Abstract
Nucleocytoplasmic trafficking of functional proteins plays a key role in regulating gene expressions in response to extracellular signals. We developed a genetically encoded bioluminescent indicator for monitoring the nuclear trafficking of target proteins in vitro and in vivo. The principle is based on reconstitution of split fragments of Renilla reniformis (Rluc) by protein splicing with a DnaE intein (a catalytic subunit of DNA polymerase III). A target cytosolic protein fused to the N-terminal half of Rluc is expressed in mammalian cells. If the protein translocates into the nucleus, the Rluc moiety meets the C-terminal half of Rluc, and full-length Rluc is reconstituted by protein splicing. We demonstrated quantitative cell-based in vitro sensing of ligand-induced translocation of androgen receptor, which allowed high-throughput screening of exo- and endogenous agonists and antagonists. Furthermore, the indicator enabled noninvasive in vivo imaging of the androgen receptor translocation in the brains of living mice with a charge-coupled device imaging system. These rapid and quantitative analyses in vitro and in vivo provide a wide variety of applications for screening pharmacological or toxicological compounds and testing them in living animals.
The control of complicated signaling networks within eukaryotic cells relies on the compartmentalization of each protein. Nucleocytoplasmic trafficking of proteins in response to extra- or intracellular stimuli is an essential step for regulating the magnitude and specificity of gene expressions. The trafficking is regulated by posttranslational modifications of proteins, which include ligand-receptor binding, protein phosphorylation, and proteolysis. The nuclear localization of those proteins is altered in the cells that are exposed to specific exogenous chemicals, of which potential effects on living animals are the major concern. Also, because various nuclear proteins are mislocalized in cancer cells, there is an intense interest in identifying small molecules that redirect the proteins to the correct compartments. Hence, development of a rapid screening system to detect the nucleocytoplasmic trafficking is essential for the discovery of novel compounds that have anticancer activity or for testing the toxicity of chemicals, from which new insights into the mechanism of nucleocytoplasmic trafficking could be provided (1–4).
A technique for monitoring the dynamics of the protein movement inside single living cells relies on the use of immunocytochemistry or optical imaging with genetically tagged GFP (5). These analyses are effective for imaging the spatial and temporal dynamics of proteins of interest within single living cells. For high-throughput analysis of the protein movement inside the cells, automated fluorescence microscopy has been developed (2). Although such technological progress is important, image acquisition can be slow and tedious. In particular, algorithms to automatically determine nuclear vs. cytoplasmic localization in an acquired image still remain imprecise and slow. The obtained results are qualitative rather than quantitative because of the limited number of analyzed cells. In addition, analyses of the protein localization in living animals require complex assay procedures such as extraction of an organ and dividing it into sliced sections, which hampers temporal and quantitative analyses.
In our previous studies, a system of split firefly luciferase reconstitution by protein splicing has been developed for detecting protein–protein interactions in cell lines and in living mice, in which the novel use of the new luminescent reporter protein was demonstrated (6, 7). We and others have also reported a split Rluc complementation system for imaging tyrosine phosphorylation and to monitor protein–protein interactions in living cells, where bioluminescence originated from Rluc complementation was sensitively detected (8, 9). A method for identifying mitochondrial proteins by using split GFP reconstitution by protein splicing has recently been developed by our group (10). These studies suggest a great potentiality of Rluc for rapid sensing of intracellular protein movements in living cells and animals.
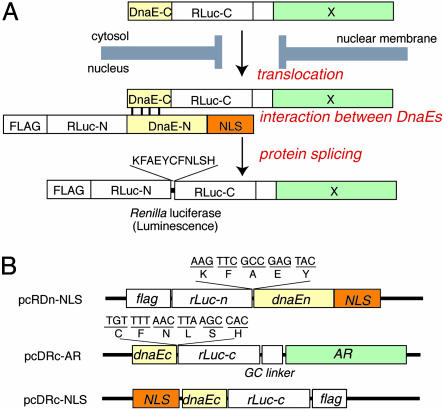
Taking advantage of the bioluminescence of Rluc with background-fluorescence-free and high-sensitive detection, we developed an indicator with general applicability for detecting transport into the nucleus of a particular protein. The indicator consists of two fragments of an Ssp. DnaE intein (a catalytic subunit of DNA polymerase III) connected with split Rluc, of which bioluminescence activity is completely lost (Fig. 1A). The N-terminal fragment is localized in the nucleus, whereas the C-terminal fragment joined to a particular protein is in the cytosol. Translocation of the protein into the nucleus results in protein splicing with DnaE, thereby producing a full-length Rluc and recovering its bioluminescence activity. To demonstrate the usefulness of the indicator, we selected a well known nuclear receptor (androgen receptor, AR), which translocates from the cytosol into the nucleus upon binding to 5α-dihydrotestosterone (DHT) (11). We herein report a method for quantitative analysis of the extent of AR translocation with various exo- and endogenous chemical compounds in vitro and inhibitory effects of these chemical compounds on the translocation of AR in the brain of living mice by using a noninvasive imaging technique. Although we demonstrate the feasibility of the present indicator using only AR translocation, it should be emphasized that the present approach can well be applied for any proteins translocating from the cytosol to the nucleus with which a rapid screening system can possibly be developed for discovering novel compounds of anticancer drugs or for testing chemicals of their toxicity.
Fig. 1.
Basic strategy for the detection of AR translocation. (A) Principle for monitoring translocation of a particular protein (X) into the nucleus using protein splicing of split-Rluc. RLuc-N (1∼229 aa) is connected with DnaE-N (1∼123 aa) and NLS [(DPKKKRKV)3], which is predominantly localized in the nucleus. DnaE-C (1∼36 aa) is connected with RLuc-C (230∼311 aa) and a protein X, which is localized in the cytosol. When the tandem fusion protein consisting of DnaE-C, Rluc-C, and protein X translocates into the nucleus, the DnaE-C interacts with DnaE-N, and protein splicing results. Rluc-N and -C are linked by a peptide bond, and the reconstituted Rluc recovers its bioluminescence activity. FLAG means epitope (DYKDDDDK). (B) Schematic structures of cDNA constructs. All italics mean the genes of their corresponding proteins. Additional sequences at the boundaries between rLuc and dnaE are shown under the bars. GC linker, cDNA sequence of amino acids GGGGSG.
Experimental Procedures
Construction of Plasmids. The cDNA-encoding N-terminal domain of Rluc (Rluc-N; 1∼229 aa) was modified by PCR to introduce the peptide (KFAEYC) to the C terminus of Rluc-N and to introduce the FLAG epitope (DYKDDDDK) to the N terminus of Rluc-N. The cDNA encoding the modified Rluc-N was fused to a cDNA encoding the N-terminal splicing domain of DnaE (DnaE-N; 1∼123 aa) with a native HindIII site. The cDNA of C terminus of DnaE-N was fused to a cDNA of a nuclear localization signal [NLS; (DPKKKRKV)3] with the NcoI site.
The cDNA-encoding C-terminal domain of Rluc (Rluc-C; 230∼311 aa) was modified by PCR to introduce the peptide, FNLSH, and the unique enzyme site, MunI, to the N terminus of Rluc-C and to introduce a linker (GGGGSG) and a unique enzyme site, NotI, to the C terminus of Rluc-C. The cDNA encoding AR (1∼918 aa) was modified by PCR to add a unique enzyme site, NotI, at its N-terminal end and XhoI at its C-terminal end. The cDNA encoding the modified Rluc-C was fused to a cDNA encoding the C-terminal splicing domain of DnaE (DnaE-C; 1∼36 aa) with the MunI site and to cDNA encoding the full AR with NotI.
The cDNAs were subcloned into the expression vector pcDNA 3.1(+) (Invitrogen) at the unique enzyme sites BamHI and XhoI. The thus subcloned vectors containing N- and C-terminal RLuc were named pcRDn-NLS and pcDRc-AR, respectively. The PCR products were sequenced to ensure fidelity with a BigDye Terminator Cycle Sequencing kit and a genetic analyzer ABI Prism310 (PE Biosystems).
Cell Culture and Transfection. COS-7 cells were cultured in DMEM supplemented with 10% steroid-free FBS (a charcoal-extracted FBS) and 1% penicillin/streptomycin at 37°C in 5% CO2. The cells seeded in 12-well culture plates were transfected with 2 μg of constructed plasmids using a lipofection reagent, lipofectAMINE2000 (Invitrogen).
Western Blot Analysis. COS-7 cells were transfected with either pcRDn-NLS or pcDRc-AR and incubated for 24 h. The cells were washed once in PBS and lysed in 200 μl of lysis buffer (1% SDS/10% glycerol/10% 2-mercaptoethanol/0.001% bromophenol blue/50 mM Tris·HCl, pH 6.8). Equal amounts of the samples were electrophoresed in 6% acrylamide gels, transferred to nitrocellulose membrane, and blotted with mouse anti-AR antibody (Santa Cruz Biotechnology) or mouse anti-β-actin antibody (Sigma). The blots were incubated with alkaline phosphatase-conjugated secondary antibodies and visualized by chemiluminescence (New England Biolabs).
Immunocytochemistry. COS-7 cells were cultured on microscope glass slides (2 × 105 cells per slide) and were transfected with the constructed plasmids. The transfected cells were fixed with a 3% paraformaldehyde solution. The cells were blocked with 0.2% fish skin gelatin and then incubated with mouse anti-AR antibody (Santa Cruz Biotechnology) or mouse anti-FLAG antibody (Sigma). The antibodies were reacted with Cy-5-conjugated secondary antibody, and then the nucleus was stained with YO-PRO-1 (Molecular Probes). The fluorescence was recorded by using a confocal laser-scanning microscope (LSM510; Zeiss) fitted with a band-pass filter (505–530 nm) for YO-PRO-1 and an LP filter (665 nm) for Cy-5.
Cell-Based in Vitro Assay. COS-7 cells were cotransfected with the plasmids pcRDn-NLS and pcDRc-AR and incubated for 12 h. The medium was replaced with DMEM supplemented with 10% FBS, and 24 h after the replacement, steroid hormones or synthetic chemicals were added to each well. After the COS-7 cells were extensively incubated for 2 hours, the cells were harvested, and luciferase activity was evaluated by using the Renilla luciferase (Rluc) assay kit (Promega) with a luminometer (Minilumat LB9506; Berthold, Nashua, NH) with an integration time of 20 sec. For cell fractionation assay, COS-7 cells cotransfected with the plasmids pcRDn-NLS and pcDRc-AR were incubated for 36 h, and DHT (10–6 M) or DMSO (0.1%, vehicle) was added to each well. The cells were incubated for 2 hours, and then the cells were harvested and suspended in a solution of 0.25 M sucrose/5 mM EDTA/20 mM Tris·HCl (pH 7.4). The cells were crushed with a tip sonicator, and the homogenate was centrifuged at 600 × g for 15 min to separate the nucleus from the cytosol. The luminescence intensity of the fractions was measured with the luminometer.
In Vivo Imaging of Living Mice. The COS-7 cells were transfected with the constructed plasmid pcRDn-NLS and pcDRc-AR, respectively, or cotransfected with pcRDn-NLS and pcDRc-AR. The cells were harvested after incubation in the DMEM with 10% FBS for 12 h after transfection. The cells were suspended in DMEM, and an aliquot of 1 × 106 cells was implanted in four different sites on the back of anesthetized BALB/c nude mice (female, 5 weeks old, ≈17 g body weight). Twelve hours after cell implantation, 100 μl of DHT (100 μg/kg of body weight) was injected i.p. Two hours after injection, 100 μl of coelenterazine (2.8 mg/kg of body weight) was injected i.p., and the mice were imaged at 2-min intervals.
For examining DHT dependence on the Rluc activity, two groups of nude mice were injected with the COS-7 cells (1 × 106 cells) cotransfected with plasmids pcRDn-NLS and pcDRc-AR directly into the backs of the mice. Of the two groups, the first mouse group was injected i.p. with 100 μl of 1.0% (vol/vol) DMSO (vehicle). The second group was injected with 100 μl of DHT (100 μg/kg of body weight). After 3 h, 100 μl of coelenterazine (1.4 mg/kg of body weight) was injected i.p., and the mice were imaged 10 min later (n = 4).
For testing inhibitory effects of the chemicals on the translocation of AR in the brains of mice, COS-7 cells (1 × 105 cells) cotransfected with plasmids pcRDn-NLS and pcDRc-AR were implanted in the forebrain of the nude mice at a depth of 3 mm through a 1-mm burrhole. Soon after implantation, the first and second mouse groups were injected i.p. with 100 μl of 1.0% DMSO. The third and fourth mouse groups were injected i.p. with 100 μl of procymidone and polychlorinated biphenyls (PCB) (10 mg/kg of body weight) dissolved in 1.0% DMSO, respectively. Two hours after injection, the second, third, and fourth groups of mice were injected i.p. with 100 μl of DHT (10 μg/kg of body weight) in 1.0% DMSO, whereas the first group of mice was injected with the same amount of DMSO. Two hours after injection, 10 μl of coelenterazine (0.14 mg/kg of body weight) was injected intracerebrally in the mice, and representatives from each mouse group (n = 3) were imaged in 2-min intervals.
All mice were imaged by using a cooled CCD camera (IVIS100 system, Xenogen, Alameda, CA). Photons emitted from the cells implanted in the mice were collected and integrated for a period of 1 min. Images were obtained by using living image software (Xenogen). To quantify the measured light, regions of interest were drawn over the cell-implanted area, and the mean luminescence intensities (photons per sec per cm2) were evaluated.
Results
Basic Concept. We have previously developed a split GFP reporter for identifying mitochondrial proteins (10). When a target protein localized in mitochondria, full-length GFP was reconstituted by protein-splicing elements of DnaE present in both split GFP-reporter constructs. This basic concept of GFP-fragment reconstitution by protein splicing was herein extended for designing a bioluminescent indicator for quantitatively evaluating protein nuclear transport with background-free high-sensitive detection. As a reporter protein, we used Rluc, because Rluc has desirable features for a monomeric protein: small size (36 kDa), and ATP is not necessary for its activity (12, 13). We also took advantage of its substrate, coelenterate luciferin (coelenterazine), which rapidly penetrates through cell membranes and produces luminescence (460–490 nm) that is applicable for in vivo imaging (14). The luciferase was dissected between G229 and K230 due to the fact that the luminescence was completely lost when Rluc was split at this particular point (8, 9). We constructed a cDNA encoding a tandem fusion protein, consisting of the N-terminal domains of Rluc (Rluc-N) and DnaE, and a NLS that localizes in the nucleus in mammalian cells (Fig. 1). The C-terminal domain of DnaE was connected with the rest of Rluc (Rluc-C) and the full length of AR, which localizes in the cytosol in the absence of DHT. For efficient splicing to occur, cDNA sequences encoding particular 11 amino acid residues (KFAEY and CFNLSH) are inserted at the splicing junctions of each plasmid. When AR is bound to DHT, it translocates into the nucleus and brings the N- and C-terminal halves of DnaEs close enough to fold correctly, thereby initiating protein splicing to link the concomitant Rluc halves with a peptide bond (Fig. 1 A). The cells containing this reconstituted Rluc allow monitoring of nuclear translocation of AR with its luminescence signals.
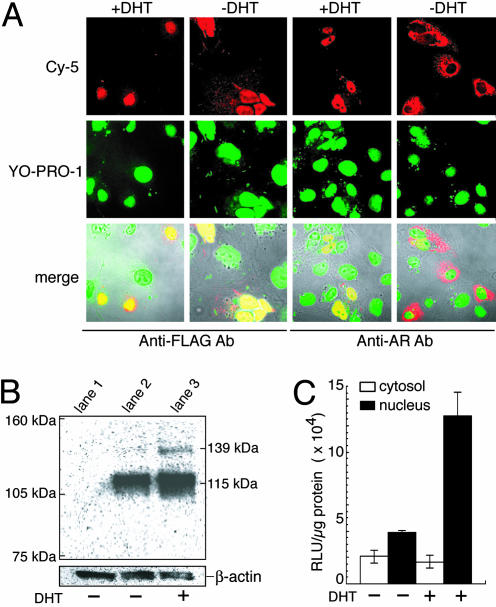
Quantitative in Vitro Sensing of AR Translocation into the Nucleus. To ensure that the fusion protein expressed with pcDRc-AR translocates from the cytosol to the nucleus upon addition of DHT and the protein expressed with pcRDn-NLS permanently resides in the nucleus, we transiently transfected COS-7 cells with either pcDRc-AR or pcRDn-NLS and examined localization of the expressed protein by immunocytochemistry. In the absence of DHT, the AR protein fused with Rluc-C and DnaE-C was predominantly in cytoplasmic, whereas addition of DHT resulted in the nuclear localization of the fusion protein (Fig. 2A). In contrast, the N-terminal fragments of Rluc and DnaE with NLS permanently localized in the nucleus both in the presence and absence of DHT. These observations were consistent with the results of Western blot analysis (Fig. 2B): in crude extracts of the cells expressed with pcRDn-NLS and pcDRc-AR in the absence of DHT, the AR antibody recognized only a specific component of an unspliced precursor, 115 kDa of AR tagged with Rluc-C and DnaE-C. In the presence of DHT, however, the AR antibody recognized 115 kDa of an unspliced precursor and 139 kDa of a polypeptide, of which electrophoretic mobility was consistent with the predicted size of the product after protein splicing. To confirm that Rluc was indeed reconstituted in the nucleus, we examined Rluc activity of each nuclear and cytoplasmic fraction (Fig. 2C). The luminescence intensity from the nucleus in the presence of DHT was found to be much higher than that of cytosol, thereby concluding that Rluc was correctly refolded and recovered its luminescence activity in the nucleus. Given all these results, we conclude that protein splicing occurred and the Rluc was reconstituted when the DHT-bound AR was translocated into the nucleus.
Fig. 2.
Characterization of the indicators in vitro.(A) The immunocytochemical images of COS-7 cells transiently transfected with pcRDn-NLS or pcDRc-AR. The COS-7 cells were cultured for 36 h, and then the cells were incubated for 2 h in the absence or presence of 1 μM DHT. (Top) The two expressed proteins were recognized by anti-AR and anti-FLAG antibodies, respectively, and stained with Cy-5-labeled secondary antibody. (Middle) The nuclei stained with YO-PRO-1; (Bottom) their merged images are shown with the transmission. (B) Western blot of protein extracts from COS-7 cells (lane 1) and from the cells cotransfected with pcRDn-NLS and pcDRc-AR in the absence (lane 2) or presence (lane 3) of 1 μM DHT. As a reference for the amounts of the electrophoresed proteins, β-actin was stained with its specific antibody. (C) Quantitative analysis of the Rluc activity for nuclear and cytoplasmic fractions. The cellular fractions were obtained from cotransfected cells with pcRDn-NLS and pcDRc-AR in the absence or presence of 1 μM DHT (n = 3).
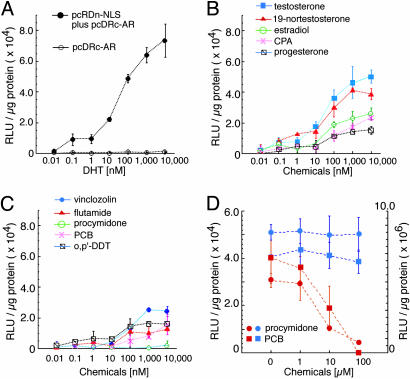
Next, to show that the Rluc reconstitution strategy works for quantitative analysis of the extent of AR translocation into the nucleus, we tested the DHT-induced translocation of AR fused to Rluc-C and DnaE-C with the in vitro cell-based assay. The COS-7 cells were transfected with both pcRDn-NLS and pcDRc-AR or only with pcDRc-AR, and differing concentrations of DHT were added to each microtiter well. The cells were harvested, and their lysates were mixed with a coelenterazine solution. The luminescence signals increased with increasing the concentration of DHT and were strong enough to discriminate them from background luminescence (Fig. 3A). The results indicate that this assay system can be used for quantitative analysis of the extent of AR translocation into the nucleus.
Fig. 3.
Quantitative analysis of AR translocation for various chemical compounds. (A) Dose–response curves for DHT based on the luminescence intensity of the reconstituted Rluc. The COS-7 cells were transiently transfected with pcDRc-AR or cotransfected with pcRDn-NLS and pcDRc-AR, and Rluc activities were tested. The mean luminescence intensities (n = 3) were determined at each DHT concentration. (B) Dose–response curves for steroid hormones based on the luminescence intensity of Rluc. The COS-7 cells were cotransfected with pcRDn-NLS and pcDRc-AR, and Rluc activities were tested upon addition of each hormone (n = 3). (C) Dose–response curves for synthetic chemical compounds based on the luminescence intensity of Rluc. (D) An inhibitory effect of procymidone or PCB on the bioluminescence developed by 0.1 μM DHT. Red dotted curves were obtained from the cells cotransfected with pcRDn-NLS and pcDRc-AR, whereas blue dotted curves were from the cells with pcRDn-NLS and pcDRc-NLS (positive control, n = 3). The circles and squares are the means of luminescence intensities obtained from the cotransfected cells stimulated with procymidone and PCB, respectively. Left and right y axes are for red and blue dotted lines, respectively.
Several endogenous hormones and synthetic chemicals were tested for demonstrating the usefulness of the assay system. Endogenous androgens, such as testosterone and 19-nortestosterone, were found to induce high luminescence intensities over their concentration range from 10–8 to 10–5 M. The relative intensities with testosterone and 19-nortestosterone with their 10–6 M concentration were 72% and 64%, respectively, of those with the same concentration of DHT (Fig. 3B). Other endogenous steroid hormones, 17β-estradiol and progesterone, a synthetic steroid hormone; cyproterone acetate (CPA); and anti-androgens, vinclozolin and flutamide, also gave slight increases in the observed luminescence intensities at their high concentrations (Fig. 3C). In contrast, procymidone did not induce any increase in its luminescence intensity over the concentration range tested. Procymidone was also found to inhibit the DHT-induced strong luminescence. The inhibitory effect of procymidone might be a consequence of inhibition of protein splicing or Rluc activity. To preclude such a possible unwanted effect, high concentrations of procimidone were added to the COS-7 cells transfected with pcRDn-NLS and pcDRc-NLS. No change in Rluc activity was observed (Fig. 3D), indicating that procymidone did not trigger the nuclear import of AR even though it was bound to AR. Of other tested chemicals, a PCB congener (Aroclor 1254), an environmental pollutant, was found to hinder nuclear translocation of AR induced with DHT, whereas 1,1,1-trichloro-2-(p-chlorophenyl)-2-(o-chlorophenyl)ethane (o,p′-DDT) directed a small quantity of AR to the nucleus. To mimic high-throughput screening, we showed the results of the luminescence intensity with a 10–6 M concentration of 13 chemicals (Fig. 6, which is published as supporting information on the PNAS web site).
In Vivo Imaging of AR Translocation in Living Mice. For further extending the usefulness of the indicator, we developed a technique for investigating the distribution of chemical compounds in the organs of living mice and quantifying their effects on AR translocation into the nucleus. It is important to know whether a drug or toxic compound reaches target organs, being left unmetabolized; whether the compound is enriched in the organs when mice are exposed to the compound for a long time period; and whether the compound affects the AR translocation into the nucleus. Such information can be provided with the present bioluminescence-imaging technique.
We implanted s.c. COS-7 cells transiently transfected with pcRDn-NLS or pcDRc-AR or cotransfected with pcRDn-NLS and pcDRc-AR on each site of the back of living mice in a depth of 1 mm. When mice were injected with DHT, an observed cooled CCD image of mice showed a significant increase in the luminescence signal only from the site implanted with the cells cotransfected with pcRDn-NLS and pcDRc-AR (Fig. 7 A and B, which is published as supporting information on the PNAS web site). The luminescence intensity obtained from the cotransfected cells reached the maximum at 15 min (Fig. 7C), which was 20 times higher than the background luminescence intensity from the cells transfected with either pcRDn-NLS or pcDRc-AR alone. The results demonstrate that high levels of bioluminescence can be detected from the implanted cells in living mice upon reconstitution of split Rluc.
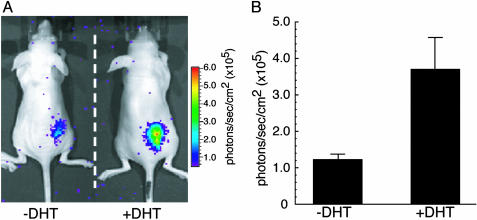
To show the DHT-dependent nuclear import of AR in living mice, COS-7 cells transiently cotransfected with pcRDn-NLS and pcDRc-AR were implanted on the back of mice. One group of mice were stimulated with 1.0% DMSO (vehicle) for 2 hours, whereas the other group of mice were stimulated with DHT (100 μg/kg of body weight) for the same period. Luminescence intensities obtained from the mice with DHT stimulation were 3.5 times higher than that with vehicle stimulation (Fig. 4). The results demonstrate that it is possible to image and quantitatively evaluate the difference in the extent of AR translocation in living mice in the presence of DHT relative to its absence.
Fig. 4.
DHT-dependent translocation of AR in living mice. (A) In vivo optical CCD imaging of mice carrying transiently transfected COS-7 cells in the presence or absence of DHT (100 μg/kg of body weight). The mice were imaged after s.c. implantation of COS-7 cells transiently cotransfected with pcRDn-NLS and pcDRc-AR. One group of mice was injected with DHT and the other group was left uninjected. (B) The average of photon counts from each implanted site in A (n = 4). The graph shows the observed photon counts for uninjected and DHT-injected group of mice with mean values over four mice, respectively. The average values of photon counts for 4 mice were (1.23 ± 0.15) × 105 (–DHT), and (3.70 ± 0.87) × 105 (+DHT) (photons per sec per cm2).
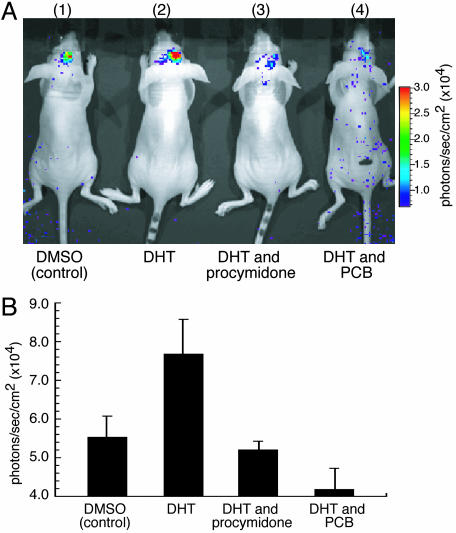
Finally, we studied the usefulness of this strategy for examining the effects of inhibitors on AR translocation into the nucleus in the mouse brain. The cells cotransfected with pcRDn-NLS and pcDRc-AR were implanted in the mouse brain at a depth of 3 mm, and i.p. injection was performed with different hormone mixtures; 1.0% DMSO (vehicle) for the first group of mice, DHT (10 μg/kg of body weight) for the second group, DHT (10 μg/kg of body weight) and procymidone (10 mg/kg of body weight) for the third group, and DHT (10 μg/kg of body weight) and PCB (10 mg/kg of body weight) for the fourth group. Induction of AR nuclear import by DHT resulted in a significant increase in photon count from the brains as compared to that with vehicle (Fig. 5). The increase in bioluminescence upon stimulation only with DHT indicates that the magnitude of bioluminescence originated from reconstituted Rluc was strong enough to be detected in the brain at a depth of 3 mm. Stimulation with DHT together with procymidone and PCB, respectively, resulted in a decrease in luminescence intensity, demonstrating that procymidone and PCB had an ability to pass through the blood–brain barrier and hindered nuclear import of AR.
Fig. 5.
An inhibitory effect of procymidone or PCB on the bioluminescence developed by DHT (10 μg/kg of body weight) in living mice. (A) The inhibitory effect of chemicals on AR translocation into the nucleus in the mouse brain. The COS-7 cells transiently cotransfected with pcRDn-NLS and pcDRc-AR were implanted in the forebrain of the nude mice at a depth of 3 mm through a 1-mm burrhole. Of mouse groups 1–4, groups 1 and 2 were stimulated with 1% DMSO, whereas groups 3 and 4 were stimulated with procymidone (10 mg/kg body weight) and PCB (10 mg/kg of body weight), respectively. Two hours after the stimulation, mouse groups 2–4 were then stimulated with DHT (10μg/kg of body weight). Two hours after DHT stimulation, the mice were imaged in 2-min intervals until reaching the maximum photon counts after intercerebral injection of coelenterazine (1.4 mg/kg of body weight). (B) The average of photon counts from each implanted site in A (n = 3). The averages of three mice were (5.53 ± 0.53) × 104 (group 1), (7.68 ± 0.91) × 104 (group 2), (5.07 ± 0.23) × 104 (group 3), and (4.18 ± 0.55) × 104 (group 4) (photons per sec per cm2).
Discussion
This study demonstrated a method of split Rluc reconstitution by protein splicing in vitro and in vivo. The method of cell-based screening with the genetically encoded indicator provided a quantitative measure of the extent of nuclear translocation of AR upon stimulation with various chemicals. Currently, high-throughput screening tools for protein translocation into the nucleus have mostly depended upon GFP- or its variant-tagged approach in combination with the fluorescence microscopy and computer-driven imaging system (2). The systems offer only semiquantitative information, because it is difficult in each cell to accurately sort out and distinguish the fluorescence of GFP-tagged proteins localized only in the nucleus from that left in the cytosol. In addition, precision of the observed fluorescence intensities from the nucleus obtained with the statistical analysis is not high, because the number of the cells examined under a fluorescence microscope is limited. On the contrary, the present method allowed us to determine the subcellular localization of AR by the luminescence signals generated only when the AR localized in the nucleus. AR remaining in the cytosol did not induce reconstitution of split Rluc, and therefore no background luminescence was observed. Moreover, the number of the cells once analyzed was ≈104 cells, which was enough to precisely evaluate the extent of AR translocation into the nucleus. Thus, this indicator enabled fluorescent-background-free, accurate, precise, and sensitive detection, which is of great advantage to quantitatively evaluate the extent of protein nuclear transport in a high-throughput manner.
This screening system was applied for the quantitative evaluation of various chemical compounds, which triggered AR translocation into the nucleus. Of 11 ligands tested, the synthetic chemicals vinclozolin and flutamide were found to induce the translocation of AR only at their higher concentrations. In addition, the nonsteroidal chemicals procymidone and PCB blocked the translocation of AR into the nucleus. PCB and procymidone have been suspected of neurotoxic and antiandrogenic effects, respectively, and possibly influence adversely hormonal activities in the brains of living animals (15–18). In the present study, we showed the usefulness of the split Rluc reporter for monitoring AR translocation into the nucleus in living mice by measuring bioluminescence with a cooled CCD camera. We thereby investigated the distribution of the chemicals in the brains of living mice. As expected, 2 hours after i.p. injection of PCB or procymidone, both chemicals were found to completely inhibit the DHT-stimulated translocation of AR. From the results, it was concluded that PCB and procymidone have the ability to pass through the blood–brain barrier in 2 hours to reach the brain and inhibit the AR signal transduction in the organ.
Protein engineering of firefly luciferase has recently been studied for the analysis of protein–protein interactions (7) or of caspase activity in apoptosis (19), as well as its use for reporter gene approaches (20–22), but there has been no report demonstrating the usefulness of the engineered Rluc in living mice. The advantage of using the engineered Rluc with the reconstitution scheme is that it has the potential to detect intracellular signaling not only in the nucleus but also anywhere in the cells, whereas conventional reporter gene approaches are limited to molecular events within the nucleus. The effects of particular chemical compounds on the AR translocation are distinguished from their effects on gene expression. In fact, it has been demonstrated that there is no transcriptional activity of AR upon its binding to flutamide or vinclozolin, even though both chemicals induce the translocation of AR into the nucleus (23). Thus, it is noted that the results obtained from the present split luciferase reporter are unique for the event of nuclear translocation of AR, and more importantly, it should be emphasized that this reporter can be generally applicable for translocation of any proteins from the cytosol to the nucleus. Using other nuclear receptors or cofactors of therapeutic targets, for example, the probes will extend their applications for screening and imaging further into other biological studies, such as sterol responsive element-binding proteins for cholesterol, mitogen-activated protein kinase for cell growth, and nuclear factor-κB for apoptosis, respectively.
In the present study, we have demonstrated ligand-dependent AR import in response to the DHT and its analogs. Procymidone and PCB, which are antagonistic chemicals, inhibited AR translocation into the nucleus. Viable targets of therapeutic and toxic chemicals are also phosphorylation-dependent nuclear import, proteolysis-induced import of a particular protein, and protein export from the nucleus to the cytosol. Such nucleocytoplasmic trafficking could also be detected if the C-terminal halves of Rluc and DnaE are connected to the target protein, and the C- and N-terminal probes are localized separately in the cytosol and nucleus, respectively. Trafficking of AR in response to an external stimulation such as DHT is a key function in the biological relevance of AR. We succeeded in demonstrating that the probe-tagged AR was indeed capable of quantitatively visualizing the key function of nuclear import of AR. Because the size of the probe (14 kDa) was much smaller than that of AR (105 kDa), unexpected repressive influence on the AR translocation was probably minimized.
We demonstrated the feasibility of the present indicator for monitoring the nuclear trafficking of target proteins. Our in vitro assay using cotransfected cells quantitatively analyzes the extent of protein trafficking simply, which allows high-throughput screening of therapeutic and toxic chemicals. When inhibitor chemicals for nuclear transport are screened, a possible effect, if any, of these chemicals on protein splicing or Rluc activity per se has to be assessed. Because the present screening system is high-throughput, this procedure is not troublesome. We showed that PCB and procymidone have an ability to pass through the blood–brain barrier and inhibit AR translocation into the nucleus. The point of such in vivo experiments in this case is that implantation of the COS-7 cells in the target organs is to probe AR-relevant chemicals in living mice for screening of their locations in various part of the organs. Although the amounts of the N- and C-terminal probes were not controlled in the present cotransfection experiments, we succeeded in imaging DHT-dependent AR translocation into the nucleus in living mice. Because the amounts of the probes in general are controllable with the conventional stable expression or viral infection techniques, the use of such techniques may improve the specification of the present method even further. The advantage of in vivo imaging is that the screened chemicals can be validated as to their location at particular target tissues or organs in living mice. In addition, the chemicals, in many cases, are metabolized or chemically modified in living mice, of which effective concentration can be evaluated noninvasively by using the present method. Therefore, the availability of this genetically encoded indicator facilitates the development of transgenic animals that express the Rluc fragments in all tissues with controllable promoters. With the animals, some specific xeno-chemicals or drugs can be noninvasively monitored as to their effects on a particular protein translocation into the nucleus in a tissue-specific manner.
Acknowledgments
This work was supported by grants from Core Research for Evolutional Science and Technology (CREST), the Precursory Research for Embryonic Science and Technology (PREST) of the Japan Science and Technology Agency, and the Ministry of Education, Science, and Culture of Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AR, androgen receptor; DHT, 5α-dihydrotestosterone; NLS, nuclear localization signal; CCD, charge-coupled device; Rluc, Renilla luciferase; Rluc-N, N-terminal domain of Rluc; Rluc-C, C-terminal domain of Rluc; PCB, polychlorinated biphenyls; DnaE-N, N-terminal splicing domain of DnaE; DnaE-C, C-terminal splicing domain of DnaE.
References
- 1.Kau, T. R. & Silver, P. A. (2003) Drug Discov. Today 8, 78–85. [DOI] [PubMed] [Google Scholar]
- 2.Kau, T. R., Way, J. C. & Silver, P. A. (2004) Nat. Rev. Cancer 4, 106–117. [DOI] [PubMed] [Google Scholar]
- 3.Rudin, M. & Weissleder, R. (2003) Nat. Rev. Drug Discov. 2, 123–131. [DOI] [PubMed] [Google Scholar]
- 4.Gray, L. E., Jr., Ostby, J., Wilson, V., Lambright, C., Bobseine, K., Hartig, P., Hotchkiss, A., Wolf, C., Furr, J., Price, M., et al. (2002) Toxicology 181–182, 371–382. [DOI] [PubMed] [Google Scholar]
- 5.Elion, E. A. (2002) Methods Enzymol. 351, 607–622. [DOI] [PubMed] [Google Scholar]
- 6.Ozawa, T., Kaihara, A., Sato, M., Tachihara, K. & Umezawa, Y. (2001) Anal. Chem. 73, 2516–2521. [DOI] [PubMed] [Google Scholar]
- 7.Paulmurugan, R., Umezawa, Y. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 15608–15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulmurugan, R. & Gambhir, S. S. (2003) Anal. Chem. 75, 1584–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaihara, A., Kawai, Y., Sato, M., Ozawa, T. & Umezawa, Y. (2003) Anal. Chem. 75, 4176–4181. [DOI] [PubMed] [Google Scholar]
- 10.Ozawa, T., Sako, Y., Sato, M., Kitamura, T. & Umezawa, Y. (2003) Nat. Biotechnol. 21, 287–293. [DOI] [PubMed] [Google Scholar]
- 11.Singh, S. M., Gauthier, S. & Labrie, F. (2000) Curr. Med. Chem. 7, 211–247. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz, W. W., McCann, R. O., Longiaru, M. & Cormier, M. J. (1991) Proc. Natl. Acad. Sci. USA 88, 4438–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews, J. C., Hori, K. & Cormier, M. J. (1977) Biochemistry 16, 85–91. [DOI] [PubMed] [Google Scholar]
- 14.Bhaumik, S. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schettler, T. (2001) Environ. Health Perspect. 109 Suppl. 6, 813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein, J., Schettler, T., Wallinga, D. & Valenti, M. (2002) J. Dev. Behav. Pediatr. 23, S13–S22. [DOI] [PubMed] [Google Scholar]
- 17.Tilson, H. A. & Kodavanti, P. R. (1998) Neurotoxicology 19, 517–525. [PubMed] [Google Scholar]
- 18.Roy, A. K., Tyagi, R. K., Song, C. S., Lavrovsky, Y., Ahn, S. C., Oh, T. S. & Chatterjee, B. (2001) Ann. N.Y. Acad. Sci. 949, 44–57. [DOI] [PubMed] [Google Scholar]
- 19.Laxman, B., Hall, D. E., Bhojani, M. S., Hamstra, D. A., Chenevert, T. L., Ross, B. D. & Rehemtulla, A. (2002) Proc. Natl. Acad. Sci. USA 99, 16551–16555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contag, C. H., Spilman, S. D., Contag, P. R., Oshiro, M., Eames, B., Dennery, P., Stevenson, D. K. & Benaron, D. A. (1997) Photochem. Photobiol. 66, 523–531. [DOI] [PubMed] [Google Scholar]
- 21.Contag, P. R., Olomu, I. N., Stevenson, D. K. & Contag, C. H. (1998) Nat. Med. 4, 245–247. [DOI] [PubMed] [Google Scholar]
- 22.Ray, P., Pimenta, H., Paulmurugan, R., Berger, F., Phelps, M. E., Iyer, M. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomura, A., Goto, K., Morinaga, H., Nomura, M., Okabe, T., Yanase, T., Takayanagi, R. & Nawata, H. (2001) J. Biol. Chem. 276, 28395–28401. [DOI] [PubMed] [Google Scholar]