ABSTRACT
Nuclear egress of herpesviruses is accompanied by changes in the architecture of the nuclear membrane and nuclear lamina that are thought to facilitate capsid access to the inner nuclear membrane (INM) and curvature of patches of the INM around the capsid during budding. Here we report the properties of a point mutant of pUL34 (Q163A) that fails to induce gross changes in nuclear architecture or redistribution of lamin A/C. The UL34(Q163A) mutant shows a roughly 100-fold defect in single-step growth, and it forms small plaques. This mutant has a defect in nuclear egress, and furthermore, it fails to disrupt nuclear shape or cause observable displacement of lamin A/C despite retaining the ability to recruit the pUS3 and PKC protein kinases and to mediate phosphorylation of emerin. Extragenic suppressors of the UL34(Q163A) phenotype were isolated, and all of them carry a single mutation of arginine 229 to leucine in UL31. Surprisingly, although this UL31 mutation largely restores virus replication, it does not correct the lamina disruption defect, suggesting that, in Vero cells, changes in nuclear shape and gross displacements of lamin A/C may facilitate but are unnecessary for nuclear egress.
IMPORTANCE Herpesvirus nuclear egress is an essential and conserved process that requires close association of the viral capsid with the inner nuclear membrane and budding of the capsid into that membrane. Access to the nuclear membrane and tight curvature of that membrane are thought to require disruption of the nuclear lamina that underlies the inner nuclear membrane, and consistent with this idea, herpesvirus infection induces biochemical and architectural changes at the nuclear membrane. The significance of the nuclear membrane architectural changes is poorly characterized. The results presented here address that deficiency in our understanding and show that a combination of mutations in two of the viral nuclear egress factors results in a failure to accomplish at least two components of lamina disruption while still allowing relatively efficient viral replication, suggesting that changes in nuclear shape and displacement of lamins are not necessary for herpes simplex virus 1 (HSV-1) nuclear egress.
INTRODUCTION
During nuclear egress, herpes simplex virus 1 (HSV-1) capsids escape from the nucleus to the cytoplasm via budding at the inner nuclear membrane (INM) and deenvelopment at the outer nuclear membrane (ONM) (1, 2). Infection is associated with changes to the architecture of the nucleus, including enlargement and convolution of the nuclear shape (3, 4). These changes are thought to reflect changes in the structure of the underlying nuclear lamina that facilitate the nuclear egress mechanism.
The nuclear lamina is a meshwork of filaments comprised of four type V intermediate filament proteins (lamins A, B1, B2, and C) that is anchored to the INM by intrinsic membrane lamin-associated proteins (LAPs). The lamina is thought to present at least two obstacles to nuclear egress. First, the mesh made by the lamin filaments may present a steric barrier for access of capsids to the INM. Second, the rigidity of the lamin network and its anchorage to the INM may prevent the tight curvature of the attached membrane necessary for capsid budding. Thus, localized disruption of the lamin filament network and its interactions with underlying LAPs is thought to be required for herpesvirus replication.
In uninfected cells, the structure of the nuclear lamina is controlled in part by phosphorylation of lamina components, with additional phosphorylation, in general, causing a loss of interactions between lamina components and consequent disruption of the lamina structure. HSV infection has been shown to induce specific structural and functional changes in components of the nuclear lamina, including (i) hyperphosphorylation and redistribution of emerin, a LEM domain LAP, from an even distribution on the INM to a punctate distribution and localization to blebs on the outer surface of the nucleus (5, 6); (ii) increased mobility of lamin B receptor (LBR) (7); (iii) masking and unmasking of lamin A (LMNA) and lamin B (LMNB) monoclonal antibody epitopes (8); (iv) redistribution of lamin A/C (LMNA/C) from a uniform lining of the INM to an uneven distribution with areas of thickening or loss of LMNA/C (3); and (v) increased acidity of LMNA/C subunits upon HSV-1 infection, suggesting phosphorylation and disruption of LMNA/C networks (9).
HSV-1 proteins, including pUL34, pUL31, and pUS3, have been shown to play a role in nuclear lamina disruption. These proteins comprise part of the larger nuclear egress complex (NEC) that is required for changes in nuclear shape and the redistribution of lamin subunits observed during infection (8). pUL34 interacts with LMNA/C proteins in vitro and can cause the disruption of LMNA/C in transfected cells (8). pUS3 regulates and mediates the phosphorylation of lamina components (3). Conserved herpesvirus protein kinases (CHPKs) have been shown to play a role in nuclear lamina disruption. Human cytomegalovirus (HCMV) pUL97 is recruited to the nuclear periphery and causes the disruption of lamin networks (10). HSV-2 pUL13 causes conformational changes in lamin A and lamin C and disrupts lamin B networks (11). Cellular proteins have also been implicated in causing nuclear lamina disruption. Protein kinases PKCα and PKCδ are recruited to the nuclear periphery upon HSV-1 infection, and they phosphorylate emerin and lamin B (12, 13). The cellular protein p32 has also been shown to be recruited to the nuclear periphery during HSV-1 infection, and it interacts with pUL31, pUL34, LBR, and other components of the NEC (14, 15).
Although much evidence has shaped the current model of HSV-1-mediated nuclear lamina disruption, direct testing of the significance of this mechanism has not been possible. Experiments to isolate the significance of this specific step of nuclear egress have not been performed due to the multiple functions of the viral proteins involved and the absence of mutants that are defective specifically in nuclear lamina disruption. For example, no study has yet shown that HSV-1-induced lamina phosphorylation actually causes dissociation of lamin subunits from the network of the nuclear lamina. In this study, we describe the characteristics of a UL34 mutant [UL34(Q163A)] that is deficient in growth, nuclear egress, and nuclear lamina disruption. Surprisingly, the growth defect can largely be suppressed by another viral mutation, without concomitant suppression of the lamina disruption defect. This allows evaluation of the importance of two of the components of nuclear architecture disruption in viral replication.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and cell lines derived from Vero cells were maintained in Dulbecco's modified Eagle medium (high glucose) (DMEM) supplemented with 5% bovine calf serum or 5% fetal calf serum. The properties of HSV-1(F), vRR1072 (TK+) (referred to here as the UL34-null virus), a bacterial artificial chromosome (BAC)-derived UL34-null virus, and the UL34-null/UL31(R229L) virus have been described previously (8, 16).
Plasmids and cell lines.
An infection-inducible UL34(Q163A)-expressing plasmid (pRR1383) was constructed. pRR1300, which contains the UL34(Q163A) gene on the background of pRR1072Rep (17), was digested with XbaI, treated with Klenow enzyme to generate a blunt end, and then further digested with NgoMIV. The resulting 1.25-kb fragment, containing the UL34 coding and 5′ promoter/regulatory sequences, was ligated into AseI/Klenow-NgoMIV-cut pTuner-IRES2 (Clontech). The resulting plasmid expresses bicistronic pUL34-IRES2-EGFP mRNA from the UL34 promoter/regulatory sequences.
The UL34(Q163A)AW clonal cell line (referred to here as Q163A-expressing cells) was constructed as previously described (16), with some modifications. Transfection of pRR1383 into Vero cells was followed by selection with G418 and isolation of clones by limiting dilution. Candidate Q163A-expressing cell clones were identified by their expression of enhanced green fluorescent protein (EGFP) 16 h after infection with HSV-1(F). Expression of pUL34(Q163A) was confirmed by an immunofluorescence assay for pUL34 16 h after infection with UL34-null virus.
A pcDNA plasmid carrying UL31-FLAG (pRR1334) was described previously (16). A pcDNA plasmid carrying UL34(Q163A), called pRR1384, was constructed by amplification of the UL34 coding sequence from pRR1300 by use of the primers 5′-ACCCAAGCTTCCATGGCGGGACTGGGC-3′ and 5′-GCCCTCTAGATTATAGGCGCGCGCCAGC-3′, digestion of the resulting PCR product with HindIII and XbaI, and ligation into HindIII-XbaI-cut pcDNA3.
Coimmunoprecipitation.
293T cells in 6-well culture plates were transfected with plasmids carrying UL34, UL34(Q163A), or FLAG-tagged UL31 (UL31-FLAG) by use of Lipofectamine reagent according to the manufacturer's instructions. Transfected cells were washed once in phosphate-buffered saline (PBS) 2 days after transfection and subsequently lysed by the addition of RIPA buffer (50 mM Tris, pH 9.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) and sonicated. Ten percent of each sample was removed and used as the input sample. The remainder was immunoprecipitated using anti-FLAG M2 resin (Sigma) according to the manufacturer's instructions, using an overnight binding step and elution with a 3×FLAG peptide. Immunoprecipitated samples were separated in 10% SDS-PAGE gels, blotted onto nitrocellulose membranes, and probed with antibodies against either FLAG or UL34.
Immunoblotting.
Nitrocellulose sheets containing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 45 min. Membranes were probed with the following primary antibodies: chicken polyclonal anti-UL34 (1:1,000) (18), mouse monoclonal anti-HSV-1 scaffolding protein (1:2,000) (Serotec), mouse monoclonal anti-emerin (Santa Cruz), and mouse monoclonal anti-FLAG M2 (Sigma). The membranes were then incubated with the respective alkaline phosphatase-conjugated secondary antibodies.
Recruitment of PKCα and PKCδ.
Overexpression constructs for MYC-tagged PKCα (a kind gift from Jae-Won Soh) and FLAG-tagged PKCδ (a kind gift from Ushio Kikkawa) were transfected into Vero cells or cell lines by use of Lipofectamine reagent according to the manufacturer's instructions. Twenty-four hours after transfection, cells were infected with HSV-1(F) or UL34-null virus at a multiplicity of infection (MOI) of 5 for 16 h. Indirect immunofluorescence assays and confocal microscopy were performed. Pearson's correlation coefficients were calculated to determine the degree of colocalization by using measurements in ImageJ.
Selection of extragenic suppressors.
Twelve-well cultures of Q163A-expressing cells were infected with 1 × 107 PFU of UL34-null virus. After 90 min, the virus inoculum was replaced with V medium (DMEM supplemented with 1% heat-inactivated bovine calf serum). Virus stocks were prepared 1 day after infection by freezing, thawing, and sonication. Infection was repeated until visually robust plaques formed, and these were subsequently isolated through 3 rounds of plaque purification on Q163A-expressing cells.
Plaque size assays.
Twelve-well tissue culture plates were seeded with 8 × 105 Vero cells the day before infection. Cells were infected at a low MOI for 1.5 h, after which the virus inoculum was replaced with a 1:250 dilution of pooled human immunoglobulin (GammaSTAN) in V medium. Cells were fixed and stained after 2 days of infection. A mouse monoclonal anti-HSV 45-kDa protein (scaffolding protein) antibody (Serotec) and Alexa Fluor 568–goat anti-mouse IgG (Invitrogen) were used as primary and secondary antibodies, respectively.
Single-step growth measurement.
Measurement of replication of the HSV-1(F), UL34-null, UL34(Q163A) selection, and UL34-null/UL31(R229L) virus strains on Vero, tUL34CX, or tUL34(Q163A)AW cells was performed as previously described (19).
Indirect immunofluorescence assay.
Cells were fixed with 4% formaldehyde for 20 min and then washed in PBS. Cells were permeabilized and blocked by incubation in IF buffer (1× PBS, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1% egg albumin, 0.01% NaN3). Primary antibodies were diluted in IF buffer as follows: chicken anti-UL34, 1:1,000; mouse anti-MYC (a kind gift from Mike Apicella), 1:500; mouse monoclonal anti-LMNA (Santa Cruz Biotechnology), 1:500; and mouse anti-emerin, 1:500. Coverslips were mounted onto glass slides, and confocal microscopy was performed using a Zeiss 510 confocal microscope. All images shown are representative of experiments performed a minimum of three times.
Transmission electron microscopy (TEM) of infected cells.
Confluent monolayers of UL34WT-expressing cells and UL34(Q613A)-expressing cells were infected with HSV-1(F) and UL34-null/UL31(R229L), respectively, at an MOI of 5 for 18 h and then fixed by incubation in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h. Cells were postfixed in 1% osmium tetroxide, washed in cacodylate buffer, embedded in Spurr's resin, and cut into 95-nm sections. Sections were mounted on grids, stained with uranyl acetate and lead citrate, and examined with a JEOL 1250 transmission electron microscope. Ten cells were used for the quantitation of viral capsids within cellular compartments. DNA- and non-DNA-containing capsids were counted in the nucleus, the perinuclear space (between the INM and the ONM), and the extranuclear compartment (within the cytoplasm or the extracellular space).
Quantitation of nuclear egress by quantitative PCR (qPCR).
Confluent monolayers of Vero cells, wild-type UL34 (UL34WT)-expressing cells, or Q163A-expressing cells were infected with UL34-null virus at an MOI of 5 for 15 h and then scraped into PBS with 1 mM EDTA. Cells were lysed by resuspension in harvest buffer (10 mM HEPES, pH 7.5, 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 0.5% Triton X-100, 1 mM dithiothreitol [DTT]), followed by incubation on ice for 5 min. Nuclear and cytoplasmic fractions were separated by centrifugation at 5,000 rpm for 10 min. Half of the nuclear fraction was saved for amplification of genomic DNA. The remainder of the nuclear fraction was sonicated for 25 s by use of a Fisher Sonic Dismembrator instrument at a power level of 1. Unencapsidated DNA in the sonicated nuclear and cytoplasmic fractions was then degraded by addition of MgCl2 to a concentration of 5 mM and digested with 50 U DNase I for 30 min at 37°C. EDTA (5 mM) was added to the nuclear and cytoplasmic fractions, and DNA was extracted from all fractions by use of a Nucleospin tissue kit (Macherey-Nagel) according to the manufacturer's instructions.
SYBR real-time PCR amplification was performed using an Mx3005P real-time PCR system (Agilent Stratagene). The primers 5′-TAACATCACCCGCACCATC-3′ and 5′-GGTCCTTCTTCTTGTCCTTCAG-3′ were used to amplify UL30 of HSV-1, and the primers 5′-CCTTCTCCCACACACATACAC-3′ and 5′-CCTAGTCCCAGGGCTTAATTT-3′ were used to amplify the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. Each experiment was repeated three times, with expression levels calculated using the 2−ΔΔCT method, where CT indicates the cycle threshold. Data were analyzed using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA), and values are presented as means ± standard errors for three replicate samples. Statistical significance was calculated by Student's t test, using results from all three experiments.
Nuclear morphology analysis.
Cross-sectional images of nuclei of Vero cells and Vero cell lines were used to measure the nuclear contour ratio. Cells were infected with the HSV-1(F), UL34-null, raw selection, or UL34-null/UL31(R229L) strain at an MOI of 5 for 15 h. The cells were fixed, and indirect immunofluorescence assay of LMNA/C was performed. The nuclear cross-sectional area and perimeter were measured using ImageJ. The nuclear contour ratio was calculated using the formula (4π × area)/perimeter2, as described by Smeulders and Dorst (20). Twenty images of each sample were used to measure the nuclear contour ratio for each experiment. Data shown are representative of one experiment that was repeated three times.
RESULTS
Construction and characterization of a UL34(Q163A)-expressing cell line.
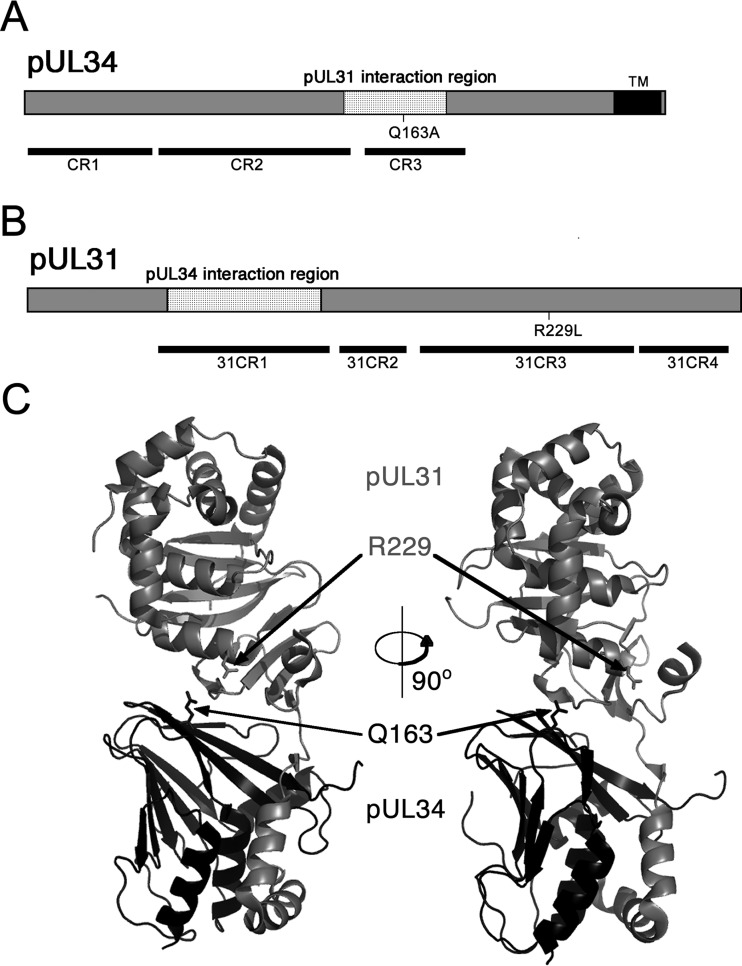
To identify amino acid residues of pUL34 that are important for HSV-1 replication, we previously described a strategy for evaluation of mutant UL34 function based on UL34-null virus infection of cell lines that express mutant pUL34 (17, 21). The Q163A mutant was evaluated as part of a larger effort to examine the function of UL34 mutants in which individual amino acids conserved among alphaherpesviruses were replaced with alanine. The UL34(Q163A) mutant was identified as having a reduced ability to complement replication of the UL34-null virus in trans, suggesting that an essential function of pUL34 had been disrupted. The Q163A mutation is located within conserved region 3 (CR3) of the protein, within the region previously shown to mediate interaction with pUL31 (22, 23) (Fig. 1A) and at the interaction interface of pUL31 and pUL34 (Fig. 1C). The glutamine at position 163 is conserved among alphaherpesviruses.
FIG 1.
Schematic designs for UL31 and UL34 showing relevant sequence features. (A) Protein sequence of pUL34 showing the location of the Q163A mutation, conserved regions, and the pUL31 interaction region. (B) Protein sequence of pUL31 showing the location of the R229L mutation, conserved regions, and the pUL34 interaction region. (C) Three-dimensional structures of pUL31 and pUL34. The locations of the Q163A mutation and the R229L mutation are indicated with arrows. Designations of conserved domains of pUL31 and pUL34 follow the nomenclature proposed by Schnee et al. (22). The protein structure of HSV-1 NEC is that previously described by Bigalke and Heldwein (31).
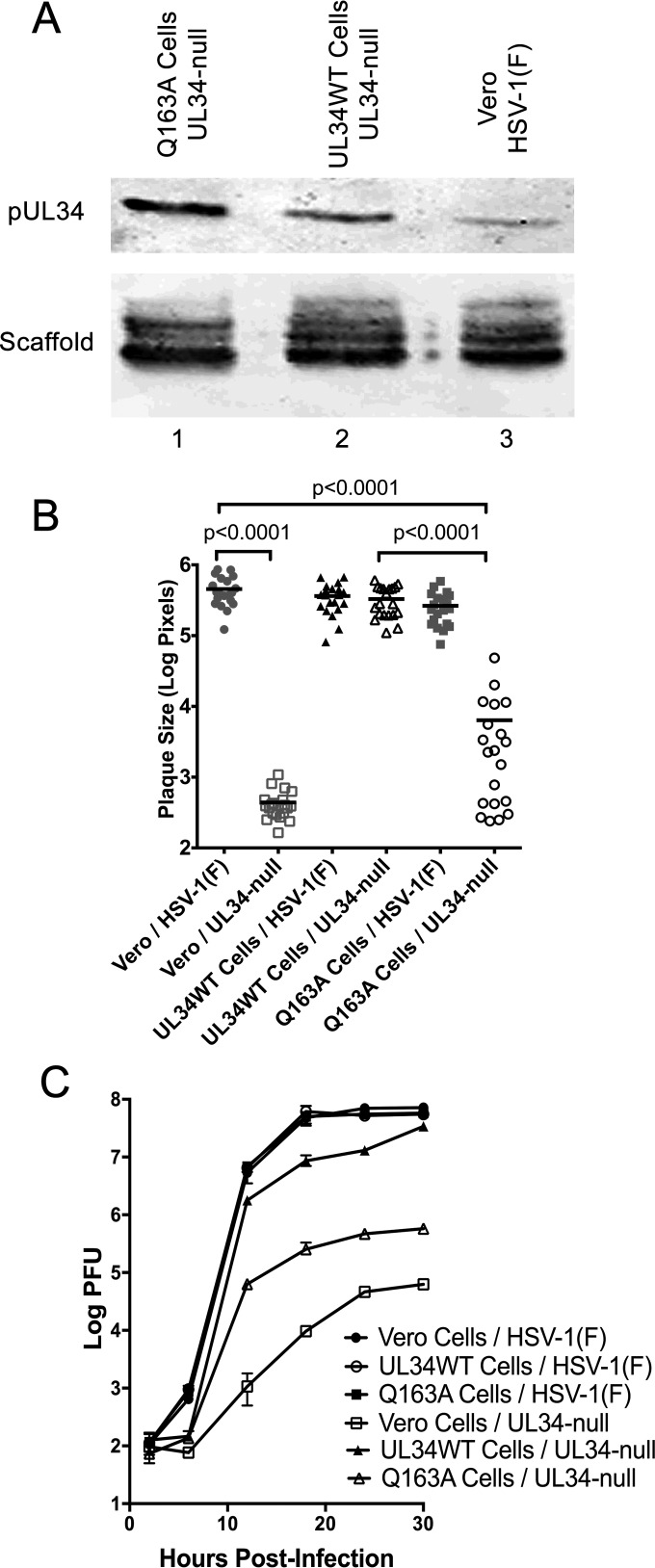
To further characterize the function of pUL34(Q163A), a stable cell line that expresses pUL34(Q163A) was created as described in Materials and Methods. Expression of pUL34 in HSV-1(F)-infected Vero cells was compared to that in UL34-null-infected Q163A-expressing and UL34WT-expressing cells by immunoblotting (Fig. 2A). An HSV-1 scaffolding protein was used as a loading control. Both the Q163A-expressing and UL34WT-expressing cells produced larger amounts of pUL34 than that produced by HSV-1(F)-infected Vero cells, ensuring that phenotypic differences between UL34WT and UL34(Q163A) cannot be due to less expression of pUL34(Q163A).
FIG 2.
Characterization of the UL34(Q163A)-expressing cell line. (A) Expression of WT and Q163A mutant UL34 by stable Vero cell lines. Vero cells (lane 3) and cells stably expressing Q163A mutant UL34 (lane 1) or WT UL34 (lane 2) were infected with HSV-1(F) (lane 3) or the UL34-null virus (lanes 1 and 2). Cell lysates were blotted for pUL34 (top) or a scaffold protein (bottom). (B) Plaque size assay of HSV-1(F) and the UL34-null virus on Vero cells, UL34WT-expressing cells, and Q163A-expressing cells. Plaque areas were measured 48 h after infection at a low MOI. Each point represents a single plaque area. Twenty plaques were measured for each condition. Statistical significance was determined by performing the Kolmogorov-Smirnov test. (C) Single-step growth of HSV-1(F) or the UL34-null virus on Vero cells, UL34WT-expressing cells, and Q163A-expressing cells. Virus yields are expressed as log PFU per milliliter. Each data point represents the mean result for three independent experiments, and error bars represent ranges. Statistical significance was determined using Student's t test.
Effects of the UL34(Q163A) mutant on virus spread and virus production.
To determine the effect of UL34(Q163A) on virus spread, a plaque size assay was performed in the presence of neutralizing antibody. HSV-1(F) and the UL34-null virus were plated on Vero cells, UL34WT-expressing cells, and Q163A-expressing cells at a low multiplicity of infection (Fig. 2B). HSV-1(F) formed large plaques of similar sizes on all three cell lines, indicating that pUL34(Q163A) does not inhibit the function of pUL34WT. As expected, the UL34-null virus formed tiny plaques of one or a few infected cells on noncomplementing Vero cells but formed large plaques on UL34WT-expressing cells that were comparable to those formed on HSV-1(F)-infected Vero cells. The UL34-null virus formed a wide range of plaque sizes on Q163A-expressing cells that were significantly smaller overall than those formed on UL34-null-infected UL34WT-expressing cells, indicating that pUL34(Q163A) results in a defect that reduces viral spread and plaque size.
To determine the effect of UL34(Q163A) on virus production, the single-step growth kinetics of HSV-1(F) and the UL34-null virus were measured on Vero cells, UL34WT-expressing cells, and Q163A-expressing cells (Fig. 2C). HSV-1(F) replicated efficiently on all three cell lines (Fig. 2C), indicating that pUL34(Q163A) does not interfere with pUL34WT function. As shown previously, the UL34-null virus replicated poorly on Vero cells (8). The UL34-null virus produced about 10-fold less virus than the amount of HSV-1(F) on UL34WT-expressing cells (Fig. 2C), most likely due to the variable expression of UL34 in each individual cell. UL34-null-infected Q163A-expressing cells produced 100-fold less virus than UL34-null-infected UL34WT-expressing cells (Fig. 2C). These data suggest that UL34(Q163A) causes a defect that results in reduced virus production.
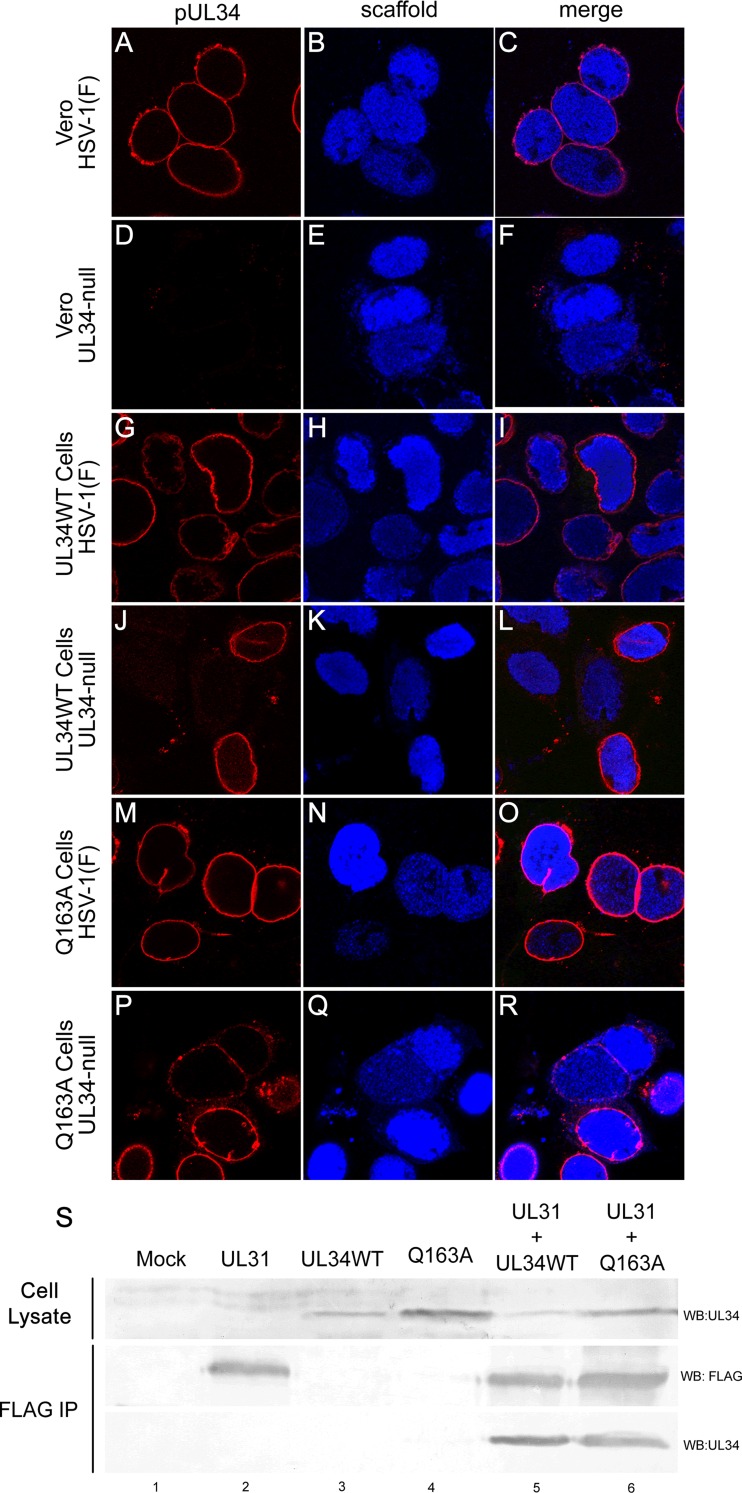
Defects in virus production caused by UL34(Q163A) may result from failure of a variety of functions, including localization to the INM, interaction with pUL31, nuclear lamina disruption, and budding. To determine the ability of pUL34(Q163A) to localize properly during infection, confocal microscopy of Vero, UL34WT-expressing, and Q163A-expressing cells infected with HSV-1(F) or the UL34-null virus was performed (Fig. 3A to R). During HSV-1(F) infection of all three cell lines, pUL34 localized to the nuclear periphery (Fig. 3A to C, G to I, and M to O), indicating that none of the cell lines causes a nonspecific defect in pUL34WT localization. pUL34(Q163A) also tightly localized to the nuclear periphery during UL34-null infection (Fig. 3P to R), suggesting that the growth and spread defects of the UL34(Q163A) mutant were not due to improper localization.
FIG 3.
UL34(Q163A) localizes to the nuclear periphery and maintains interaction with pUL31-FLAG. (A to R) Representative confocal images of pUL34 and scaffold localization. Vero cells (A to F), UL34WT-expressing cells (G to L), or Q163A-expressing cells (M to R) were infected with HSV-1(F) (A to C, G to I, and M to O) or the UL34-null virus (D to F, J to L, and P to R). Cells were stained for pUL34 (left column) or a scaffold protein (center column) at 16 h postinfection (hpi). (S) Coimmunoprecipitation of UL31-FLAG and UL34WT or UL34(Q163A). UL31-FLAG was purified from untransfected 293T cells (lane 1) or cells transfected with UL31-FLAG (lane 2), UL34WT (lane 3), UL34(Q163A) (lane 4), UL31-FLAG and UL34WT (lane 5), or UL31-FLAG and UL34(Q163A) (lane 6). Western blots (WB) of cell lysates (top) and magnetic bead-purified samples (bottom) were probed for UL34 or FLAG. IP, immunoprecipitation.
Proper recruitment of pUL34(Q163A) to the nuclear periphery during infection suggests that mutant pUL34 interacts normally with pUL31. To confirm this, a coimmunoprecipitation assay of pUL31-FLAG and pUL34(Q163A) was performed (Fig. 3S). 293T cells were transfected with the UL31-FLAG, UL34WT, or UL34(Q163A) construct, and pUL31-FLAG was immunoprecipitated by use of anti-FLAG magnetic beads. Consistent with their proper localization during infection, pUL34WT and pUL34(Q163A) could be coprecipitated efficiently with pUL31-FLAG, suggesting a normal pUL31-pUL34 interaction.
UL34(Q163A) causes a nuclear egress defect.
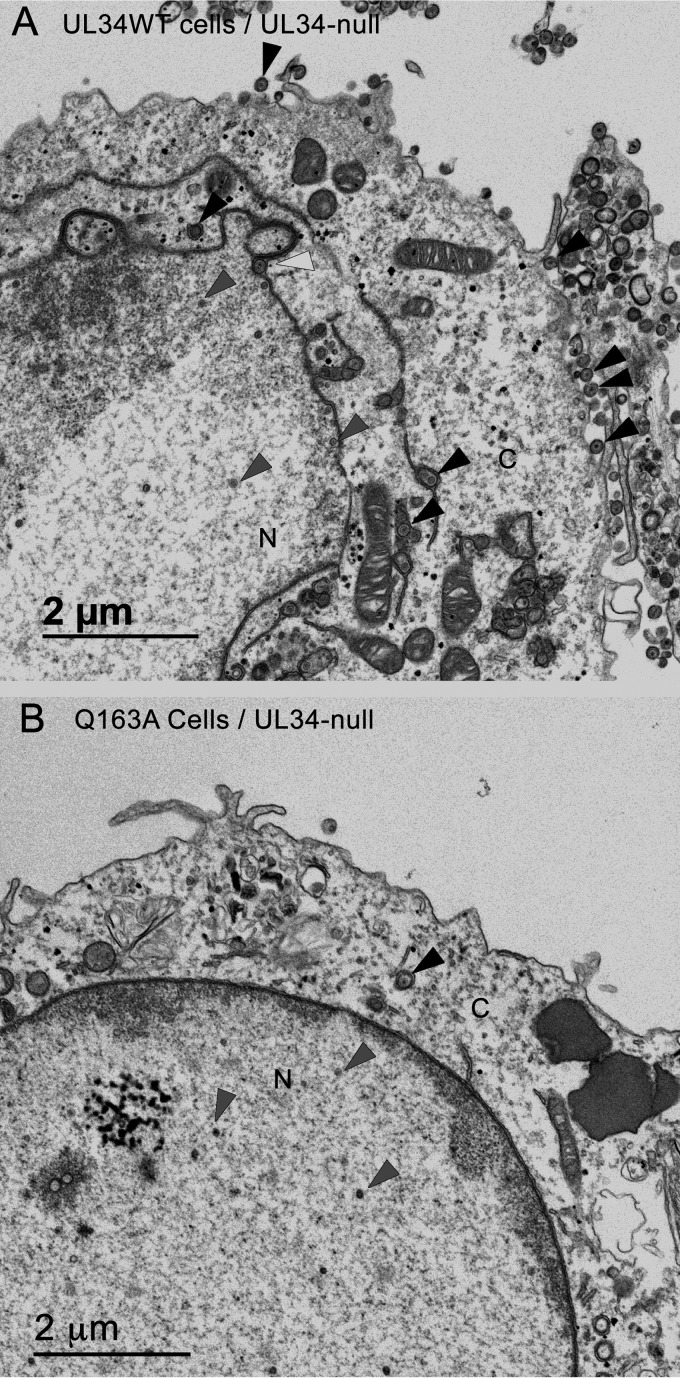
In order to characterize the cause of the defect of the UL34(Q163A) mutant, UL34WT-expressing cells or Q163A-expressing cells were infected with HSV-1(F) or the UL34-null virus at an MOI of 10 for 20 h and then prepared for analysis by TEM (Fig. 4). HSV-1(F)-infected UL34WT-expressing cells produced a large number of extranuclear virions and nuclear egress intermediates (Fig. 4A; Table 1), and the nuclear membrane acquired a convoluted shape (Fig. 4A). UL34-null-infected Q163A-expressing cells produced few extranuclear virions (Fig. 4B; Table 1), and the nuclear membrane retained a uniform ovoid structure similar to that of an uninfected cell. The sequestration of capsids in the nucleus and the inability to cause changes in nuclear shape in UL34-null-infected Q163A-expressing cells suggested that UL34(Q163A) causes a nuclear egress defect.
FIG 4.
UL34(Q163A) causes a nuclear egress defect. Digital micrographs show UL34WT-expressing (A) and Q163A-expressing (B) cells infected with the UL34-null virus for 20 h. Black arrowheads point to extranuclear capsids, white arrowheads point to perinuclear capsids, and gray arrowheads point to nuclear capsids. N, nucleus; C, cytoplasm.
TABLE 1.
Distributions of capsids in cellular compartments
| Cell typea | Virus | % of capsids |
||
|---|---|---|---|---|
| Extranuclearb | Perinuclearc | Intranucleard | ||
| UL34WT-expressing cells | HSV-1(F) | 43.35 | 3.19 | 53.46 |
| UL34-null | 42.66 | 1.21 | 56.13 | |
| Q163A-expressing cells | HSV-1(F) | 28.76 | 0.67 | 70.56 |
| UL34-null | 7.18 | 0 | 92.81 | |
Ten cell sections for each condition were used for quantitation.
Extranuclear capsids are defined as DNA- or non-DNA-containing capsids located within the cytoplasm or the extracellular space.
Perinuclear capsids are defined as enveloped capsids between the INM and the ONM.
Intranuclear capsids are defined as DNA- or non-DNA-containing capsids within the nucleus.
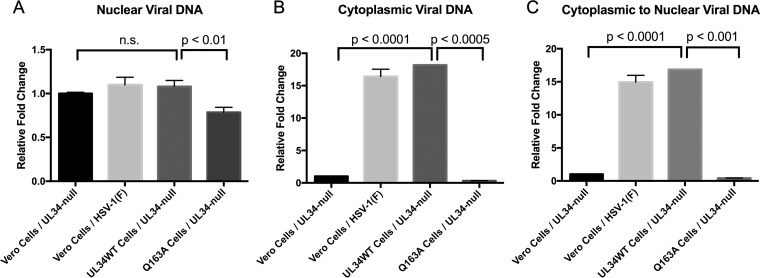
qPCR analysis was performed to further determine if UL34(Q163A) causes a nuclear egress defect. Vero, UL34WT-expressing, and Q163A-expressing cells were infected with HSV-1(F) or the UL34-null virus, and encapsidated viral DNA was extracted from the nucleus and the cytoplasm. The amounts of viral nuclear DNA were relatively the same under all conditions (Fig. 5A), indicating no DNA packaging or nucleocapsid assembly defects. HSV-1(F)-infected Vero cells and UL34-null-infected UL34WT-expressing cells contained significantly more cytoplasmic viral DNA than the nuclear egress-deficient UL34-null-infected Vero cells (Fig. 5B). Similarly, UL34-null-infected Q163A-expressing cells contained significantly smaller amounts of viral DNA than the UL34-null-infected UL34WT-expressing cells, indicating a nuclear egress defect (Fig. 5B). To account for any variation in infection under any of the tested conditions, the ratio of cytoplasmic to nuclear viral DNA was calculated. UL34-null-infected Vero cells had a significantly lower cytoplasmic/nuclear viral DNA ratio than that of UL34-null-infected UL34WT-expressing cells, showing the ability of qPCR to determine nuclear egress defects (Fig. 5C). UL34-null-infected Q163A-expressing cells had a cytoplasmic/nuclear viral DNA ratio similar to that of infected Vero cells, adding further evidence that UL34(Q163A) causes a nuclear egress defect.
FIG 5.
Characterization of a nuclear egress defect by qPCR. (A) Amounts of nuclear encapsidated viral DNA in UL34-null- or HSV-1(F)-infected Vero cells, UL34-null-infected UL34WT-expressing cells, and UL34-null-infected Q163A-expressing cells. (B) Amounts of cytoplasmic encapsidated viral DNA. (C) Ratios of cytoplasmic to nuclear viral encapsidated DNA. The graphs shown are representative of three replicates of each experiment performed a total of three times. Relative fold changes were calculated by the 2−ΔΔCT method. Statistical significance was determined by Student's t test, using all three replicate values for three separate experiments.
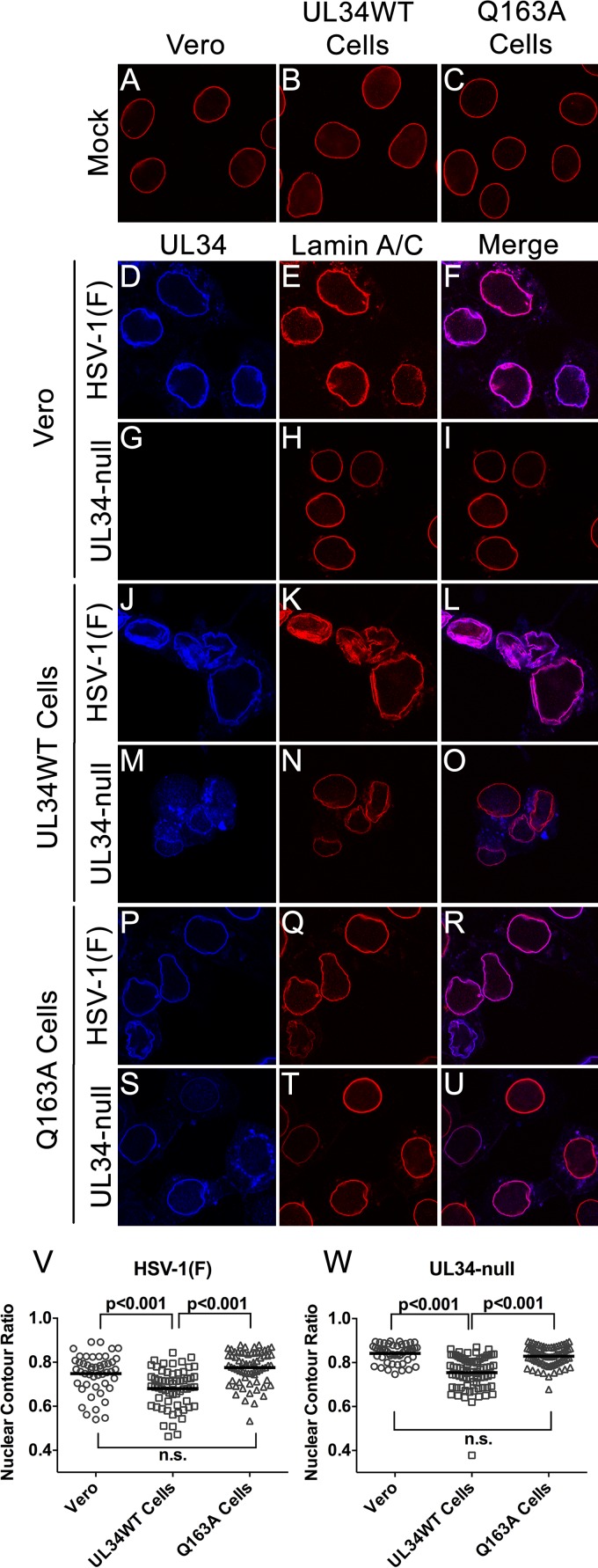
UL34(Q163A) is unable to alter nuclear shape and disrupt lamin A/C localization.
Disruption of the nuclear lamina is thought to be essential for capsid access to the INM during nuclear egress, and this disruption is correlated with alterations of nuclear shape and the distribution of lamin proteins. The regular contour of the nuclear membrane seen in UL34-null-infected Q163A-expressing cells (Fig. 4) suggested the hypothesis that the nuclear egress defect seen might result from a failure to disrupt the nuclear lamina. To test this, confocal microscopy of Vero, UL34WT-expressing, and Q163A-expressing cells infected with HSV-1(F) or the UL34-null virus was performed (Fig. 6). In uninfected cells, lamin A/C (LMNA/C) localized uniformly to the nuclear periphery, creating a smooth outline of the nucleus (Fig. 6A to C). As expected for HSV-1(F) infection of Vero cells and UL34-null infection of UL34WT-expressing cells, LMNA/C was redistributed such that there were areas of aggregation and areas of thinning or complete absence of LMNA/C at the nuclear periphery (Fig. 6). HSV-1(F) infection of UL34WT-expresssing cells caused increased LMNA/C aggregation and nuclear convolution (Fig. 6), most likely due to increased pUL34 from expression from both the viral and cellular genomes. HSV-1(F)-infected Q163A-expressing cells similarly caused a redistribution of LMNA/C and nuclear convolutions, indicating that pUL34(Q163A) does not have a dominant negative effect on the lamina disruption activity of pUL34WT (Fig. 6P to R). UL34-null-infected Vero cells maintained a uniform localization of LMNA/C at the nuclear periphery and an ovoid nuclear shape, showing the inability of pUL34(Q163A) to disrupt the LMNA/C network (Fig. 6G to I). UL34-null-infected Q163A-expressing cells were also unable to cause observable redistribution of this network, and they maintained a phenotype similar to that of uninfected cells (Fig. 6S to U).
FIG 6.
UL34(Q163A) causes an inability to disrupt the nuclear lamina. (A to U) Representative confocal images of UL34 and lamin A/C in infected and uninfected cells. (A to C) Uninfected Vero cells, UL34WT-expressing cells, and Q163A-expressing cells were fixed and stained for lamin A/C. Vero cells (D to I), UL34WT-expressing cells (J to O), or Q163A-expressing cells (P to U) were infected with HSV-1(F) (D to F, J to L, and P to R) or the UL34-null virus (G to I, M to O, and S to U). Cells were fixed at 16 hpi and stained for pUL34 or lamin A/C. (V and W) Nuclear contour ratios for cells infected with HSV-1(F) (V) or the UL34-null virus (W).
To provide a quantitative analysis of nuclear morphology, the nuclear contours of cross-sectional images of nuclei were calculated (Fig. 6V and W). A nuclear contour ratio of 1 indicates a perfect circle, and the score decreases with an increasingly convoluted nuclear shape. HSV-1(F)-infected Vero and Q163A-expressing cells produced similar ranges of contour ratio scores, whereas UL34WT-infected cells produced significantly lower scores (Fig. 6V). This correlates with the confocal microscopy data showing increased wrinkling of nuclei when viral and cellular expression data for pUL34 were combined. UL34-null-infected UL34WT-expressing cells produced lower scores than infected Vero cells, showing that pUL34WT expression is required for a change in nuclear morphology (Fig. 6W). UL34-null-infected Q163A-expressing cells produced scores that were not significantly different from those of infected Vero cells (Fig. 6W), showing that pUL34(Q163A) is unable to mediate changes in nuclear shape and LMNA/C distribution. This supports the hypothesis that the nuclear egress defect of pUL34(Q163A) is caused by the inability to disrupt LMNA/C networks.
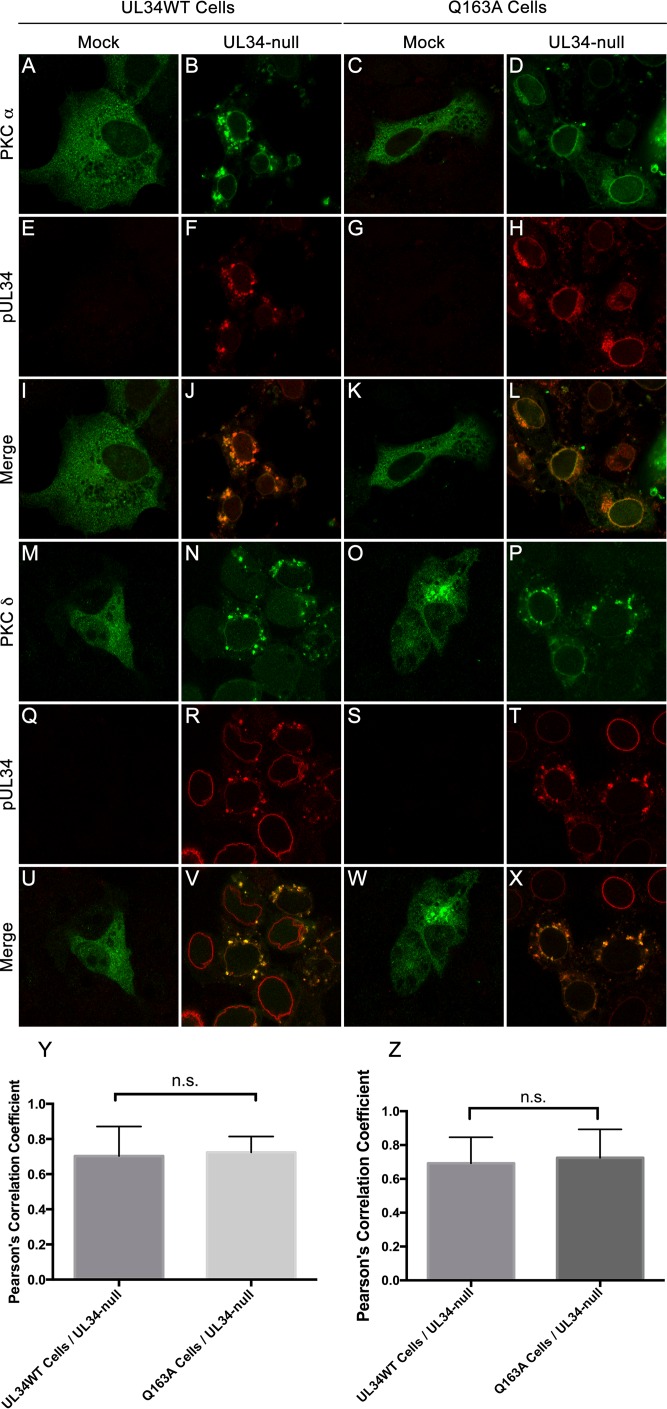
The inability to alter the lamin A/C distribution is not due to an absence of recruitment of cellular kinases or pUS3.
Protein kinases, including the cellular kinases PKCα and PKCδ and the viral protein pUS3, are recruited to the nuclear periphery and are thought to play a role in nuclear egress by mediating lamina disruption. To determine if the inability of UL34(Q163A) to disrupt the LMNA/C network was due to the inability to recruit PKC isoforms to the nuclear periphery, UL34WT-expressing and Q163A-expressing cells were transfected with either PKCα or PKCδ and then infected with the UL34-null virus (Fig. 7). Uninfected UL34WT-expressing and Q163A-expressing cells showed diffuse cytoplasmic localization of both PKCα and PKCδ (Fig. 7A, C, M, and O). As expected, PKCα and PKCδ were recruited to the nuclear periphery and colocalized with pUL34 in UL34-null-infected UL34WT-expressing cells (Fig. 7B, F, J, N, R, and V). UL34-null-infected Q163A-expressing cells maintained the ability to recruit PKCα and PKCδ to sites where pUL34(Q163A) was present (Fig. 7D, H, L, P, T, and X). Pearson's correlation coefficients were calculated to determine the amount of colocalization between pUL34 or pUL34(Q163A) and PKCα or PKCδ. Scores for UL34-null-infected UL34WT-expressing and Q163A-expressing cells were not significantly different from each other and were both above 0.6, indicating colocalization of the two proteins.
FIG 7.
PKCα and PKCδ are recruited to sites containing UL34(Q163A). (A to L) Confocal images of PKCα and pUL34 in infected and uninfected cells. UL34WT-expressing cells (A, B, E, F, I, and J) and Q163A-expressing cells (C, D, G, H, K, and L) were transfected with an HA-PKCα overexpression construct and subsequently infected with no virus (A, C, E, G, I, and K) or the UL34-null virus (B, D, F, H, J, and L). Cells were fixed at 16 hpi and stained for HA-PKCα (A to D) or pUL34 (E to H). (M to X) Representative confocal images of PKCδ and pUL34 in infected and uninfected cells. UL34WT-expressing cells (M, N, Q, R, U, and V) and Q163A-expressing cells (O, P, S, T, W, and X) were transfected with MYC-tagged PKCδ and subsequently infected with no virus (M, O, Q, S, U, and W) or the UL34-null virus (N, P, R, T, V, and X). (Y) Colocalization of HA-PKCα and pUL34 was determined by calculating Pearson's correlation coefficient. Statistical significance was determined by one-way analysis of variance (ANOVA). (Z) Colocalization of PKCδ and UL34 was determined by calculating Pearson's correlation coefficient. Statistical significance was determined by one-way ANOVA. n.s., not significant.
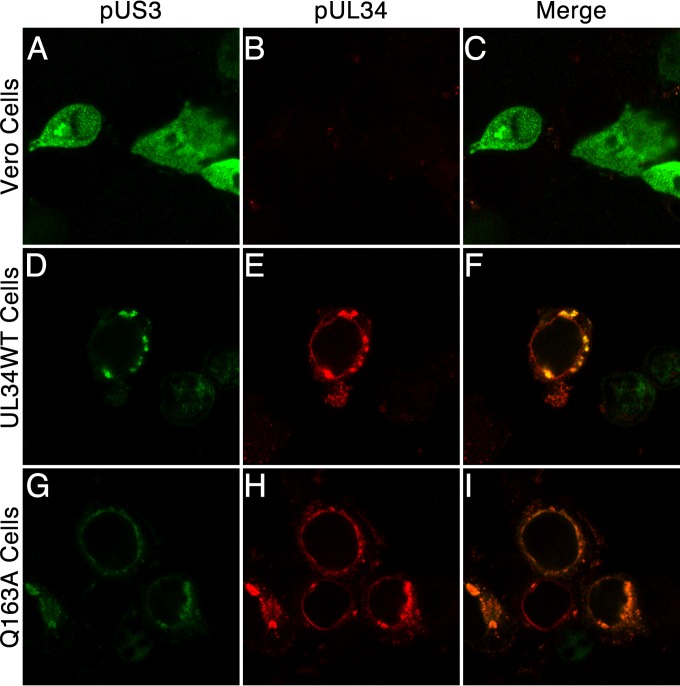
To determine whether recruitment of pUS3 to the nuclear periphery was impaired by the Q163A mutation, cells expressing no pUL34, WT pUL34, or pUL34(Q163A) were infected with the UL34-null virus, and the localization of pUL34 and pUS3 was determined by immunofluorescence 14 h after infection (Fig. 8). In Vero cells infected with the UL34-null virus, pUS3 was diffusely distributed in the cytoplasm and the nucleus, whereas in cells that expressed either pUL34WT or pUL34(Q163A), infection resulted in recruitment of pUS3 to sites of pUL34 localization.
FIG 8.
pUS3 is recruited to sites containing UL34(Q163A). Confocal images show Vero cells (A to C), UL34WT-expressing cells (D to F), or Q163A-expressing cells (G to I) infected with 5 PFU/cell UL34-null virus for 14 h and stained using polyclonal antibodies directed against pUS3 (green) and pUL34 (red).
The inability to alter the lamin A/C distribution is not due to an inability to phosphorylate emerin.
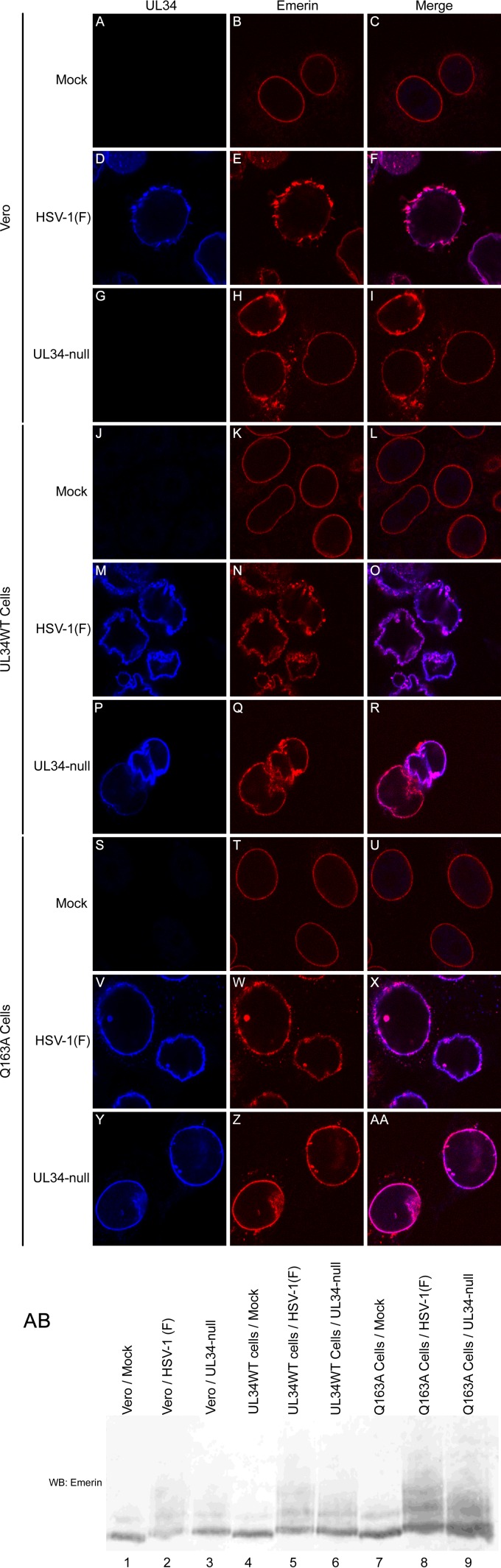
Disruption of the nuclear lamina is thought to involve a loss of interaction between individual lamin subunits and between lamin proteins and integral membrane lamin-associated proteins (LAPs) that anchor the lamin network to the membrane. pUL34 and pUS3 have previously been shown to play a role in hyperphosphorylation of one of these LAPS, emerin, in HSV-1 infection (5). To determine the effect of UL34(Q163A) on the localization of emerin, Vero, UL34WT-expressing, and Q163A-expressing cells were either left uninfected or infected with HSV-1(F) or the UL34-null virus, and confocal microscopy was performed (Fig. 9). In uninfected cells, emerin localized to the nuclear periphery, creating a smooth and uniform stain around the nucleus (Fig. 9A to C, J to L, and S to U). In HSV-1(F)-infected Vero, UL34WT-expressing, and Q163A-expressing cells, emerin localization became punctate at the nuclear periphery and colocalized with pUL34 (Fig. 9D to F, M to O, and V to X). This is thought to be due to hyperphosphorylated emerin losing its interaction with lamin subunits at the INM but still being retained at the nuclear membrane through interaction with the INM integral protein pUL34 (5). In UL34-null-infected Vero cells, a portion of emerin localized to vesicular membranes in the cytoplasm (Fig. 9G to I). Hyperphosphorylated emerin is unable to maintain connection to the nuclear lamina, and with pUL34 absent, it was unable to be retained at the nuclear membrane. Emerin maintained a somewhat punctate localization at the nuclear periphery, with little emerin located on cytoplasmic membranes, in UL34-null-infected Q163A-expressing cells (Fig. 9Y to AA). Emerin located at the nuclear periphery colocalized with pUL34(Q163A). This suggests that pUL34(Q163A) is able to interact with emerin, but with a lower affinity than that of pUL34WT.
FIG 9.
Emerin localization. (A to AA) Confocal images of pUL34 (left column) and emerin (center column) in infected and uninfected cells. Vero (A to I), UL34WT-expressing (J to R), and Q163A-expressing (S to AA) cells were infected with no virus (A to C, J to L, and S to U), HSV-1(F) (D to F, M to O, and V to X), or the UL34-null virus (G to I, P to R, and Y to AA). Cells were fixed at 16 hpi and stained for pUL34 or emerin. (AB) Western blot of Vero (lanes 1 to 3), UL34WT-expressing (lanes 4 to 7), and Q163A-expressing (lanes 7 to 9) cells. Cells were left uninfected (lanes 1, 4, and 7) or infected with HSV-1(F) (lanes 2, 5, and 8) or the UL34-null virus (lanes 3, 6, and 9). Cell lysates were collected at 15 hpi, and Western blotting was performed as described in Materials and Methods, with probing for emerin.
To determine if pUL34(Q163A) is able to interfere with the phosphorylation of emerin, Western blotting of cell lysates from HSV-1(F)- or UL34-null-infected cells was performed (Fig. 9AB). Hyperphosphorylation of emerin retards the movement of the protein during SDS-PAGE, causing an upward shift of emerin moieties. HSV-1(F) infection of all three cell lines resulted in an upward mobility shift of the lowest emerin band and the occurrence of multiple bands of higher molecular weights. For UL34-null-infected Vero cells, a mobility shift of the bottom band occurred, but fewer bands of higher molecular weights were present. In the absence of pUL34, emerin is phosphorylated at lower levels by pUS3. In UL34-null-infected Q163A-expressing cells, there was no difference in the mobility shift pattern compared to that for the HSV-1(F)-infected samples, indicating that pUL34(Q163A) maintains the ability to cause hyperphosphorylation of emerin. Together, these data suggest that the nuclear egress defect of UL34(Q163A) is not due to an inability to recruit PKCα and PKCδ to the nuclear periphery or an inability to phosphorylate emerin.
An extragenic suppressor of UL34(Q163A) maps to UL31.
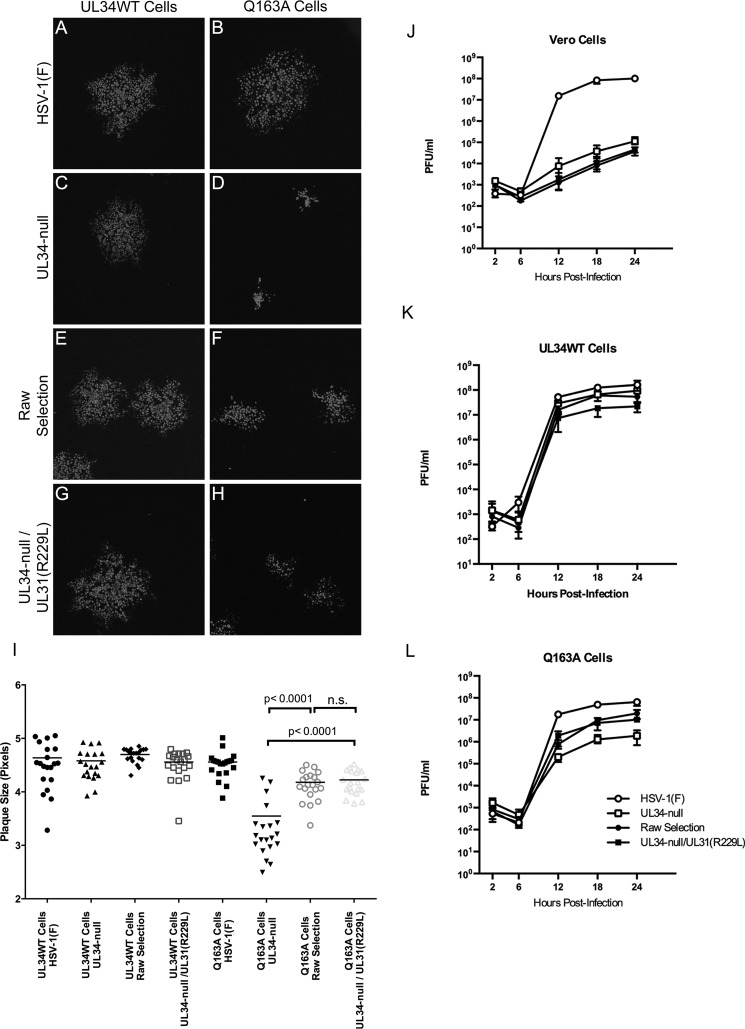
The transcomplementation system used for characterization of UL34(Q163A) is well suited for selection of extragenic suppression of point mutants, as there is no opportunity for intragenic selection or WT reversion. Twenty putative suppressor viruses were isolated from selections performed as described in Materials and Methods and were sequenced in the UL31 region. All 20 isolated viruses contained a mutation at amino acid position 229, from an arginine to a lysine [UL31(R229L)]. The R229L mutation is located in CR3 of the protein, outside but near the pUL31-pUL34 interaction interface (Fig. 1B and C). To test whether the suppressor phenotype observed for the variants was due to the R229L mutation, the spread and single-step growth activities of one of these variants (termed the “raw selection” virus) and a recombinant UL34-null/UL31(R229L) virus were compared.
In spread assays, the raw selection and recombinant UL34-null/UL31(R229L) viruses produced plaques on Q163A-expressing cells that were similar in size to each other and significantly larger than those produced by the UL34-null virus (Fig. 10D, F, and H), showing that the R229L mutation is sufficient to confer the suppressor phenotype for virus spread. The plaques formed by the R229L viruses on cells expressing pUL34(Q163A) were, however, significantly smaller than those formed on cells expressing WT pUL34 (Fig. 10E to H), indicating that phenotypic suppression was incomplete.
FIG 10.
UL31(R229L) partially rescues the virus spread and virus production defects of UL34(Q163A). (A to H) Representative images of plaques produced by HSV-1(F) (A and B), UL34-null (C and D), the raw selection virus (E and F), or the UL34-null/UL31(R229L) recombinant virus (G and H) on UL34WT-expressing cells (left column) and Q163A-expressing cells (right column). (I) Plaque size assay under the above-mentioned conditions. Each data point represents a single plaque. Twenty plaques were measured for each condition. Statistical significance was determined by performing the Kolmogorov-Smirnov test. (J to L) Single-step growth analysis of HSV-1(F), UL34-null, the raw selection virus, and the UL34-null/UL31(R229L) virus on Vero cells (J), UL34WT-expressing cells (K), or Q163A-expressing cells (L). Each data point represents the mean for three independent experiments. Error bars represent ranges. Statistical significance was determined by performing Student's t test for each condition and time point.
In single-step growth assays (Fig. 10J to L), both the raw selection and recombinant UL34-null/UL31(R229L) viruses grew roughly 10-fold better than the UL34-null virus on Q163A-expressing cells. The R229L mutant viruses were not significantly different from the UL34-null virus in growth on Vero or UL34WT-expressing cells, indicating that the R229L mutation specifically suppresses the UL34(Q163A) phenotype. These mutant viruses produced similar amounts of virus, indicating that the R229L mutation is solely responsible for the suppressive phenotype. While the R229L viruses did not produce as much virus as HSV-1(F) on Q163A-expressing cells, virus production was comparable to that seen on cells expressing WT pUL34, suggesting that the UL31(R229L) mutation largely suppresses the virus production defect of the UL34(Q163A) mutant.
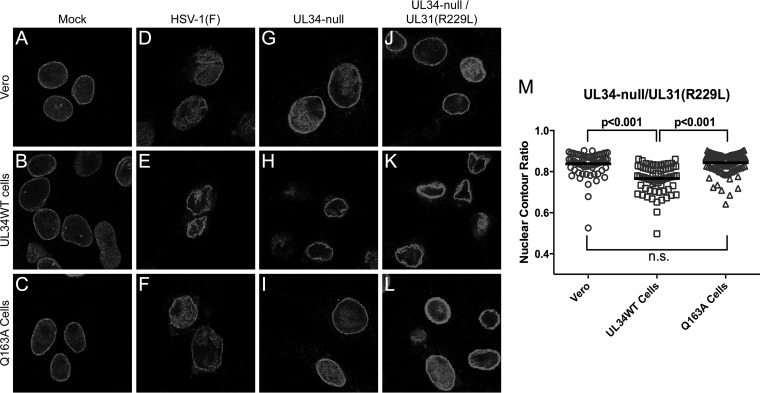
UL31(R229L) does not regain the ability to alter nuclear shape or displace lamins.
Since the R229L mutation was able to increase viral spread and growth, we expected that the mutant would regain the ability to alter nuclear morphology and disrupt the LMNA/C network. To test this, confocal microscopy was performed on Vero, UL34WT-expressing, and Q163A-expressing cells infected with HSV-1(F), UL34-null, or UL34-null/UL31(R229L) (Fig. 11). As expected, HSV-1(F)-infected cells and UL34-null- and UL34-null/UL31(R229L)-infected UL34WT-expressing cells caused aggregation of LMNA/C and changes to nuclear morphometry (Fig. 11D to F, H, and K). Surprisingly, UL34-null/UL31(R229L)-infected Q163A-expressing cells did not show an altered nuclear shape, as indicated by the nuclear contour ratio (Fig. 11M), or produce a change in LMNA/C distribution at the nuclear periphery (Fig. 11L), indicating no observable redistribution of the LMNA/C network.
FIG 11.
UL31(R229L) does not regain the ability to disrupt the nuclear lamina. (A to L) Representative confocal images of lamin A/C in uninfected and infected cells. Vero cells (top row), UL34WT-expressing cells (middle row), and Q163A-expressing cells (bottom row) were either left uninfected (A to C) or infected with HSV-1(F) (D to F), the UL34-null virus (G to I), or the UL34-null/UL31(R229L) virus (J to L). Cells were fixed at 18 hpi and stained for lamin A/C. (M) Nuclear contour ratios for UL34-null/UL31(R229L)-infected cells. One hundred nuclei were used to calculate the nuclear contour ratio for each condition. Statistical significance was determined by using Student's t test.
DISCUSSION
In this study, we characterized a mutation in UL34 that causes viral growth and spread defects. These defects were not due to the inability of UL34(Q163A) to properly localize to the nuclear periphery or due to a loss of interaction with UL31. TEM analysis showed retention of viral capsids in the nucleus, indicating a nuclear egress deficiency. Immunofluorescence and nuclear contour analyses showed an inability of UL34(Q163A) to cause the changes in nuclear shape and redistribution of LMNA/C normally seen in wild-type infection. This mutant was, however, able to recruit cellular kinases and to cause hyperphosphorylation of emerin. We originally hypothesized that the nuclear egress and growth defects of UL34(Q163A) were due to the inability to cause displacement of lamins and convolution of the nuclear membrane; however, the extragenic suppressor UL31(R229L) was able to substantially recover the replication and plaque formation defects without restoring disruptive effects on the nuclear lamina. This strongly suggests that redistribution of LMNA/C and accompanying changes in nuclear shape are not essential for relatively efficient nuclear egress.
The Q163 residue in pUL34 is located at the UL31/UL34 heterodimer interaction interface and is buried within the heterodimer structure (Fig. 1). Mutation of Q163 to alanine would be expected to destabilize interaction between these two proteins. Our results show undiminished interaction between UL31 and UL34(Q163A), demonstrating that while the UL34(Q163A) mutation may reduce the affinity of interaction between pUL31 and pUL34, this reduction is insufficient to prevent or greatly reduce heterodimer formation. Nonetheless, a complex containing the Q163A mutation is unable to mediate LMNA/C network disruption, changes in nuclear morphology, or nuclear egress of viral capsids, further suggesting that this mutation causes some significant change in the conformation of the NEC that renders it unable to orchestrate nuclear egress.
Disruption of the nuclear lamina during herpesvirus infection is thought to result from recruitment and activation of viral and cellular protein kinases at the nuclear envelope, where they phosphorylate lamin subunits and lamina-associated proteins, such as emerin, causing dissociation of lamin subunits from the network and from the LAPs that anchor the network to the INM. Recruitment of the PKC α, β, and δ isoforms to the nuclear membrane by the NECs of alpha- and betaherpesviruses has been reported (12, 13, 24), although the significance of these kinases for lamina disruption and nuclear egress in infected cells is not yet defined. In HSV-infected cells, for example, inhibition of PKCα and PKCβ does not appreciably affect lamin distribution, emerin hyperphosphorylation, or viral replication. The PKCδ inhibitor rottlerin inhibits emerin hyperphosphorylation and nuclear egress, but rottlerin's inhibitory activity affects an unknown spectrum of other kinases (5, 13). Virus-encoded kinases, on the other hand, have clearly demonstrated significance for phosphorylation of lamin subunits in alpha-, beta-, and gammaherpesviruses (3, 5, 25–27). However, in HSV, deletion of US3 does not prevent lamina disruption and has a relatively small effect on virus replication. The UL34(Q163A) mutant is not impaired in the ability to recruit pUS3 in infected cells or PKC isoforms in transfected cells to itself, and consistent with this, it is also able to mediate normal hyperphosphorylation of emerin in infected cells. This further suggests that failure to mediate LMNA/C redistribution results from an inability either to recruit an as yet unidentified kinase or to complete some lamina disruption function not related to kinase recruitment. It also suggests that disruption of the lamina in infected cells may occur as two separable events: dissociation and displacement of lamin subunits and disconnection of the lamin network from the LAPs that connect it to the INM. Our data suggest the possibility that only the first of these events is impaired by the UL34(Q163A) mutation, whereas the second is not.
Disconnection of the lamin filament network from the underlying INM might allow for relatively efficient capsid budding even without disruption of the filament network. Lamin subunits are continually being redistributed within the nucleus (28, 29). This addition and subtraction of lamin subunits may result in temporary perforations in the nuclear lamina meshwork that are large enough to accommodate the passage of viral capsids. An HSV-1 virion of approximately 125 nm in diameter would require a membrane area of roughly 400 nm2 for envelopment (calculated as the area of a sphere with a 125-nm diameter). Recent studies have shown that there is a large range in size of areas of the lamin networks in mammalian cells that are devoid of lamins, termed faces (28). These faces can be as large as 500 nm2, and combined with decreased anchorage of the lamin network to the INM by the hyperphosphorylation and redistribution of emerin (and perhaps other LAPs), this may allow for both capsid access to the INM and flexibility of an area of the INM sufficient for curvature around the capsid.
Although we have shown through immunofluorescence and nuclear contour analyses that UL34(Q163A) does not cause overall LMNA/C reorganization during infection, lamin displacement may still occur at levels undetectable by the methods used in this study. A recent study showed through superresolution microscopy that lamins A, C, B1, and B2 create distinct yet connected network structures (30). These individual networks have similar properties and are most likely influenced by the presence of other lamin subunits. Although we have shown that UL34(Q163A) does not cause reorganization of LMNA/C networks, there may be a disruption of LMNB networks that might increase the size of lamin-free faces and thereby facilitate passage of viral capsids to the cytoplasm. However, this is not considered likely, as disruptions of one subunit's network have been shown to influence the organization of other subunit networks, resulting in gross changes in nuclear shape (30). Since changes in nuclear morphology were not observed in any infections containing UL34(Q163A), the disruption of LMNB networks with the maintenance of LMNA/C networks is not expected.
The UL31(R229L) mutant was previously isolated in a screen for extragenic suppressors of a charge cluster mutation (CL04) of UL34 defective in completing membrane curvature of the INM during nuclear egress (16). The R229L mutation is located outside the UL31/UL34 interaction interface (Fig. 1), so it is not thought to directly stabilize the interaction between these two proteins. The R229L mutation may, however, stabilize UL31/UL34 heterodimer interaction to allow the complex to regain the ability to perform membrane curvature of the INM.
Due to the ability of UL31(R229L) to rescue membrane curvature defects and its ability to only partially recover the viral growth and spread defects of UL34(Q163A), it is possible that UL34(Q163A) causes two nuclear egress defects that contribute to its diminished replication. We hypothesize that the UL31(R229L) mutant is able to suppress a membrane curvature defect caused by UL34(Q163A) and that this defect is masked by the inability of UL34(Q163A) to disrupt the lamin network, as this step precedes lamina disruption in the nuclear egress pathway. Under this hypothesis, the membrane curvature defect would contribute to a 10-fold reduction in viral growth and about a 1.6-fold reduction in viral spread, and the inability to cause dispersal of lamins and consequent nuclear shape changes would result in only a 5-fold reduction in viral growth and about a 1.5-fold reduction in viral spread. It therefore seems likely that HSV-1-mediated dispersal of lamins is not essential for viral replication and that hindering it results in only a nominal reduction in overall viral production.
ACKNOWLEDGMENTS
We thank Alison Haugo, Erika Feutz, and Michael Feiss for critical readings of the manuscript and Keith Jarosinski for invaluable discussions.
This work was funded by a University of Iowa Department of Microbiology development grant and by NIH NIAID grants AI41478 and AI099597. A.V. was supported by NIAID grant T32 AI007533.
REFERENCES
- 1.Mettenleiter TC, Muller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol 15:170–178. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 3.Bjerke SL, Roller RJ. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J Virol 79:12840–12851. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leach N, Bjerke SL, Christensen DK, Bouchard JM, Mou F, Park R, Baines J, Haraguchi T, Roller RJ. 2007. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J Virol 81:10792–10803. doi: 10.1128/JVI.00196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris JB, Hofemeister H, O'Hare P. 2007. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J Virol 81:4429–4437. doi: 10.1128/JVI.02354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott ES, O'Hare P. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J Virol 75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds AE, Liang L, Baines JD. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J Virol 78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mou F, Forest T, Baines JD. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J Virol 81:6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marschall M, Marzi A, aus dem Siepen P, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J Biol Chem 280:33357–33367. doi: 10.1074/jbc.M502672200. [DOI] [PubMed] [Google Scholar]
- 11.Cano-Monreal GL, Wylie KM, Cao F, Tavis JE, Morrison LA. 2009. Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology 392:137–147. doi: 10.1016/j.virol.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park R, Baines JD. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J Virol 80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leach NR, Roller RJ. 2010. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology 406:127–137. doi: 10.1016/j.virol.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Yang Y, Wu S, Pan S, Zhou C, Ma Y, Ru Y, Dong S, He B, Zhang C, Cao Y. 2014. p32 is a novel target for viral protein ICP34.5 of herpes simplex virus type 1 and facilitates viral nuclear egress. J Biol Chem 289:35795–35805. doi: 10.1074/jbc.M114.603845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Kato A, Oyama M, Kozuka-Hata H, Arii J, Kawaguchi Y. 2015. Role of host cell p32 in herpes simplex virus 1 de-envelopment during viral nuclear egress. J Virol 89:8982–8998. doi: 10.1128/JVI.01220-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roller RJ, Bjerke SL, Haugo AC, Hanson S. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J Virol 84:3921–3934. doi: 10.1128/JVI.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J Virol 77:7601–7610. doi: 10.1128/JVI.77.13.7601-7610.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol 74:117–129. doi: 10.1128/JVI.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smeulders AW, Dorst L. 1990. An introduction to image processing, p 77–104. In Baak JPA. (ed), Manual of quantitative pathology in cancer diagnosis and prognosis. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 21.Haugo AC, Szpara ML, Parsons L, Enquist LW, Roller RJ. 2011. Herpes simplex virus 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J Virol 85:7203–7215. doi: 10.1128/JVI.00262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnee M, Ruzsics Z, Bubeck A, Koszinowski UH. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J Virol 80:11658–11666. doi: 10.1128/JVI.01662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lotzerich M, Ruzsics Z, Koszinowski UH. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J Virol 80:73–84. doi: 10.1128/JVI.80.1.73-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 25.Krosky PM, Baek MC, Coen DM. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J Virol 77:905–914. doi: 10.1128/JVI.77.2.905-914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CP, Huang YH, Lin SF, Chang Y, Chang YH, Takada K, Chen MR. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J Virol 82:11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mou F, Wills EG, Park R, Baines JD. 2008. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J Virol 82:8094–8104. doi: 10.1128/JVI.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moir RD, Yoon M, Khuon S, Goldman RD. 2000. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J Cell Biol 151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerace L, Comeau C, Benson M. 1984. Organization and modulation of nuclear lamina structure. J Cell Sci Suppl 1:137–160. [DOI] [PubMed] [Google Scholar]
- 30.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. 2015. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol Biol Cell 26:4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]