
- 133.Gesser RM, Koo SC. 1996. Oral inoculation with herpes simplex virus type 1 infects enteric neuron and mucosal nerve fibers within the gastrointestinal tract in mice. J Virol 70:4097–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thompson RL, Sawtell NM. 2000. Replication of herpes simplex virus type 1 within trigeminal ganglia is required for high frequency but not high viral genome copy number latency. J Virol 74:965–974. doi: 10.1128/JVI.74.2.965-974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Song HS, Back JH, Jin DK, Chung PW, Moon HS, Suh BC, Kim YB, Kim BM, Woo HY, Lee YT, Park KY. 2008. Cardiac troponin T elevation after stroke: relationships between elevated serum troponin T, stroke location, and prognosis. J Clin Neurol 4:75–83. doi: 10.3988/jcn.2008.4.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hasirci B, Okay M, Agircan D, Kocer A. 2013. Elevated troponin level with negative outcome was found in ischemic stroke. Cardiovasc Psychiatry Neurol 2013:953672. doi: 10.1155/2013/953672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang DW, Yoo SH, Chun S, Kwon KY, Kwon SU, Koh JY, Kim JS. 2009. Inflammatory and hemostatic biomarkers associated with early recurrent ischemic lesions in acute ischemic stroke. Stroke 40:1653–1658. doi: 10.1161/STROKEAHA.108.539429. [DOI] [PubMed] [Google Scholar]
- 138.Wirrell E, Hill MD, Jadavji T, Kirton A, Barlow K. 2004. Stroke after varicella vaccination. J Pediatr 145:845–847. doi: 10.1016/j.jpeds.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 139.van Velzen M, Laman JD, Kleinjan A, Poot A, Osterhaus AD, Verjans GM. 2009. Neuron-interacting satellite glial cells in human trigeminal ganglia have an APC phenotype. J Immunol 183:2456–2461. doi: 10.4049/jimmunol.0900890. [DOI] [PubMed] [Google Scholar]
- 140.Trudler D, Farfara D, Frenkel D. 2010. Toll-like receptors expression and signaling in glia cells in neuro-amyloidogenic diseases: towards future therapeutic application. Mediators Inflamm 2010:497987. doi: 10.1155/2010/497987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 142.Arvin AM, Kushner JH, Feldman S, Baehner RL, Hammond D, Merigan TC. 1982. Human leukocyte interferon for the treatment of varicella in children with cancer. N Engl J Med 306:761–765. doi: 10.1056/NEJM198204013061301. [DOI] [PubMed] [Google Scholar]
- 143.Meyer C, Dewane J, Kerns A, Haberthur K, Barron A, Park B, Messaoudi I. 2013. Age and immune status of rhesus macaques impact simian varicella virus gene expression in sensory ganglia. J Virol 87:8294–8306. doi: 10.1128/JVI.01112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Verweij MC, Wellish M, Whitmer T, Malouli D, Lapel M, Jonjic S, Haas JG, DeFilippis VR, Mahalingam R, Fruh K. 2015. Varicella viruses inhibit interferon-stimulated JAK-STAT signaling through multiple mechanisms. PLoS Pathog 11:e1004901. doi: 10.1371/journal.ppat.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Clarner T, Janssen K, Nellessen L, Stangel M, Skripuletz T, Krauspe B, Hess FM, Denecke B, Beutner C, Linnartz-Gerlach B, Neumann H, Vallieres L, Amor S, Ohl K, Tenbrock K, Beyer C, Kipp M. 2015. CXCL10 triggers early microglial activation in the cuprizone model. J Immunol 194:3400–3413. doi: 10.4049/jimmunol.1401459. [DOI] [PubMed] [Google Scholar]
- 146.Muller M, Carter S, Hofer MJ, Campbell IL. 2010. Review: the chemokine receptor CXCR3 and its ligands CXCL9, CXCL10 and CXCL11 in neuroimmunity—a tale of conflict and conundrum. Neuropathol Appl Neurobiol 36:368–387. doi: 10.1111/j.1365-2990.2010.01089.x. [DOI] [PubMed] [Google Scholar]
- 147.Whitaker-Dowling PA, Wilcox DK, Widnell CC, Youngner JS. 1983. Interferon-mediated inhibition of virus penetration. Proc Natl Acad Sci U S A 80:1083–1086. doi: 10.1073/pnas.80.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Campadelli-Fiume G, Menotti L. 2007. Entry of alphaherpesviruses into the cell. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 149.Fazakerley JK, Walker R. 2003. Virus demyelination. J Neurovirol 9:148–164. doi: 10.1080/13550280390194046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martin JR. 1984. Intra-axonal virus in demyelinative lesions of experimental herpes simplex type 2 infection. J Neurol Sci 63:63–74. doi: 10.1016/0022-510X(84)90109-6. [DOI] [PubMed] [Google Scholar]
- 151.Hood C, Cunningham AL, Slobedman B, Arvin AM, Sommer MH, Kinchington PR, Abendroth A. 2006. Varicella-zoster virus ORF63 inhibits apoptosis of primary human neurons. J Virol 80:1025–1031. doi: 10.1128/JVI.80.2.1025-1031.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Malmquist SJ, Abramsson A, McGraw HF, Linbo TH, Raible DW. 2013. Modulation of dorsal root ganglion development by ErbB signaling and the scaffold protein Sorbs3. Development 140:3986–3996. doi: 10.1242/dev.084640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kennedy PG, Major EO, Williams RK, Straus SE. 1994. Down-regulation of glial fibrillary acidic protein expression during acute lytic varicella-zoster virus infection of cultured human astrocytes. Virology 205:558–562. doi: 10.1006/viro.1994.1679. [DOI] [PubMed] [Google Scholar]
- 154.Jones M, Dry IR, Frampton D, Singh M, Kanda RK, Yee MB, Kellam P, Hollinshead M, Kinchington PR, O'Toole EA, Breuer J. 2014. RNA-seq analysis of host and viral gene expression highlights interaction between varicella zoster virus and keratinocyte differentiation. PLoS Pathog 10:e1003896. doi: 10.1371/journal.ppat.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jones JO, Arvin AM. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J Virol 77:1268–1280. doi: 10.1128/JVI.77.2.1268-1280.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.