ABSTRACT
SERINC3 (serine incorporator 3) and SERINC5 are recently identified host cell inhibitors of HIV-1 particle infectivity that are counteracted by the viral pathogenesis factor Nef. Here we confirm that HIV-1 Nef, but not HIV-1 Vpu, antagonizes the particle infectivity restriction of SERINC5. SERINC5 antagonism occurred in parallel with other Nef activities, including cell surface receptor downregulation, trans-Golgi network targeting of Lck, and inhibition of host cell actin dynamics. Interaction motifs with host cell endocytic machinery and the Nef-associated kinase complex, as well as CD4 cytoplasmic tail/HIV-1 protease, were identified as essential Nef determinants for SERINC5 antagonism. Characterization of antagonism-deficient Nef mutants revealed that counteraction of SERINC5 occurs in the absence of retargeting of the restriction factor to intracellular compartments and reduction of SERINC5 cell surface density is insufficient for antagonism. Consistent with virion incorporation of SERINC5 being a prerequisite for its antiviral activity, the infectivity of HIV-1 particles produced in the absence of a SERINC5 antagonist decreased with increasing amounts of virion SERINC5. At low levels of SERINC5 expression, enhancement of virion infectivity by Nef was associated with reduced virion incorporation of SERINC5 and antagonism-defective Nef mutants failed to exclude SERINC5 from virions. However, at elevated levels of SERINC5 expression, Nef maintained infectious HIV particles, despite significant virion incorporation of the restriction factor. These results suggest that in addition to virion exclusion, Nef employs a cryptic mechanism to antagonize virion-associated SERINC5. The involvement of common determinants suggests that the antagonism of Nef to SERINC5 and the downregulation of cell surface CD4 by Nef involve related molecular mechanisms.
IMPORTANCE HIV-1 Nef critically determines virus spread and disease progression in infected individuals by incompletely defined mechanisms. SERINC3 and SERINC5 were recently identified as potent inhibitors of HIV particle infectivity whose antiviral activity is antagonized by HIV-1 Nef. To address the mechanism of SERINC5 antagonism, we identified four molecular determinants of Nef antagonism that are all linked to the mechanism by which Nef downregulates cell surface CD4. Functional characterization of these mutants revealed that endosomal targeting and cell surface downregulation of SERINC5 are dispensable and insufficient for antagonism, respectively. In contrast, virion exclusion and antagonism of SERINC5 were correlated; however, Nef was also able to enhance the infectivity of virions that incorporated robust levels of SERINC5. These results suggest that the antagonism of HIV-1 Nef to SERINC5 restriction of virion infectivity is mediated by a dual mechanism that is related to CD4 downregulation.
INTRODUCTION
Nef is a myristoylated 25- to 34-kDa protein that, in addition to human immunodeficiency virus type 1 (HIV-1), is encoded by HIV-2 and simian immunodeficiency virus (SIV). While dispensable for virus replication in cultured cells, Nef potently increases virus replication and thus serves as a pathogenicity factor that accelerates disease progression in the infected host (1–3). Nef does not bear enzymatic activity but mediates its functions through a large set of interactions with cellular proteins. By virtue of this adaptor function, Nef affects many central processes in HIV target cells. This includes modulation of cellular transport pathways leading to downregulation of an array of receptors from the surface of infected cells (4–6), which, e.g., prevents superinfection (7, 8) and lysis of productively infected cells by cytotoxic T or NK cells (9, 10). HIV-1 Nef also alters the response of CD4 T lymphocytes to stimulation via the T cell receptor (TCR), and modulation of the resulting cellular signaling pathways is thought to increase virus replication in the infected host (11–17). This involves the retargeting of the TCR-proximal Src kinase Lck from the plasma membrane to the trans-Golgi network (12, 14, 18) as well as the inhibition of actin reorganization induced upon TCR engagement (19, 20). Inhibition of host cell actin remodeling by Nef also occurs upon chemokine stimulation of T lymphocytes and translates into a potent block to cell migration (21–24).
Comparison of nef-deficient HIV-1 variants with wild-type (WT) HIV-1 revealed that Nef elevates the infectivity of HIV particles by 2- to 10-fold in single rounds of infection (25–27; reviewed in reference 28). This effect requires the presence of Nef in the producer but not the target cell or the virion itself and is due to an effect on the infectivity of HIV particles and not the amount of particles released from the producer cells (26, 27, 29, 30). SERINC5 (serine incorporator 5) was recently identified as a host cell factor that restricts HIV particle infectivity up to 100-fold and is counteracted by Nef (31, 32). Knockdown/knockout and overexpression approaches established SERINC5 as necessary and sufficient for the HIV particle infectivity restriction. SERINC5 is a member of the SERINC protein family that is conserved from yeast to mammals and contains 5 members that are predicted to contain 10 to 12 transmembrane domains. SERINC protein function has not been studied in detail, but SERINC proteins were reported to facilitate the incorporation of serine in the biosynthesis of sphingolipids and phosphatidylserine when ectopically expressed in Escherichia coli, yeast, and COS-7 cells (33). The mechanisms by which SERINC5 impairs HIV-1 particle infectivity and how Nef counteracts this restriction remain to be defined (34), but HIV particles incorporate significant amounts of the restriction factor when produced in the absence of Nef, which appears to reduce the fusogenicity of HIV particles and possibly early postentry steps. The presence of Nef in the producer cell markedly reduces virion incorporation of SERINC5, which is paralleled by an overall reduction of SERINC5 cell surface exposure and a redistribution of the restriction factor from the plasma membrane into an intracellular Rab7-positive membrane compartment (31, 32). Here we set out to define molecular determinants of SERINC5 antagonism in HIV-1 Nef and to assess which of the reported alterations to SERINC5 localization are required to release the restriction to HIV particle infectivity.
MATERIALS AND METHODS
Cells, plasmids, and reagents.
Jurkat (T antigen [TAg] and CCR7) T lymphocytes were cultivated in RPMI 1640 plus GlutaMAX-I supplemented with 10% fetal calf serum (FCS) and 1% penicillin-streptomycin (all from Invitrogen). 293T and TZM-bl cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with the same reagents described for RPMI medium. Jurkat TAg and CCR7 T cells were electroporated with 10 to 25 μg of total plasmid DNA per 0.5 × 107 to 1 × 107 cells (at 250 V and 950 and 850 μF, respectively, with a Bio-Rad Gene Pulser). The expression plasmid for the C-terminally hemagglutinin (HA)-tagged version of SERINC5 (pBJ6_SERINC5.HA) as well as pcDNA-based expression plasmids of SERINC5 and constructs for the expression of green fluorescent protein (GFP) fusion proteins of SERINC5 were recently described (31). pBJ5-based constructs for the expression of HA-tagged SERINC5 (pBJ5_SERINC5.intHA or empty control vector) are a kind gift from Heinrich Göttlinger (32). The expression constructs for GFP fusion proteins of HIV-1SF2 Nef wild-type (Nef wt) and mutant proteins, as well as for red fluorescent protein (Nef.RFP) and Vpu.GFP are described elsewhere (4, 20, 35, 36). The proviral plasmids pHIV-1NL4-3 WT (NL4-3 Nef wt), pHIV-1NL4-3 Nef stop (ΔNef), and pHIV-1NL4-3 Vpu stop (ΔVpu) were kindly provided by Frank Kirchhoff. The proviral chimera pHIV-1NL4-3 SF2 Nef WT (WT) and the Nef mutant panel that originated thereof are described previously (37). In short, SF2 nef variants were introduced into the genetic background of the HIV-1 strain NL4-3 by replacing the nef gene of HIV-1NL4-3 with that of HIV-1SF2. The following antibodies and reagents were used: allophycocyanin (APC)-conjugated mouse anti-human CD4 (RPA-T4), CD81 (JS-81), and HLA-ABC (major histocompatibility complex class I [MHC-I]; G46-2.6) antibody (all from BD Biosciences), mouse anti-HA or biotinylated mouse anti-HA (HA.11, clone 16B11; Biolegend), mouse anti-Lck (3A5; Santa Cruz), mouse anti-transferrin receptor/CD71 clone H68.4 (Thermo Fisher), polyclonal anti-Vpu (Biozol), sheep anti-Nef (arp444; NIH AIDS repository), rabbit anti-Nef, rat anti-GFP (Chromothek), and sheep anti-HIV-1 p24CA antiserum (from Barbara Müller). Phalloidin-tetramethyl rhodamine isocyanate (TRITC) was purchased from Sigma-Aldrich, and stromal-derived factor 1α (SDF-1α) was obtained from Immunotools.
Flow cytometry.
In order to quantify the surface expression levels of different cell surface receptors in cells expressing GFP or Nef.GFP in the presence or absence of SERINC5.intHA, the above-mentioned antibodies against CD4, CD81, or HLA-ABC were used for cell surface staining of Jurkat TAg T cells. For this, 5 × 106 cells were cotransfected with 10 μg plasmid DNA each for expression of GFP or Nef.GFP fusion proteins and internally HA-tagged SERINC5 proteins or an empty control vector via electroporation (950 μF, 300 V; Bio-Rad GenePulser). Forty-eight hours posttransfection, cells were stained with 0.5 μg of the respective antibody in fluorescence-activated cell sorter (FACS) buffer (0.1% fetal calf serum [FCS] in phosphate-buffered saline [PBS]) in a V-bottom 96-well plates for 30 min on ice. For intracellular staining, cells were washed with FACS buffer, fixed in 3% paraformaldehyde (PFA) in PBS for 10 min, permeabilized with 0.1% Triton X-100 in FACS buffer for 5 min, washed again, and stained for intracellular SERINC5.intHA using a biotinylated HA-conjugated antibody (1:100 in FACS buffer) for 1 h followed by a second staining step with phycoerythrin (PE)-labeled streptavidin for 45 min. Cells expressing SERINC5.intHA were gated, and surface-exposed levels of CD4, CD81 and major histocompatibility complex class I (MHC-I) on HA-positive cells were analyzed by flow cytometry (FACSVerse with BD CellQuest Pro 4.0.2 software (BD Pharmingen; FlowJo analyzing software 10.0.8). For control cells without SERINC5 expression, surface receptor levels were determined on all living cells. Within one sample, the surface receptor levels (geometric mean of mean fluorescence intensity [MFI]) of medium- to high-GFP-expressing cells were compared to surface receptor levels of non-GFP-expressing cells as described before (4, 7). Data were processed with Microsoft Office Excel 2007 and GraphPad Prism 5.0 software.
In order to quantify the surface levels of SERINC5 in p24-positive cells, surface-exposed SERINC5 molecules were stained using an HA-conjugated antibody (1:100 in FACS buffer) for 1 h followed by a second staining step with APC-labeled anti-mouse secondary antibody for 45 min prior to being fixed with 3% PFA in PBS for 90 min. Fixed cells were permeabilized with 0.1% Triton X-100 in FACS buffer for 5 min, washed and stained for intracellular p24 (1:100 p24-FITC, clone KC57), and analyzed by flow cytometry. Within one sample, the surface levels of SERINC5 of p24-positive cells were compared to surface SERINC5 levels of p24-negative cells and the relative expression levels were calculated, which is presented as the percentage of HIV ΔNef plus SERINC5. To determine intracellular SERINC5 levels, cells were first fixed in 3% PFA for 90 min, and then permeabilized and stained for intracellular SERINC5 using the above-described antibodies, followed by staining of p24. The MFI of the HA stain specific to SERINC5.intHA in p24-positive cells was used to calculate the intracellular SERINC5 levels relative to ΔNef (in percentage).
Immunofluorescence microscopy.
293T cells growing on cover glasses (Marienfeld) were transfected with the proviral HIV-1NL4-3 constructs together with expression plasmids coding for an internally HA-tagged version of SERINC5 (pBJ5_ SERINC5.intHA) or a vector control. Forty-eight hours posttransfection, the cells were fixed with 4% PFA for at least 90 min, permeabilized for 5 min with 0.1% Triton X-100 in PBS, blocked for 1 h with 5% milk in PBS at room temperature, stained for SERINC5.HA using the mouse anti-HA antibody (1:1,000 in 5% milk–PBS) followed by staining with an appropriate secondary Alexa Fluor-labeled antibody (Invitrogen), mounted in Mowiol, and analyzed with a Leica TCS SP5 confocal microscope with a 100× Plan-Apo objective lens. Single-plane images were recorded with the Leica LAS AF (Leica Application Suite for Advanced Fluorescence) software and processed with ImageJ 1.50e and GIMP 2.8.14. For SERINC5 localization and Lck accumulation studies in Jurkat TAg T cells, microscope cover glasses were coated with 0.01% poly-l-lysine (Sigma) solution for 1 h at room temperature and washed with 1× PBS. One day posttransfection with the indicated expression plasmids (via electroporation as described above), Jurkat TAg cells were plated onto the cover glasses (3 × 105 cells/cover glass) and fixed after 10 min at 37°C with PBS–3% PFA for 15 to 30 min. Samples with fluorescent protein tags were analyzed directly. For indirect immunofluorescence of endogenous Lck, cells were permeabilized with 0.1% Triton X-100 for 2 min and blocked with PBS–1% BSA for 30 min. The primary mouse anti-Lck (1:50) antibody was applied for 2 h, followed by the secondary goat anti-mouse Alexa Fluor 568 for 1 h. After being washed with PBS, cover glasses were mounted in Mowiol. Images were taken by using a 100× oil immersion objective lens (Olympus IX81 SIF-3 microscope) and processed by using Adobe Photoshop. Image quantification was performed as described previously (12).
Virus production, infectivity measurements, and quantification of HIV-1 particles in cell culture supernatants.
Virus was generated by transfection (via Metafectene [Biontex] or calcium phosphate precipitation) of proviral HIV-1NL4-3 plasmids into 293T cells. For most assays, 12-well plates were seeded with 293T cells (1.5 × 105/well) 1 day before cotransfection of proviral DNA and plasmid DNA for expression of different versions of HA-tagged SERINC5 (pBJ6_SERINC5.HA or pBJ5_SERINC5.intHA). Two days posttransfection, culture supernatants were harvested and investigated for particle release and infectivity. Release of viral particles was determined using a one-step SYBR green I-based product-enhanced reverse transcriptase assay (SG-PERT) (38). For that, 5 μl of virus-containing supernatant was lysed in 2× virus lysis buffer for 10 min and diluted with 90 μl PCR buffer, and 10 μl thereof was used for the reverse transcriptase PCR (RT-PCR). Absolute values of the virus suspensions tested were retrieved from a standard curve created using serial dilutions of a previously characterized virus stock. To measure the infectivity of virus particles, 25 μl of the culture supernatants was used to infect TZM-bl reporter cells cultured in a 96-well format, and the infectivity was determined 48 h after infection by analysis of firefly luciferase activity as described previously (39, 40).
T cell chemotaxis and actin dynamics.
Analyses of SDF-1α-mediated T lymphocyte chemotaxis and membrane ruffling, as well as analysis of the phosphorylation state of serine 3 of cofilin, were carried out as described previously (20, 24, 41).
Western blot analysis.
Cells were either lysed directly in 2× SDS sample buffer containing 50 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP; 0.5 M stock, pH 7.0 [Sigma]) or using a radioimmunoprecipitation assay (RIPA) lysis buffer containing 1% n-dodecyl β-d-maltoside (DDM [Sigma]) as detergent. Cell lysates were sonicated using a Branson Sonifier 450. For Western blot analysis of virions, virus particles were concentrated via 20% sucrose cushion ultracentrifugation (28,000 rpm for 2 h in an LE80 Optima or 44,000 rpm for 45 min in a TL100 ultracentrifuge), resuspended in 1× PBS, and lysed in 6× SDS sample buffer containing 50 mM TCEP (0.5 M stock, pH 7.0 [Sigma]). Proteins were separated via 12.5% SDS-PAGE and blotted to nitrocellulose membranes. Membranes were blocked in 5% milk in PBS-Tween 20 (PBS-T) and probed with the following primary antibodies: mouse anti-HA (1:1,000 [Biolegend]), sheep anti-HIV-1 p24CA antiserum (1:5,000), rabbit polyclonal anti-Vpu (1:1,000), sheep anti-HIV-1 Nef serum (1:2,000), or rabbit anti-Nef (1:2,000). Secondary antibodies were conjugated to horseradish peroxidase for enhanced chemiluminescence (ECL)-based detection. SERINC5 levels in cell lysates as well as virions were quantified by densitometry using the software QuantityOne (Bio-Rad). For this, areas of interests were defined (one size for all signals on the same membrane) and background values were subtracted (global background subtraction method). SERINC5 levels are presented relative to HIV-1 ΔNef plus SERINC5 (in percentage).
Statistical analysis.
Statistical analysis of data sets was carried out using Microsoft Excel and GraphPad Prism. Statistical significance of parametrically or not normally distributed data sets was analyzed by unpaired two-tailed Student t test.
RESULTS
Antagonism of SERINC5 does not compete with other Nef functions.
It was recently reported that expression of SERINC5 in HIV-1-producing 293T cells potently suppresses the infectivity of released HIV-1 virus in the case of nef-negative HIV-1 (ΔNef), while Nef expression by wild-type HIV-1 (WT) antagonizes the particle infectivity restriction by SERINC5 (31, 32). Consistently, expression of SERINC5 during virus production reduced the relative infectivity of HIV-1 ΔNef 32-fold, while the infectivity of HIV-1 WT remained unaffected by SERINC5 expression (Fig. 1A, upper panel). As judged by quantification of reverse transcriptase (RT) activity in the cell culture supernatant, HIV-1 particle production was unaffected by coexpression of SERINC5 under these experimental conditions (data not shown). While only small amounts of SERINC5 were detectable in HIV particles produced in the presence of Nef (WT), virion incorporation of the restriction factor was markedly increased in the case of ΔNef (Fig. 1A, lower panel). To test whether the detection of SERINC5 in virion pellets reflects exclusively its association with virus particles or includes substantial contamination with SERINC5-containing extracellular vesicles, culture supernatants of cells transfected with expression constructs for SERINC5 in the absence of HIV-1 proviral DNA were subjected to the pelleting procedure used to concentrate HIV particles (Fig. 1B). Under these conditions, SERINC5 was not detected in these pellets, irrespectively of whether we analyzed low cellular expression levels (using the pBJ6-based construct also used in Fig. 1A) or strongly overexpressed SERINC5 (using a pcDNA-based expression construct). Similar results were obtained upon coexpression of Nef, which has been reported to increase the release of extracellular vesicles (42). Under the experimental conditions used here, incorporation of SERINC5 in extracellular vesicles unrelated to HIV therefore does not significantly contribute to the detection of the restriction factor in HIV particle pellets.
FIG 1.
Overexpression of SERINC5 reduces the infectivity of HIV-1 particles but does not interfere with other Nef functions. (A) Relative infectivity of HIV-1 particles produced in 293T cells transfected with pBJ6-SERINC5.HA or a control vector. Infectivity was measured by infection of TZM-bl reporter cells with virus-containing supernatant and is normalized to virus release. Depicted are mean values ± standard deviations from three independent experiments. Representative Western blot analyses of the producer cell lysate and virions are shown below. Immunodetection of SERINC5.HA, Nef, and HIV-1 Gag (p55, p41, and p24) is shown. (B) Analysis of cell culture supernatants for the presence of SERINC5 in extracellular vesicles. 293T cells were transfected to express SERINC5.HA either from pBJ6- or pcDNA-based plasmids or with an empty control vector. Extracellular vesicles were pelleted similar to HIV-1 particle preparations via 20% sucrose cushions and together with cell lysates subjected to SDS-PAGE followed by Western blotting using an anti-HA antibody to detect SERINC5.HA. Asterisks to the right denote unspecific signals. Shown are representative Western blots from two independent experiments (C) Analysis of subcellular localization of endogenous Lck in Jurkat TAg T lymphocytes cotransfected with expression constructs for GFP or Nef.GFP and either RFP or SERINC5.RFP. Cells were plated on poly-l-lysine-coated cover glasses, fixed, permeabilized, and stained for endogenous Lck. Depicted is the quantification of the frequency of transfected cells with perinuclear Lck accumulation. Values are the means from 3 independent experiments ± standard deviations in which at least 100 cells were analyzed per condition. (D) Cell surface levels of CD4, CD81, and MHC-I in Jurkat TAg T lymphocytes cotransfected with expression constructs for GFP or Nef.GFP and pBJ5-SERINC5.intHA or an empty control vector. Forty-eight hours posttransfection, cells were stained for receptor surface expression with allophycocyanin (APC)-conjugated antibodies against CD4, CD81, or MHC-I, fixed, permeabilized, stained with a biotin-conjugated antibody against the HA tag followed by a second staining step with R-phycoerythrin (R-PE)-conjugated streptavidin, and analyzed by flow cytometry. The ratio of the MFI of transfected and untransfected cells stained for the surface receptor was calculated and set to 100% for GFP-transfected cells. Shown are the means from three independent experiments ± standard deviations. (E to G) Analysis of Jurkat CCR7 T lymphocytes transiently transfected with expression constructs for GFP or Nef.GFP and SERINC5.HA (pcDNA_SERINC5.HA) or a vector control (pcDNA). Depicted are mean values ± standard deviations from two experiments performed in triplicate with at least 100 cells analyzed per condition. (E) Chemotaxis toward SDF-1α. Cells were allowed to migrate through a 5-μm-pore porous Transwell filter toward SDF-1α (50 ng/ml) for 2 h. Migrated cells were quantified by flow cytometry and data plotted relative to the corresponding GFP control, which was set to 100%. (F) Frequency of cells with high phosphorylated cofilin (p-cofilin) levels. Cells were plated onto coverslips, fixed, permeabilized, and stained for p-cofilin. Shown are the frequencies of cells showing high levels of phosphorylated cofilin. (G) Inhibition of chemokine-induced F-actin-rich membrane ruffling by Nef. Cells were seeded onto coverslips 24 h posttransfection, stimulated with 200 ng/ml SDF-1α for 20 min, fixed, permeabilized, and stained with phalloidin-TRITC to visualize F-actin. Shown are the frequencies of cells with chemokine-induced F-actin-rich membrane ruffles. Statistical significance was assessed by Student's t test. ns, not significant; **, P < 0.001.
Since Nef function has been characterized predominately in cell systems that lack expression of SERINC5 to levels sufficient for its antiviral activity (31, 32), we wondered whether Nef can exert other activities simultaneously to SERINC5 antagonism. We therefore transiently transfected human T cell lines with expression constructs for GFP or Nef.GFP together with an empty control plasmid (control) or the SERINC5 expression construct (Fig. 1C to G). In these analyses, coexpression of SERINC5 in Jurkat T cells did not affect the ability of Nef.GFP to induce intracellular accumulation of the Src kinase Lck (14, 20) (Fig. 1C) or to reduce cell surface levels of CD4, CD81, and MHC-I (4) (Fig. 1D). Similarly, inhibition of chemotaxis of Jurkat CCR7 cells toward SDF-1α (Fig. 1E), phosphorylation and thus inactivation of the actin-severing factor cofilin (Fig. 1F) (24, 43), and interference with chemokine-induced formation of F-actin rich plasma membrane ruffles (24, 43) (Fig. 1G) by Nef.GFP were unaffected by coexpression of SERINC5. We conclude that Nef exerts “classical” activities irrespectively of SERINC5 expression, indicating that high levels of SERINC5 do not compete with Nef functions unrelated to virion infectivity enhancement.
Determinants in HIV-1 Nef for antagonism of SERINC5.
We next sought to define in more detail which molecular determinants in HIV-1 Nef are involved in antagonism of SERINC5. To this end, we tested a well-characterized panel of HIV-1 proviral constructs encoding mutant HIV-1SF2 Nef proteins with defects in individual protein interaction surfaces (Fig. 2A) (37). Most Nef variants antagonized SERINC5 and thus enhanced virion infectivity with similar efficiency to WT Nef (Fig. 2B). This included mutations of lysines 4 and 7, which are required for incorporation of Nef into detergent-resistant membrane microdomains (KKAA), of four N-terminal arginines that facilitate membrane association (R4A4), and the combination of both (KR) (44, 45). Three mutants displayed intermediate antagonistic activity, indicating that efficient membrane interactions (including G2A Nef in which the myristoylation acceptor glycine is replaced by alanine and membrane association is reduced [44]) and interactions with SH3 domain-containing proteins (including AxxA and V78A, in which residues of the proline-rich SH3 interaction motif are replaced [46]) contribute to SERINC5 antagonism by Nef. Notably, four Nef variants failed to display any SERINC5 antagonism. Expectedly (31, 32), these included mutations in the dileucine (LLAA Nef mutant) and diacidic (EDAA Nef mutant) interaction motifs in the C-terminal flexible loop of Nef that connect the viral protein to the endocytic machinery (47, 48). In addition, deletion of a CAW motif required for interactions of Nef with the cytoplasmic tail of CD4 as well as for cleavage of Nef by the viral protease (ΔCAW Nef mutant) (49, 50) or of the interaction site for the Nef-associated kinase complex (NAKC) via which Nef influences HIV transcription and release of extracellular vesicles (Δ12–39 Nef mutant) (42, 51) abrogated SERINC5 antagonism by Nef. Of note, all four molecular determinants essential for antagonism of SERINC5 are also essential for downregulation of cell surface CD4 by Nef in HIV-1-infected cells (37).
FIG 2.
Characterization of determinants in HIV-1 Nef for antagonism of SERINC5. (A) Schematic domain organization of HIV-1SF2 Nef. Highlighted residues mediate interactions with the host cell proteins indicated above. (B) Relative infectivity of HIV-1 virions produced in 293T cells transfected with HIV-1NL4-3 proviral DNA expressing the indicated HIV-1SF2 Nef proteins and the pBJ6 SERINC5.HA expression plasmid or a vector control. Forty-eight hours posttransfection, cell culture supernatants were used to infect TZM-bl reporter cells to determine the infectivity of released virions (measured as relative luciferase light units [RLU]). The amount of virus particles in the supernatant was determined by quantification of RT activity (microunits of RT), and relative infectivity was calculated as RLU per unit of RT activity (RLU/microunit of RT). Depicted are mean values ± standard deviations from two independent experiments, each performed in triplicate. Shown below are representative Western blots for expression levels of Nef and HIV-1 Gag (p55 and p24) in producer cell lysates.
Endosomal targeting is dispensable for SERINC5 antagonism.
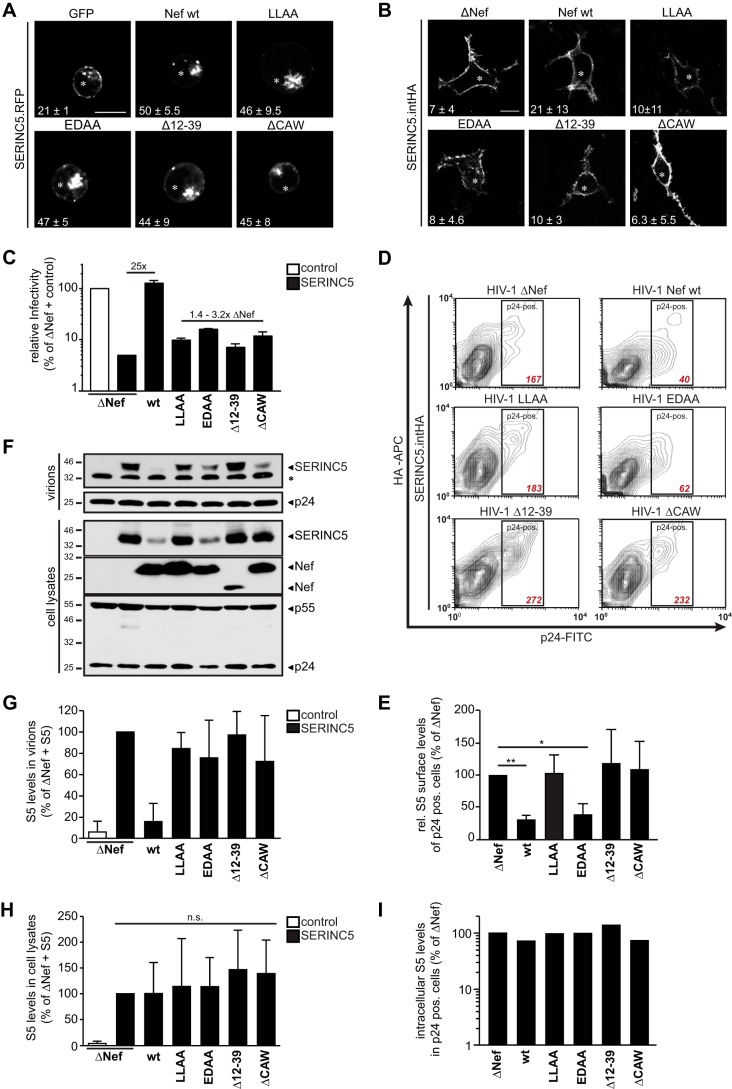
We next addressed which of the proposed effects of Nef on SERINC5 contributed to antagonism of the particle infectivity restriction. To address the role of endosomal targeting of SERINC5 for its antagonism by Nef, we first recapitulated that upon isolated expression of Nef.GFP in Jurkat T cells (31, 32), SERINC5 relocalized from the plasma membrane to an intracellular compartment that likely includes endosomes (Fig. 3A). However, such retargeting was only observed in approximately half of the Nef.GFP-expressing cells analyzed and all Nef mutants deficient for SERINC5 antagonism retargeted the restriction factor with similar efficacy. Moreover, in 293T cells, in which expression of SERINC5 leads to reduced particle infectivity unless counteracted by Nef, SERINC5 resided at the plasma membrane without appreciable targeting to intracellular compartments upon coexpression of WT or mutant Nef from HIV-1 proviral plasmids (Fig. 3B). Parallel analysis of the infectivity of the virus particles released from these producer cells confirmed that SERINC5 potently suppressed infectivity of HIV-1 ΔNef, WT Nef efficiently antagonized this restriction, and the four mutants were deficient in SERINC5 antagonism (Fig. 3C). Nef-mediated antagonism of SERINC5 thus can be fully efficient in the absence of endosomal retargeting of the restriction factor.
FIG 5.
Exclusion of SERINC5 from virions correlates with the enhancement of infectivity, but downregulation from the cell surface and endosomal accumulation of SERINC5 are not sufficient for antagonism by Nef. (A) Localization of SERINC5 in Jurkat TAg cells transiently expressing SERINC5.RFP and GFP, Nef.GFP, or the indicated GFP-tagged Nef mutants. Twenty-four hours posttransfection, poly-l-lysine-coated cover glasses were seeded with cells. Shown are representative confocal images from three independent experiments. Asterisks denote GFP-positive cells. White numbers represent the mean frequency of double positive cells with intracellular SERINC5 accumulation ± standard deviation. Scale bar, 10 μm. (B) Localization of SERINC5.intHA in 293T cells transfected with the indicated proviral HIV-1 plasmids and the SERINC5.intHA expression constructs. Shown are representative confocal microscopy pictures of cells fixed 48 h posttransfection following permeabilization and staining for SERINC5.intHA and HIV-1 CA (p24). Asterisks denote p24-positive cells. White numbers represent the mean frequency of double positive cells with intracellular accumulation of SERINC5 ± standard deviation of three independent experiments. Scale bar, 10 μm. (C) Relative infectivity of HIV-1 particles produced in 293T cells analyzed in panel B. Values for ΔNef plus control vector were set to 100%. Shown are means from 3 independent experiments ± standard deviations. (D) SERINC5 surface levels of virus-producing 293T cells described in panel B. Surface-exposed SERINC5.intHA proteins were stained with an anti-HA antibody, followed by an intracellular p24-FITC stain and analysis by flow cytometry. Shown are representative contour plots with the mean fluorescence intensity (MFI) of the SERINC5 stain in the p24-positive cell (gate) depicted in red. (E) Quantification of SERINC5.intHA cell surface levels as analyzed in panel D. Shown are means from 3 independent experiments ± standard deviations relative to ΔNef (set to 100%). (F) Western blot analysis of lysates of the producer cells and purified virions transfected as described in panels B to E. The asterisk to the right denotes an unspecific band. Immunodetection of SERINC5, HIV-1 Gag (p24 and p55), and Nef is shown. (G and H) Quantification of SERINC5 levels in virions (G) and cell lysates (H) determined by densitometry of Western blot analysis as shown in panel F. Depicted are means ± standard deviations from 3 independent experiments. (I) Quantification of intracellular SERINC5.intHA levels determined by flow cytometry (MFI of p24+ cells, with ΔNef plus SERINC5 set to 100%). Cells were transfected as described in panel B but derive from independent experiments. Shown are results from one representative out of two independent experiments. n.s., not significant; *, P < 0.05; **, P < 0.01.
Reducing cell surface exposure of SERINC5 is insufficient for SERINC5 antagonism.
In parallel, we assessed SERINC5 cell surface levels on these producer cells (Fig. 3D and E). As reported (32), Nef significantly reduced the levels of SERINC5 at the cell surface. The antagonism-deficient LLAA, Δ12–39, and ΔCAW Nef mutants failed to reduce cell surface exposure of SERINC5. In contrast, Nef EDAA retained full SERINC5 downregulation potential, demonstrating that cell surface downregulation of SERINC5 alone is not sufficient to antagonize the restriction factor.
Reduction of SERINC5 virion incorporation by Nef.
We also analyzed the amounts of SERINC5 that were incorporated into virions produced from these cells (Fig. 3F and G) as well as the corresponding cell-associated SERINC5 levels (Fig. 3F and H). Western blot analysis revealed efficient incorporation of SERINC5 into HIV particles produced in the absence of Nef, while the restriction factor was virtually undetectable in virions produced in the presence of WT Nef. In contrast, significant amounts of SERINC5 were detected in particles produced in the presence of each of the antagonism-defective Nef variants (Fig. 3F, upper panel). The extent of virion incorporation of the restriction factor in the presence of antagonism-deficient Nef mutants varied and ranged between 35 and 220% of that observed in HIV-1 ΔNef particles (see, e.g., Nef mutants EDAA and ΔCAW in the experiments shown in Fig. 3F). Over all experiments analyzed, the four Nef mutants displayed consistent defects in virion exclusion of SERINC5 (Fig. 3G). In the case of Nef EDAA, significant amounts of the restriction factor were incorporated into progeny virions despite efficient downregulation of SERINC5 from the cell surface (Fig. 3D and E). Western blot analysis of cell-associated SERINC5 levels gave variable results (Fig. 3F, lower panel; see detection of reduced levels of cell-associated SERINC5 for Nef WT and EDAA in the experiment shown), presumably due to intrasample variation in SERINC5 aggregation/solubility. Quantification of cell-associated SERINC5 levels in all experiments analyzed revealed that overall, Nef or its mutants did not significantly affect expression of the restriction factor (Fig. 3H). Consistently, quantification of total cellular SERINC5 levels by intracellular FACS, which we found to be more robust and reliable than Western blotting, in independent experiments indicated that SERINC5 expression was unaltered by coexpression of WT or mutant Nef (Fig. 3I), and treatment of these cells with proteasome or lysosome inhibitors did not increase SERINC5 levels (data not shown). These results indicate that under the experimental conditions used, the ability of Nef to antagonize SERINC5 is associated with efficient exclusion of the restriction factor from HIV particles and suggest that SERINC5 virion levels lower than those typically observed in HIV-1 ΔNef particles are sufficient to significantly inhibit particle infectivity.
HIV-1 Vpu does not antagonize SERINC5.
Since HIV-1 Nef shares several activities with the accessory HIV-1 protein Vpu, in particular with regard to downregulation of cell surface receptors (4–6), we asked next whether Vpu is able to antagonize SERINC5 (Fig. 4). We produced virus particles in the presence of SERINC5 for the HIV-1 WT, ΔNef, or an HIV-1NL4-3 variant that lacks expression of Vpu (ΔVpu). Expectedly, expression of SERINC5 during virus production did not affect the HIV-1 WT but markedly reduced the infectivity of ΔNef virions (Fig. 4A). This reduction in infectivity of ΔNef was observed despite the robust expression of Vpu (Fig. 4B). In contrast, expression of Nef from the ΔVpu provirus and thus in the absence of Vpu during virus production was sufficient to antagonize the SERINC5 infectivity restriction. In the context of the HIV-1NL4-3 strain used for these analyses, Vpu therefore does not contribute to antagonism of SERINC5. However, ΔVpu particles were slightly less infectious and contained more SERINC5 than HIV-1 WT particles. Since these two viruses differ in the Nef allele expressed and differences in the antagonistic potential of the Nef proteins of HIV-1SF2 and HIV-1NL4-3 were previously described (31, 32), we tested directly if these differences were due to allelic variation in Nef. Indeed, when expressed from the identical proviral backbone, NL4-3 Nef antagonized SERINC5 less efficiently than SF2 Nef (Fig. 4C) (13-fold versus 30-fold over HIV-1 ΔNef, respectively). This partial antagonism was paralleled by an intermediate reduction of SERINC5 virion incorporation that resulted in virions containing approximately 50% of SERINC5 of that detected in HIV-1 ΔNef particles (Fig. 4D). Correlating relative infectivity and SERINC5 virion levels as analyzed above emphasized that high levels of SERINC5 in virions are associated with a marked loss of infectivity, and complete removal of the restriction factor from virus particles coincides with full infectivity (Fig. 4E). However, in some instances in which Nef was present during virus production, robust infectivity could be observed despite a SERINC5 virion load in the range of 30 to 50% of that of HIV-1 ΔNef particles.
FIG 4.
HIV-1 Vpu is dispensable for antagonism of SERINC5. (A) Relative infectivity of HIV-1 particles produced in 293T cells expressing different variants of HIV-1NL4-3 (SF2 Nef WT, ΔNef, and ΔVpu) and pBJ6-SERINC5.HA or a vector control. Infectivity values are normalized to virus release and depicted as percentage of ΔNef plus control. Shown are the mean values from three independent experiments performed in triplicate ± standard deviations. (B) Western blot analyses of producer cell lysates and virions, respectively. Shown is immunodetection of SERINC5.HA, Nef, and HIV-1 Gag (p55 and p24) and quantification of virion-associated SERINC5.HA levels (ΔNef plus SERINC5.HA set to 100%). (C) Relative infectivity of HIV-1 particles from 293T cells transfected with SERINC5.HA or a control vector and the proviral plasmids HIV-1NL4-3 ΔNef, SF2 Nef, or NL4-3 Nef. Shown are means from three independent experiments ± standard deviation relative to ΔNef (set to 100%). (D) Western blot analysis of the producer cell lysate and purified virions as described in panel C. (E) Correlation of relative infectivity of virus particles and amounts of virion-associated SERINC5 protein levels from Fig. 1A and 3C and G, as well as panels A to D in this figure. All values shown are relative to ΔNef plus SERINC5.HA, which was set to 100%. ns, not significant.
Nef desensitizes HIV particles against infectivity inhibition of virion-associated SERINC5.
The above results suggested that, at least in the presence of an antagonist, the relationship between SERINC5 virion levels and virion infectivity is nonlinear. However, Fig. 4E pools data from independent experiments and was difficult to interpret due to the variability in SERINC5 protein detection and quantification by Western blotting. To assess the relationship between amounts of virion SERINC5 and particle infectivity more directly, we therefore titrated the amounts of SERINC5 expression plasmid in virus-producing cells and assessed virion infectivity and SERINC5 load in parallel (Fig. 5). The infectivity of HIV-1 ΔNef particles was already sensitive to the smallest amounts of SERINC5 tested, and virion infectivity linearly decreased with increasing amounts of expression and virion incorporation of SERINC5 (Fig. 5A). This suggests that in the absence of an antagonist, a direct correlation exists between the amounts of SERINC5 in virions and their infectivity. Expression of Nef from HIV-1 WT during virus production significantly increased virion infectivity. Importantly, the difference between HIV-1 WT and HIV-1 ΔNef increased with higher SERINC5 expression levels (6-fold versus 64-fold) (Fig. 5A). While detection of cell-associated levels by Western blotting was relatively insensitive (Fig. 5B), intracellular flow cytometry revealed a dose-dependent increase of SERINC5 expression with increasing amounts of expression plasmid used (Fig. 5C). These levels tended to be slightly reduced when Nef was present during virus production; however, this effect was subtle and did not correlate with the magnitude of virion infectivity enhancement. While SERINC5 was efficiently incorporated in HIV-1 ΔNef virions in a dose-dependent manner, Nef reduced but did not fully prevent virion incorporation of the restriction factor. Virion exclusion by Nef was efficient at low levels of SERINC5 expression (0.01 and 0.03 μg plasmid), but increasing amounts of virion SERINC5 were detected despite the presence of Nef at higher SERINC5 expression during virus production. Importantly, HIV-1 WT particles maintained robust infectivity irrespective of significant SERINC5 incorporation and displayed a more than 10-fold-higher infectivity than HIV-1 ΔNef virions with comparable incorporation of the restriction factor (e.g., HIV-1 WT with 0.3 μg plasmid approximately 12-fold more infectious than HIV-1 ΔNef with 0.1 μg plasmid with comparable amounts of virion SERINC5). Transfection of even larger amounts of SERINC5 expression plasmid overwhelmed the ability of Nef to antagonize the restriction factor, and SERINC5 levels in virions became too high for accurate quantification (data not shown). These results suggest that in addition to its ability to reduce virion incorporation of SERINC5 at low levels of expression, Nef also interferes with the antiviral potency of SERINC5 molecules that were incorporated into virus particles produced from cells with high SERINC5 expression. We conclude that with virion exclusion and counteraction of virion-associated SERINC5, Nef can employ two mechanisms for antagonism of SERINC5.
FIG 5.
Dose response of HIV-1 WT and ΔNef infectivity to SERINC5 expression. (A) 293T cells were transfected to produce HIV-1SF2 Nef or ΔNef in the absence (control) or presence of increasing amounts of SERINC5.intHA (0.01 to 0.3 μg plasmid DNA transfected). Relative infectivity of virus particles was determined and is presented relative to the WT plus control (in percentage). Shown are means ± standard deviations from three independent experiments performed in triplicate. (B) Western blot analysis of cell lysates and purified virus particles of producer cells described in panel A. Shown is immunodetection of SERINC5.intHA, Nef, and HIV-1 Gag (p55 and p24), with TfR as a loading control, including quantification of virion-associated SERINC5.intHA levels as determined by densitometry. The asterisks to the right and left denote unspecific bands. (ΔNef plus 0.3 μg SERINC5.intHA was set to 100%.) (C) Quantification of intracellular SERINC5.intHA levels in p24-positive cells analyzed by flow cytometry. Forty-eight hours posttransfection, cells were fixed, permeabilized, and stained for SERINC5.intHA and CA p24. Intracellular SERINC5 levels are presented relative to ΔNef plus 0.3 μg SERINC5.intHA (set to 100%). Shown are results from one representative out of two independent experiments.
DISCUSSION
Endosomal targeting, cell surface downregulation, and virion exclusion have been suggested to contribute to antagonism of SERINC5 by HIV-1 Nef (31, 32). In this study, we identified four molecular determinants in HIV-1 Nef required for antagonism of SERINC5. Notably, each of these determinants is also essential for downregulation of CD4 by Nef (37). Our parallel analysis of subcellular localization, virion incorporation, and infectivity restriction within the same experiment allowed us to exclude an essential role of endosomal targeting of SERINC5 for antagonism by Nef. These results also revealed that reducing cell surface exposure is not sufficient for antagonism, which, however, does not exclude that the reduction of cell surface density contributes to antagonism by Nef. In contrast, reduction of SERINC5 virion levels below the detection limit of our Western blot analysis was strictly associated with full recovery of virion infectivity. This was observed at low levels of SERINC5 expression, where Nef efficiently prevented virion incorporation of SERINC5. However, Nef also increased the infectivity of HIV-1 particles produced from cells with high SERINC5 expression that led to the robust incorporation of SERINC5 in virions. Finally, the reduction in infectivity of HIV-1 particles produced in the absence of a SERINC5 antagonist was correlated to the increasing amounts of restriction factor incorporated in virions. Together, these results suggest that (i) incorporation of SERINC5 into virions is a central aspect of its antiviral activity in the absence of an antagonist, (ii) Nef, in addition to excluding the restriction factor from virions at low levels of expression, can antagonize the SERINC5 infectivity barrier by inactivation of the antiviral activity of SERINC5 molecules that are associated with virus particles, and (iii) antagonism of SERINC5 by Nef is likely mediated by mechanisms related to downregulation of cell surface CD4. Whether these aspects also apply to SERINC3, the second SERINC family member with anti-HIV activity (31, 32), remains to be determined using a Nef protein with more pronounced antagonistic activity against SERINC3 than the SF2 Nef studied herein.
One main conclusion from our data is that, in the absence of an antagonist, the abundance of SERINC5 in virus particles is a key parameter that determines its inhibitory effect on virion infectivity. This supports the current view that SERINC5 reduces fusion as well as early postentry events via its physical presence in HIV-1 particles (31, 32, 34). The mechanism underlying this antiviral activity of virion-associated SERINC5 remains to be determined, but the specificity of the restriction and Nef antagonism thereof for specific viral glycoproteins suggests that this effect is related to the subcellular site and kinetics of the viral entry process and may involve the functional impairment of sensitive glycoproteins (31, 32, 52). It is conceivable that such a mechanism would depend on the amount of SERINC5, and our results demonstrate that in the absence of a SERINC5 antagonist, cell-associated levels of SERINC5 expression correlate well with its incorporation into virions as well as with its antiviral activity.
In contrast, no linear relationship between the efficiency of SERINC5 virion incorporation and particle infectivity was observed when HIV particles were produced in the presence of Nef. At low levels of expression, Nef expression was associated with efficient exclusion of the restriction factor from virions, which therefore displayed full infectivity. At higher levels of SERINC5 expression, however, a substantial load of virion SERINC5 was observed, which reduced particle infectivity much less than when virions were produced in the absence of Nef. Nef therefore antagonizes the inhibition of particle infectivity even when substantial amounts of SERINC5 are incorporated into virions. This identifies experimental conditions under which virion exclusion by Nef is not sufficient for antagonism of SERINC5 and illustrates that Nef is able to inactivate the antiviral activity of virion-associated SERINC5. Future studies will be required to address whether virion exclusion of low levels of SERINC5 are a by-product of the mechanism used by Nef to inactivate virion-associated SERINC5 or whether these two activities are uncoupled. With respect to virion exclusion of SERINC5, it will be important to assess whether the reduction of SERINC5 levels in virions by Nef is partial but homogenous among all produced particles or whether Nef generates a subpopulation of virions that gains infectivity because SERINC5 is fully excluded. Inactivation of virion-associated SERINC5 by Nef may reflect, e.g., alterations of critical posttranslational modifications of the restriction factor or its topology in virions. Which of these scenarios are physiologically relevant will depend on the SERINC5 protein levels in cells in which this particle infectivity restriction is present in vivo. Definition of the precise stoichiometry of Nef and SERINC5 in restriction and antagonism as well as the analysis of SERINC5 incorporation into individual virions will be important steps toward dissection of the molecular mechanisms involved.
While our results do not identify how Nef antagonizes SERINC5, they emphasize a similarity between the mechanisms used by Nef to downregulate cell surface CD4 and to antagonize SERINC5. Since CD4 is not present in the 293T cell system used, this does not reflect direct involvement of CD4 in SERINC5 antagonism but likely indicates that Nef targets similar host cell machinery to downregulate CD4 and antagonize SERINC5. CD4 downregulation by Nef largely relies on enhancing internalization rates from the plasma membrane via an AP-2- and clathrin-dependent pathway that targets CD4 for degradation (47, 53–58). Similarly, infectivity enhancement of virions produced from SERINC5-expressing cells by Nef requires AP-2 as well as dynamin (31, 32, 59, 60), and it seems plausible that Nef reduces cell surface exposure of SERINC5 by this connection to the host cell endocytic machinery. However, the results obtained with the Nef EDAA mutant reveal that cell surface downregulation may contribute to but is not sufficient for antagonism of SERINC5. Similar to the dileucine motif disrupted in the Nef mutant LLAA, the diacidic motif mutated in the Nef EDAA mutant is involved in the direct interaction between Nef and AP-2, and both motifs are essential for triggering internalization of plasma membrane CD4 (56, 61, 62). It was therefore surprising that Nef EDAA retained full SERINC5 downregulation ability, and detailed mechanistic studies are warranted to address the molecular basis of the respective roles of the dileucine and diacidic motifs in this process. The finding that downregulation of cell surface SERINC5 is not sufficient for antagonism implies that additional, yet to be discovered, mechanisms are critical for this Nef activity. This may, e.g., include alterations of biosynthetic transport routes of the restriction factor or prevention of lateral recruitment of SERINC5 to virus assembly sites at the plasma membrane. Whether virion exclusion and inactivation of virion-associated SERINC5 are mediated by the same molecular determinants in Nef also remains to be determined, and the antagonism-defective Nef mutants identified herein will likely be useful tools for such investigations. Irrespective of the precise molecular mechanism of Nef antagonism, the fact that overexpression of SERINC5 did not result in a reduction of unrelated Nef activities suggests that even at high levels of expression, the restriction factor does not physically sequester the viral protein with an efficiency sufficient to suppress Nef's biological properties. This may hint at a scenario where Nef may not need to physically interact with SERINC5 for antagonism, and in line with this model, we were unable to demonstrate association of both proteins by coimmunoprecipitation (data not shown). Similar to its effects on the trafficking of a wide range of cell surface receptors and peripheral membrane proteins (4, 12, 63), the effects of Nef on SERINC5 may therefore be indirect by acting on cellular pathways that control subcellular localization and function of the restriction factor. While the mechanisms of SERINC5 restriction and Nef antagonism require further investigation, the results presented here indicate that Nef can antagonize SERINC5 by exclusion from virions as well as inactivation of virion-associated SERINC5 and define specific Nef mutants as useful tools for future mechanistic studies.
ACKNOWLEDGMENTS
We are grateful to Heinrich Göttlinger for the gift of reagents, to Nadine Tibroni and Ina Ambiel for technical assistance, and to Barbara Müller for helpful discussion.
O.T.F. is a member of the Cellnetworks Cluster of Excellence (EXC81).
Funding Statement
This project is supported by the Deutsche Forschungsgemeinschaft (TRR83 project 15 and SPP1923 project FA378/17-1 to O.T.F.) and by Caritro Ricerca Biomedica grant no. 2013.0248 to M.P.
REFERENCES
- 1.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, Lawson VA, Crowe S, Maerz A, Sonza S, Learmont J, Sullivan JS, Cunningham A, Dwyer D, Dowton D, Mills J. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 2.Kestler HW III, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651–662. doi: 10.1016/0092-8674(91)90097-I. [DOI] [PubMed] [Google Scholar]
- 3.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 4.Haller C, Muller B, Fritz JV, Lamas-Murua M, Stolp B, Pujol FM, Keppler OT, Fackler OT. 2014. HIV-1 Nef and Vpu are functionally redundant broad-spectrum modulators of cell surface receptors, including tetraspanins. J Virol 88:14241–14257. doi: 10.1128/JVI.02333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lambele M, Koppensteiner H, Symeonides M, Roy NH, Chan J, Schindler M, Thali M. 2015. Vpu is the main determinant for tetraspanin downregulation in HIV-1-infected cells. J Virol 89:3247–3255. doi: 10.1128/JVI.03719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matheson NJ, Sumner J, Wals K, Rapiteanu R, Weekes MP, Vigan R, Weinelt J, Schindler M, Antrobus R, Costa AS, Frezza C, Clish CB, Neil SJ, Lehner PJ. 2015. Cell surface proteomic map of HIV infection reveals antagonism of amino acid metabolism by Vpu and Nef. Cell Host Microbe 18:409–423. doi: 10.1016/j.chom.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michel N, Allespach I, Venzke S, Fackler OT, Keppler OT. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr Biol 15:714–723. doi: 10.1016/j.cub.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 8.Benson RE, Sanfridson A, Ottinger JS, Doyle C, Cullen BR. 1993. Downregulation of cell-surface CD4 expression by simian immunodeficiency virus Nef prevents viral super infection. J Exp Med 177:1561–1566. doi: 10.1084/jem.177.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matusali G, Potesta M, Santoni A, Cerboni C, Doria M. 2012. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol 86:4496–4504. doi: 10.1128/JVI.05788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 11.Fortin JF, Barat C, Beausejour Y, Barbeau B, Tremblay MJ. 2004. Hyper-responsiveness to stimulation of human immunodeficiency virus-infected CD4+ T cells requires Nef and Tat virus gene products and results from higher NFAT, NF-kappaB, and AP-1 induction. J Biol Chem 279:39520–39531. doi: 10.1074/jbc.M407477200. [DOI] [PubMed] [Google Scholar]
- 12.Pan X, Rudolph JM, Abraham L, Habermann A, Haller C, Krijnse-Locker J, Fackler OT. 2012. HIV-1 Nef compensates for disorganization of the immunological synapse by inducing trans-Golgi network-associated Lck signaling. Blood 119:786–797. doi: 10.1182/blood-2011-08-373209. [DOI] [PubMed] [Google Scholar]
- 13.Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, Bailes E, Roques P, Sodora DL, Silvestri G, Sharp PM, Hahn BH, Kirchhoff F. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067. doi: 10.1016/j.cell.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Thoulouze MI, Sol-Foulon N, Blanchet F, Dautry-Varsat A, Schwartz O, Alcover A. 2006. Human immunodeficiency virus type-1 infection impairs the formation of the immunological synapse. Immunity 24:547–561. doi: 10.1016/j.immuni.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Schrager JA, Marsh JW. 1999. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc Natl Acad Sci U S A 96:8167–8172. doi: 10.1073/pnas.96.14.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham L, Fackler OT. 2012. HIV-1 Nef: a multifaceted modulator of T cell receptor signaling. Cell Commun Signal 10:39. doi: 10.1186/1478-811X-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauter D, Hotter D, Van Driessche B, Sturzel CM, Kluge SF, Wildum S, Yu H, Baumann B, Wirth T, Plantier JC, Leoz M, Hahn BH, Van Lint C, Kirchhoff F. 2015. Differential regulation of NF-kappaB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep 10:586–599. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haller C, Rauch S, Fackler OT. 2007. HIV-1 Nef employs two distinct mechanisms to modulate Lck subcellular localization and TCR induced actin remodeling. PLoS One 2:e1212. doi: 10.1371/journal.pone.0001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham L, Bankhead P, Pan X, Engel U, Fackler OT. 2012. HIV-1 Nef limits communication between linker of activated T cells and SLP-76 to reduce formation of SLP-76-signaling microclusters following TCR stimulation. J Immunol 189:1898–1910. doi: 10.4049/jimmunol.1200652. [DOI] [PubMed] [Google Scholar]
- 20.Haller C, Rauch S, Michel N, Hannemann S, Lehmann MJ, Keppler OT, Fackler OT. 2006. The HIV-1 pathogenicity factor Nef interferes with maturation of stimulatory T-lymphocyte contacts by modulation of N-Wasp activity. J Biol Chem 281:19618–19630. doi: 10.1074/jbc.M513802200. [DOI] [PubMed] [Google Scholar]
- 21.Choe EY, Schoenberger ES, Groopman JE, Park IW. 2002. HIV Nef inhibits T cell migration. J Biol Chem 277:46079–46084. doi: 10.1074/jbc.M204698200. [DOI] [PubMed] [Google Scholar]
- 22.Janardhan A, Swigut T, Hill B, Myers MP, Skowronski J. 2004. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate Rac and inhibit lymphocyte chemotaxis. PLoS Biol 2:E6. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stolp B, Imle A, Coelho FM, Hons M, Gorina R, Lyck R, Stein JV, Fackler OT. 2012. HIV-1 Nef interferes with T-lymphocyte circulation through confined environments in vivo. Proc Natl Acad Sci U S A 109:18541–18546. doi: 10.1073/pnas.1204322109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stolp B, Reichman-Fried M, Abraham L, Pan X, Giese SI, Hannemann S, Goulimari P, Raz E, Grosse R, Fackler OT. 2009. HIV-1 Nef interferes with host cell motility by deregulation of cofilin. Cell Host Microbe 6:174–186. doi: 10.1016/j.chom.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol 68:2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiken C, Trono D. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol 69:5048–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz O, Marechal V, Danos O, Heard JM. 1995. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol 69:4053–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basmaciogullari S, Pizzato M. 2014. The activity of Nef on HIV-1 infectivity. Front Microbiol 5:232. doi: 10.3389/fmicb.2014.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laguette N, Benichou S, Basmaciogullari S. 2009. Human immunodeficiency virus type 1 Nef incorporation into virions does not increase infectivity. J Virol 83:1093–1104. doi: 10.1128/JVI.01633-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi M, Aiken C. 2008. Nef enhances HIV-1 infectivity via association with the virus assembly complex. Virology 373:287–297. doi: 10.1016/j.virol.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inuzuka M, Hayakawa M, Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 34.Fackler OT. 2015. Spotlight on HIV-1 Nef: SERINC3 and SERINC5 identified as restriction factors antagonized by the pathogenesis factor. Viruses 7:6730–6738. doi: 10.3390/v7122970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rauch S, Pulkkinen K, Saksela K, Fackler OT. 2008. Human immunodeficiency virus type 1 Nef recruits the guanine exchange factor Vav1 via an unexpected interface into plasma membrane microdomains for association with p21-activated kinase 2 activity. J Virol 82:2918–2929. doi: 10.1128/JVI.02185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meuwissen PJ, Stolp B, Iannucci V, Vermeire J, Naessens E, Saksela K, Geyer M, Vanham G, Arien KK, Fackler OT, Verhasselt B. 2012. Identification of a highly conserved valine-glycine-phenylalanine amino acid triplet required for HIV-1 Nef function. Retrovirology 9:34. doi: 10.1186/1742-4690-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fackler OT, Moris A, Tibroni N, Giese SI, Glass B, Schwartz O, Krausslich HG. 2006. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology 351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 38.Pizzato M, Erlwein O, Bonsall D, Kaye S, Muir D, McClure MO. 2009. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J Virol Methods 156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Fritz JV, Tibroni N, Keppler OT, Fackler OT. 2012. HIV-1 Vpu's lipid raft association is dispensable for counteraction of the particle release restriction imposed by CD317/Tetherin. Virology 424:33–44. doi: 10.1016/j.virol.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Keppler OT, Allespach I, Schuller L, Fenard D, Greene WC, Fackler OT. 2005. Rodent cells support key functions of the human immunodeficiency virus type 1 pathogenicity factor Nef. J Virol 79:1655–1665. doi: 10.1128/JVI.79.3.1655-1665.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph JM, Eickel N, Haller C, Schindler M, Fackler OT. 2009. Inhibition of T-cell receptor-induced actin remodeling and relocalization of Lck are evolutionarily conserved activities of lentiviral Nef proteins. J Virol 83:11528–11539. doi: 10.1128/JVI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muratori C, Cavallin LE, Kratzel K, Tinari A, De Milito A, Fais S, D'Aloja P, Federico M, Vullo V, Fomina A, Mesri EA, Superti F, Baur AS. 2009. Massive secretion by T cells is caused by HIV Nef in infected cells and by Nef transfer to bystander cells. Cell Host Microbe 6:218–230. doi: 10.1016/j.chom.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Stolp B, Abraham L, Rudolph JM, Fackler OT. 2010. Lentiviral Nef proteins utilize PAK2-mediated deregulation of cofilin as a general strategy to interfere with actin remodeling. J Virol 84:3935–3948. doi: 10.1128/JVI.02467-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giese SI, Woerz I, Homann S, Tibroni N, Geyer M, Fackler OT. 2006. Specific and distinct determinants mediate membrane binding and lipid raft incorporation of HIV-1(SF2) Nef. Virology 355:175–191. doi: 10.1016/j.virol.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Geist MM, Pan X, Bender S, Bartenschlager R, Nickel W, Fackler OT. 2014. Heterologous Src homology 4 domains support membrane anchoring and biological activity of HIV-1 Nef. J Biol Chem 289:14030–14044. doi: 10.1074/jbc.M114.563528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manninen A, Hiipakka M, Vihinen M, Lu W, Mayer BJ, Saksela K. 1998. SH3-domain binding function of HIV-1 Nef is required for association with a PAK-related kinase. Virology 250:273–282. doi: 10.1006/viro.1998.9381. [DOI] [PubMed] [Google Scholar]
- 47.Craig HM, Pandori MW, Guatelli JC. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci U S A 95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X, Yu H, Liu SH, Brodsky FM, Peterlin BM. 1998. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8:647–656. doi: 10.1016/S1074-7613(00)80569-5. [DOI] [PubMed] [Google Scholar]
- 49.Schorr J, Kellner R, Fackler O, Freund J, Konvalinka J, Kienzle N, Krausslich HG, Mueller-Lantzsch N, Kalbitzer HR. 1996. Specific cleavage sites of Nef proteins from human immunodeficiency virus types 1 and 2 for the viral proteases. J Virol 70:9051–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mangasarian A, Piguet V, Wang JK, Chen YL, Trono D. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol 73:1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baur AS, Sass G, Laffert B, Willbold D, Cheng-Mayer C, Peterlin BM. 1997. The N-terminus of Nef from HIV-1/SIV associates with a protein complex containing Lck and a serine kinase. Immunity 6:283–291. doi: 10.1016/S1074-7613(00)80331-3. [DOI] [PubMed] [Google Scholar]
- 52.Usami Y, Gottlinger H. 2013. HIV-1 Nef responsiveness is determined by Env variable regions involved in trimer association and correlates with neutralization sensitivity. Cell Rep 5:802–812. doi: 10.1016/j.celrep.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, Greene WC. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol 8:1235–1238. doi: 10.1016/S0960-9822(07)00517-9. [DOI] [PubMed] [Google Scholar]
- 54.Burtey A, Rappoport JZ, Bouchet J, Basmaciogullari S, Guatelli J, Simon SM, Benichou S, Benmerah A. 2007. Dynamic interaction of HIV-1 Nef with the clathrin-mediated endocytic pathway at the plasma membrane. Traffic 8:61–76. doi: 10.1111/j.1600-0854.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 55.Greenberg M, DeTulleo L, Rapoport I, Skowronski J, Kirchhausen T. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol 8:1239–1242. doi: 10.1016/S0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 56.Ren X, Park SY, Bonifacino JS, Hurley JH. 2014. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. eLife 3:e01754. doi: 10.7554/eLife.01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 58.Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pizzato M, Helander A, Popova E, Calistri A, Zamborlini A, Palu G, Gottlinger HG. 2007. Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef. Proc Natl Acad Sci U S A 104:6812–6817. doi: 10.1073/pnas.0607622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Usami Y, Popov S, Gottlinger HG. 2014. The Nef-like effect of murine leukemia virus glycosylated gag on HIV-1 infectivity is mediated by its cytoplasmic domain and depends on the AP-2 adaptor complex. J Virol 88:3443–3454. doi: 10.1128/JVI.01933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindwasser OW, Smith WJ, Chaudhuri R, Yang P, Hurley JH, Bonifacino JS. 2008. A diacidic motif in human immunodeficiency virus type 1 Nef is a novel determinant of binding to AP-2. J Virol 82:1166–1174. doi: 10.1128/JVI.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geyer M, Yu H, Mandic R, Linnemann T, Zheng YH, Fackler OT, Peterlin BM. 2002. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J Biol Chem 277:28521–28529. doi: 10.1074/jbc.M200522200. [DOI] [PubMed] [Google Scholar]
- 63.Pan X, Geist MM, Rudolph JM, Nickel W, Fackler OT. 2013. HIV-1 Nef disrupts membrane-microdomain-associated anterograde transport for plasma membrane delivery of selected Src family kinases. Cell Microbiol 15:1605–1621. doi: 10.1111/cmi.12148. [DOI] [PubMed] [Google Scholar]