ABSTRACT
One of the goals of HIV-1 vaccine development is the elicitation of neutralizing antibodies against vulnerable regions on the envelope glycoprotein (Env) viral spike. Broadly neutralizing antibodies targeting the Env glycan-V3 region (also called the N332 glycan supersite) have been described previously, with several single lineages each derived from different individual donors. We used a high-throughput B-cell culture method to isolate neutralizing antibodies from an HIV-1-infected donor with high serum neutralization breadth. Clonal relatives from three distinct antibody lineages were isolated. Each of these antibody lineages displayed modest breadth and potency but shared several characteristics with the well-characterized glycan-V3 antibodies, including dependence on glycans N332 and N301, VH4 family gene utilization, a heavy chain complementarity-determining region 2 (CDRH2) insertion, and a longer-than-average CDRH3. In contrast to previously described glycan-V3 antibodies, these antibodies preferentially recognized the native Env trimer compared to monomeric gp120. These data indicate the diversity of antibody specificities that target the glycan-V3 site. The quaternary binding preference of these antibodies suggests that that their elicitation likely requires the presentation of a native-like trimeric Env immunogen.
IMPORTANCE Broadly neutralizing antibodies targeting the HIV-1 glycan-V3 region with single lineages from individual donors have been described previously. Here we describe three lineages from a single donor, each of which targets glycan-V3. Unlike previously described glycan-V3 antibodies, these mature antibodies bind preferentially to the native Env trimer and weakly to the gp120 monomer. These data extend our knowledge of the immune response recognition of the N332 supersite region and suggest that the mode of epitope recognition is more complex than previously anticipated.
INTRODUCTION
During the course of natural human immunodeficiency virus type 1 (HIV-1) infection, most individuals generate neutralizing antibodies (NAbs) to the infecting viral strain, but viral evolution and escape allow persistent viral replication despite this antibody (Ab) response (1–3). As HIV-1 infection progresses over months to years, the neutralizing antibody response often broadens, and it is estimated that sera from about 50% of HIV-1-infected donors can neutralize about 50% of diverse HIV-1 strains (4, 5). This observation leads to some optimism that a fuller understanding of the characteristics and evolution of such NAbs could provide insights for the design of vaccine immunogens and immunization strategies that would elicit such cross-reactive NAbs.
The most potent and broadly reactive neutralizing monoclonal antibodies (bNAbs) were isolated from highly selected donors whose sera had unusually potent or broad neutralizing activity. These antibodies form the basis for our current understanding of the major neutralization epitopes on the HIV-1 envelope glycoprotein (Env). Most of these antibodies can be grouped into five categories depending on the epitope that they target on the HIV-1 envelope (6, 7). Of particular interest for this study is a category of antibodies that target a high-mannose patch of gp120 centered around the glycan at position 332 (N332). These glycan-V3-directed antibodies, also called N332 glycan supersite-directed antibodies, are represented by three well-known clonal families, PGT121-123, PGT125-131, and PGT135-137 (8), and the recently described PCDN antibodies (9) that mediate potent neutralization of diverse HIV-1 strains. Passive administration of monoclonal antibody (MAb) PGT121 protects against simian-human immunodeficiency virus (SHIV) challenge (10) and suppresses SHIV replication when used alone (11) or in combination with other bNAbs (12). In infected humans, such antibodies appear early, at a high frequency, and contribute greatly to the serum neutralization activity (13, 14). These antibodies target the HIV envelope glycan shield, which the virus normally uses as a defense mechanism to evade immune recognition.
Structural studies of the glycan-V3 Abs revealed that they target an antigenic region of the HIV-1 envelope that is highly accessible and allows multiple binding modes with various angles of approach (9, 15–18). The breadth and potency of the PGT antibodies are associated with the N-glycan at residue N332 (8) that lies within a broad glycan-V3 supersite composed of six conserved core residues (N295, N301, N332, N339, N385, and N392) (9, 15–18). The epitope typically involves multiple glycosylated residues, including unprocessed high-mannose and complex-type glycans, as well as the underlying protein surface. Several studies have shown that such antibodies can alternate contacts between glycan residues to neutralize viruses that shift the glycan back and forth between residues 332 and 334 during immune escape (19, 20). Importantly, the N332 glycan is necessary but not sufficient for neutralization by this class of antibodies (19).
The PGT121, PGT128, and PGT135 families share some common features in their genetics. They are highly affinity matured, with ∼20% nucleotide mutation from heavy chain germ line genes (8). This frequency is substantially higher than the average mutation frequency for CD19+ B cells, which average 4% mutation from the germ line (21), or CD27+ IgD− switched memory B cells, which have an average 8.5% mutation frequency (22), but is much lower than the average 30% mutation frequency seen in VRC01 class CD4 binding site-directed antibodies (23, 24) and occasional memory B cells from healthy individuals. Not all of the mutations are required for potent antiviral activity: evolutionary studies of the PGT121 antibodies with 454 pyrosequencing demonstrate that the putative intermediates with 50% fewer mutations than the mature PGT antibodies are still capable of neutralizing 40 to 80% of the viruses (25). Insertions and deletions (indels), which are associated with increased germinal center activity, are present in some of these antibodies, but they are not required for neutralization breadth and potency (9). Unlike the VRC01 class antibodies, they do not show a strict requirement for a particular VH gene, although VH4 family genes are commonly used. The PCDN antibodies are similar but with a lower frequency of mutation from the germ line and no indels (9). Structural studies revealed the importance of a moderately long heavy chain complementarity-determining region 3 (CDRH3) loop for penetration of the glycan shield and binding to conserved gp120 peptide residues at the base of the V3 loop as well as contacting the glycans themselves (9, 15–18).
Thus, the frequency of glycan-V3 antibodies, their promiscuous mode of recognition, and the diverse VDJ repertoire suggest that they may be easier to elicit than other broadly neutralizing antibodies if an appropriately designed immunogen is available. A better understanding of the minimum features required for glycan-V3 antibody breadth and potency will provide important information for immunogen design.
To date, only a limited number of broadly neutralizing glycan-V3 antibodies have been identified, all from just four select individuals. It is not known how commonly the well-characterized features of PGT-like antibodies are found in the glycan-V3 response in other individuals. By using a high-throughput B-cell culture method (26), we isolated four antibody lineages from a single donor that partially recapitulated serum breadth and potency. Three of the lineages were glycan-V3 specific and shared multiple features with previously described glycan-V3 antibodies. These lineages have unique characteristics, including a preference for quaternary epitopes on the Env trimer, demonstrating the great variety of possible modes of recognition of the glycan-V3 supersite.
MATERIALS AND METHODS
Donor PBMCs.
Donor N170 is a participant in the NIAID protocol Evaluation of Viral Factors and Immune Parameters To Study HIV-Specific Immunity in the neutralizing antibody arm and gave informed consent (5, 27). The serum and peripheral blood samples used in this study were obtained in 2010. On the basis of the locations of current and former residences, donor N170 is presumed to be infected with a clade B virus and at the time of sampling had a viral load of 7,892 copies/ml and 664 CD4 cells/μl, was 45 years old, and had been chronically infected for at least 11 years without antiretroviral treatment.
Isolation of human B cells.
Frozen peripheral blood mononuclear cells (PBMCs) were thawed and stained with Live/Dead aqua blue (Invitrogen) to exclude dead cells and IgM-phycoerythrin (PE)-Cy5, CD3-allophycocyanin (APC)-Cy7, (BD Pharmingen), IgD-PE (Dako), and CD19-QD585 and CD16-PacBlue (VRC in-house conjugates) for negative selection of viable memory B cells on a FACSAria II instrument using FACSDiva software (BD Biosciences). IgM− IgD− B cells are considered to be class switched and therefore memory cells. Preparations were typically >95% pure.
Memory B-cell culture and immunoglobulin gene amplification and cloning.
As described previously (26), bulk-sorted memory B cells were aliquoted by limiting dilution to a density of 2 or 4 B cells/well into 384-well plates with a Biomek NXP instrument. B cells were cocultured in 55 μl/well Iscove's modified Dulbecco's medium (IMDM)–10% fetal calf serum (FCS)–1× Mycozap (Lonza) for 14 days with 1 × 105 irradiated NIH 3T3 fibroblasts/ml expressing murine CD40L, 100 U/ml interleukin-2 (IL-2) (Roche), and 50 ng/ml IL-21 (Invitrogen). After 14 days of culture, 40 μl of the culture supernatant was removed for microneutralization screening, and B cells were frozen in lysis buffer containing 0.25 μl of an RNase inhibitor (New England BioLabs), 0.3 μl of 1 M Tris (pH 8), and 19.45 μl diethyl pyrocarbonate (DEPC)-treated H2O.
The high-throughput NVITAL automated microneutralization qualitative assay was developed from the well-characterized TZM-bl HIV serum assay utilizing pseudotyped virus expressing the envelope antigen and the luciferase reporter gene (28). Neutralization activity is quantitated as the relative decrease in the luciferase activity compared to infection of TZM-bl cells in the absence of samples. Single wells of the culture supernatant were evaluated at a volume of 20 μl/well against each of two viruses. Twenty microliters of diluted pseudovirus was added to each well, and the mixture was incubated at 37°C for 45 to 90 min. Following incubation, TZM-bl cells were added at a total of 3 × 103 cells per well in 20 μl. The plates were incubated at 37°C with 5% CO2 for 48 to 54 h. Luciferase activity was quantitated with a Molecular Devices Paradigm luminometer following the addition of the Britelite Plus substrate (PerkinElmer). The relative percent neutralization of each sample was determined based upon the signal compared to that of the virus controls (signal without the addition of samples). Samples with a neutralization capacity greater than the selected thresholds (typically 50%) were further evaluated. For each set of culture plates, two pseudoviruses were used for the screening of supernatants.
B-cell culture supernatants were frozen until they were used for screening of the viruses most sensitive to neutralization by donor N170 serum. Control JRCSF viruses and SIV were utilized by Monogram Biosciences, San Francisco, CA, to screen 30-μl supernatant aliquots from 14 384-well plates. In a second experiment, BaL.01 and JRCSF were used by the NIAID NVITAL laboratory to screen 20-μl aliquots from 60 384-well plates. In a third experiment, Q23.17 and 769.h5 were used by NVITAL to screen 20-μl aliquots from 64 384-well plates. Wells were selected for reverse transcriptase PCR (RT-PCR) on the basis of a 50% reduction in neutralization of at least one test virus.
cDNA was synthesized from a 15-μl aliquot of the B-cell lysate from the neutralization-positive wells by the addition of 1 μl Superscript III RT (Life Technologies), 5 μl of 5× buffer, 1.25 μl 5 mM dithiothreitol (DTT), 0.38 μl of 150 ng of a random hexamer, 2 μl of 100 mM deoxynucleoside triphosphate (dNTP) mix, 0.5 μl of an RNase inhibitor, and 0.0625 μl of Igepal (Sigma). The thermocycle program used for reverse transcription was 42°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 94°C for 5 min. The cDNA was diluted in 25 μl distilled water (dH2O) before proceeding with heavy chain and light chain amplification as described previously (23). Briefly, the variable regions were amplified with 50 cycles of multiplex nested PCR, followed by amplification with gene-specific primers containing restriction sites for subcloning into the corresponding IgG1, kappa, and lambda vectors. Immunoglobulin sequences were analyzed with JoinSolver (http://joinsolver.niaid.nih.gov/) (29), IMGT/VQuest (http://www.imgt.org/IMGT_vquest/vquest?livret=0&Option=humanIg) (30), and HTJOINSOLVER (https://dcb.cit.nih.gov/HTJoinSolver/) (31). The heavy and light chains were expressed by cotransfection in 293 FreeStyle cells using Fugene (Promega) and 293fectin (Invitrogen). Antibodies were purified by using a recombinant protein A column (GE Healthcare) as described previously (32–34).
Env-pseudotyped virus neutralization and luciferase reporter cell assays.
To assess serum and isolated antibodies, neutralization was determined by measuring the reduction in the luminescence of TZM-bl cells infected with HIV-1 Env-pseudotyped viruses, limited to a single round of replication (32–34). A nonlinear-regression 5-parameter Hill slope equation was used to calculate the 50% or 80% inhibitory concentration (IC50 or IC80) in micrograms per milliliter for the MAbs and to calculate the 50% or 80% inhibitory dilution (ID50 or ID80) for serum. The neutralization breadth of donor N170 serum was determined by using a panel of 29 geographically and genetically diverse Env-pseudotyped viruses representing subtypes A, B, and C. Mutant viruses were generated by using a QuikChange mutagenesis kit (Invitrogen) or by GeneImmune, Inc. (New York, NY). Amino acid numbering is based on the sequence of HIV-1 HXB2. Point mutants were used for epitope mapping; mutants matched the wild type at all residues excluding the indicated amino acid, with the exception of JRCSF, as follows. Neutralization panels used a pseudovirus designated JRCSF.JB, made from an Env expression plasmid that was derived from a sequence matching the sequence reported under GenBank accession number AY426125. The JRCSF point mutants were made in an expression plasmid with a sequence matching that reported under GenBank accession number AY669726, which differs from the sequence of JRCSF.JB by a single point mutation, D167N.
Peptide inhibition of neutralization.
Peptide competition neutralization assays were done in the same assay format as the one used for the neutralization assay. Competition neutralization was determined by the addition of 25 μg/ml of a peptide competitor to a 5-fold serial dilution of the serum or MAb for 30 min before the addition of JRCSF.JB virus. The two V3 peptides used were a 23-mer clade B V3.01 peptide (TRPNNNTRKSIHIGPGRAFYTTG) and an irrelevant peptide that does not block V3 antibodies, V3.02 (YTTGEIIGDIRQAHC). The potent anti-V3 monoclonal antibody 447-52D was used as a positive control. The IC50 was determined, and fold inhibition was calculated by using the value for the mock peptide as a baseline. Competition was observed when the addition of the peptide resulted in a >3-fold decrease in the neutralizing capacity of the antibody.
Neutralization fingerprinting.
Neutralization fingerprints are the rank order of neutralization potencies for an antibody against a set of diverse viral strains, calculated as described previously (35). Composite vectors for bNAbs targeting the same epitope (e.g., CD4 binding site-directed bNAbs) are defined as epitope-specific signatures and are used for comparison with sera. The serum signal is deconvoluted to components that match the epitope-specific signatures.
Env soluble trimer capture enzyme-linked immunosorbent assay (ELISA).
For JRFL soluble trimers (JRFL E168K SOSIP gp140) (36), MaxiSorp plates (Thermo) were coated overnight at 4°C with 1 μg/ml of a mouse anti-His tag monoclonal antibody (R&D Systems) in phosphate-buffered saline (PBS) (pH 7.5). The next day, the plates were incubated at 4°C in blocking buffer (2% [wt/wt] milk powder plus 5% [vol/vol] fetal bovine serum in PBS [pH 7.5]) for 2 h and then washed three times in 0.05% Tween 20–PBS (pH 7.4). The Env soluble trimer, which bears a His tag, was added to the plates (100 μl per well) at a concentration of 2 μg/ml in blocking buffer, and the plates were incubated at 4°C for 1 h. The plates were washed three times in 0.05% Tween 20–PBS (pH 7.4). The primary antibodies (human HIV bNAbs) were added to the plates at a maximum concentration of 5 μg/ml and serially diluted 1:5 in blocking buffer. The plates were incubated at 4°C for 1 h and then washed three times in 0.05% Tween 20–PBS (pH 7.4). A secondary antibody (peroxidase-conjugated goat anti-human IgG antibody; Jackson ImmunoResearch Labs) diluted 5,000-fold in blocking buffer was added to the plates, and the plates were incubated at 4°C for 30 min. The plates were then washed three times in 0.05% Tween 20–PBS (pH 7.4). The substrate solution (TMB [3,3′,5,5-tetramethyl benzidine] chromogen solution; Invitrogen) was added to the plates (100 μl per well), and the plates were incubated at room temperature for 5 min. One hundred microliters per well of 1 N sulfuric acid was added to stop the reaction, and the plates were read at 450 nm.
For BG505.T332N SOSIP trimers (37), plates were coated with 2 μg/ml of a sheep anti-gp120 C5 antibody, D7324 capture antibody (Cliniqa Corp., Fallbrook, CA), in PBS overnight at 4°C. The plates were then blocked with 200 μl/well of 5% milk in PBS for 1 h at 37°C, followed by incubation with the BG505 SOSIP.664 trimer with a D7324 tag at 2 μg/ml in 10% PBS for 2 h at 37°C. Plates were further incubated with diluted test antibodies and then incubated with goat anti-human IgG conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Plates were washed 6 times between each step. Plates were incubated with 100 μl TMB (Kirkegaard & Perry Laboratories [KPL]) for 10 min. One hundred microliters per well of 1 N sulfuric acid was added to stop the reaction, and the plates were read at 450 nm.
The area under the curve (AUC) was calculated by using Prism software (GraphPad). To normalize data across assays, the AUC for the VRC01 run in the same assay was set to 100%, and the AUC for other antibodies was expressed as a percentage of the VRC01 value.
ELISA for gp120.
For BaL gp120 and BG505 gp120, Reacti-Bind plates (catalog number 15041; Pierce) were coated with 2 μg/ml of the probe diluted in PBS at 4°C overnight. Plates were blocked with 200 μl/well of B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine serum albumin [BSA], 0.07% Tween 20, 0.02% thimerosal) at 37°C for 1 h. Samples were incubated on plates at 37°C for 1 h, with one row serving as a blank that was loaded with B3T buffer containing no primary antibody. Goat anti-human Fcγ secondary antibody (catalog number 109-035-098; Jackson ImmunoResearch Laboratories) was diluted 1:10,000 and incubated on plates at 37°C for 1 h. The TMB substrate (SureBlue, catalog number 52-00-03; KPL, Gaithersburg, MD) was equilibrated to ambient temperature and incubated in wells for 10 min. The reaction was stopped with 1 N sulfuric acid, and plates were read at 450 nm. Plates were washed 6 times between each step, except for the substrate incubation step and the addition of 1 N sulfuric acid. All volumes were 100 μl/well except where otherwise noted.
For JRFL gp120, which bears a His tag, ELISA conditions were the same as those described above for JFRL soluble trimers.
Biolayer light interferometry (BLI) binding analysis and kinetics.
Kinetic measurements were obtained with an Octet Red instrument by immobilizing trimeric JRFL SOSIP and monomeric gp120 as ligands on HIS2 sensors (FortéBio). Fabs VRC29.03 and VRC22.01 were serially diluted (1:2) from 8 μM down to 125 nM in PBS to measure their binding affinities as analytes in solution. The JRFL-derived ligands were in contact with the Fab analytes for 5 min to measure on-rates and were allowed to dissociate for 10 min to measure off-rates. Data analysis was done by using FortéBio analysis software version 7.1 (FortéBio), and the kinetic parameters were calculated by using a 1:1 global fit model for applicable MAbs. VRC01 was run in parallel to verify that the proteins were intact.
Cell surface Env binding and cell surface competition assays.
Binding to cell surface-expressed Env was assessed as described previously (38, 39). HEK293T cells were transfected with BaL.01 gp160 dCT or JRFL gp160 E168K dCT by using Truefect Max (United Biosystems) and harvested 2 days later. Envelope-expressing cells were washed with sterile PBS before staining with Violet Live/Dead (Invitrogen). Serial dilutions of antibodies were added to the cells, and the cells were incubated for 30 min at room temperature. Anti-human IgG-phycoerythrin secondary antibody was added, followed by a 30-min incubation. Cells were fixed in 2% paraformaldehyde, and binding was analyzed by flow cytometry.
Binding of all competitors in a binding assay was demonstrated before use in a competition assay. Binding competitions were performed at least twice on cells expressing JRFL gp160 E168K dCT by adding titrating amounts of competitor monoclonal antibodies simultaneously with 8 μg/ml biotinylated VRC29.03. All competitors except VRC28.01 and VRC22.01 had at least the same level of binding, as demonstrated by this assay. Phycoerythrin-labeled streptavidin (Invitrogen) was added to measure binding. Cells were interrogated on an LSR II instrument (BD Biosciences), and the data were analyzed by using FlowJo software. Percent competition was calculated by taking the mean fluorescence intensity (MFI) value of the point with 100 μg/ml of the competitor and dividing it by the average MFI of the anti-influenza virus negative control.
Accession number(s).
Newly determined sequences were deposited in GenBank under accession numbers KX912935 to KX912950.
RESULTS
Serum characterization.
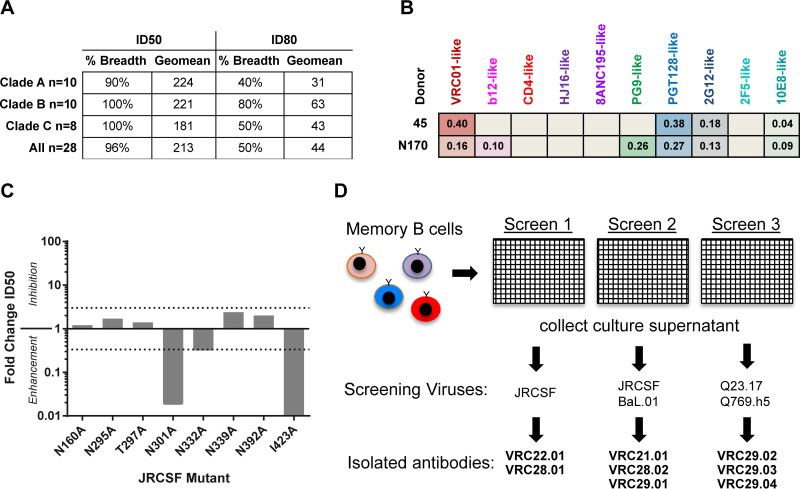
Sera from chronically HIV-1-infected donors in an NIAID, NIH, cohort were screened with a panel of genetically diverse strains of HIV-1 to identify donors with neutralization breadth and potency (5). Donor N170 serum was found to have potent and broad activity, neutralizing 27/28 isolates in clades A, B, and C with a geometric mean ID50 of 212 (Fig. 1A). To further characterize the polyclonal antibodies that were contributing to serum neutralization, we used a recently developed algorithm that utilizes the well-defined neutralization pattern of known broadly neutralizing antibodies (bNAbs) to generate a neutralization fingerprint for undefined sera (35). Using this algorithm, the neutralization fingerprint of donor N170 serum correlated best with those of the PG9-like antibodies (V1/V2 epitope) and the PGT128-like antibodies (glycan-V3 epitope), suggesting that donor N170 serum was a likely candidate for isolating similar glycan-reactive antibodies (Fig. 1B). While serum from donor 45, from whom VRC01 was isolated, displayed strong signals for VRC01- and PGT128-like epitopes, the fingerprint of donor N170 serum revealed a weaker dominance of any single antibody specificity but rather general glycan reactivity. To further characterize the serum, we tested the neutralization of a panel of Env-pseudotyped viruses with mutations in residues that contribute to the glycan-V3 supersite and the V1V2 site (8, 15) (Fig. 1C). Interestingly, none of the individual glycan mutants resulted in a >3-fold decrease in serum neutralization, and rather than inhibiting serum neutralization, the N301A glycan knockout mutant enhanced serum neutralization more than 10-fold. Of note, it is known that the removal of the N301 glycan can increase antibody accessibility to both the V3 loop region and the CD4 binding site (40, 41), and this can confound the interpretation of these mutant data. The nonglycan I423A mutant also enhanced neutralization. While the specific role for I423 in epitope specificity has not been defined, the I423A mutation may be playing a similar role of increasing epitope accessibility with the removal of a hydrophobic residue that is likely buried inside a protein core.
FIG 1.
Donor N170 serum mapping and B-cell culture. (A) Breadth and potency of serum from donor N170. Neutralization was assessed by single-round-of-infection pseudoviruses on TZMbl cells. Breadth is defined as the percentage of viruses neutralized at an ID50 of >40 or an ID80 of >40. Geomean, geometric mean. (B) Neutralization fingerprint analysis of serum from donor N170. Data for serum from donor 45, from whom antibody VRC01 was isolated, are shown for comparison. (C) Effect of mutations in JRCSF on neutralization by donor N170 serum. Neutralization activity is reported as fold changes in ID50 values relative to JRCSF using the equation ID50 of the wild type/ID50 of the mutant. Values of <1 indicate enhanced neutralization. Threefold differences are considered significant, indicated by dotted lines. (D) Schematic for isolation of monoclonal antibodies from donor N170. In three separate experiments, memory B cells were bulk sorted and cultured at 2 to 4 B cells/well for 14 days in 50 ml of culture medium containing IL-2 and IL-21 to stimulate differentiation and IgG secretion. The supernatant was removed for microneutralization screening by viruses, and B cells were frozen for subsequent RT-PCR amplification of immunoglobulin genes and subcloning to express the monoclonal antibodies.
Since serum mapping and neutralization fingerprinting failed to demonstrate a strong association with any one particular epitope specificity, we could not define an antigen-specific probe to isolate a particular category of antibodies by flow cytometry. Therefore, we chose to use high-throughput B-cell culture, in which peripheral B cells are isolated without using a sorting probe. B cells are cultured at limiting dilution, and supernatants from these cultures are then screened for virus neutralization. In contrast with screening by a binding assay, this functional assay avoids the bias that may be introduced by the choice of antigen. This approach allows the isolation of any antibodies that display neutralization activity, including those with quaternary epitopes (39, 42). We chose four viruses that displayed high sensitivity to serum neutralization (JRCSF, BaL.01, Q23, and Q769.h5) and used these viruses to screen the B-cell culture supernatants for neutralizing activity (Fig. 1D).
Isolation and genetics of naturally occurring neutralizing MAbs from donor N170.
Peripheral blood CD19+ IgM− IgD− memory B cells from donor N170 were isolated in three separate experiments using frozen PBMC samples collected during the same year of chronic infection (>10 years after HIV diagnosis). Memory B cells were cultured at a low density (2 to 4 B cells per well) to facilitate the isolation of naturally occurring heavy and light chain immunoglobulin pairs by PCR. After 14 days of in vitro B-cell stimulation, expansion, and differentiation in the presence of CD40L, IL-21, and IL-2, the culture supernatants containing secreted IgG and IgA were collected and screened for virus neutralization (Fig. 1D). The antibodies that the B cells secreted were recovered by reverse transcription-PCR of the antibody variable region, subcloned, and reconstituted as IgG1 antibodies (26). After culturing over 83,000 B cells, we isolated 8 MAbs with neutralizing activity. The antibodies are named by the convention donor-lineage.clone. In the first experiment, the screening virus was JRCSF, and N170-VRC22.01 and N170-VRC28.01 were isolated. In the second experiment, BaL.01 and JRCSF were used to screen supernatants, and N170-VRC21.01, N170-VRC21.02, and N170-VRC29.01 were isolated. In the third experiment, Q23.17 and Q769.h5 were used, and N170-VRC29.02 to -04 were isolated.
Genetic analysis of the 8 antibodies revealed that they belonged to four distinct lineages and had VH somatic mutation levels of 7% to 19% (Tables 1 and 2). Three of the lineages utilized VH4 family genes and a variety of JH genes, generating rearrangements resulting in relatively long CDRH3s (19 to 26 amino acids [aa]). Of note, these two characteristics (VH4 and a long CDRH3) are found among the PGT121-123, PGT125-131, PGT135-137, and PCDN lineages (8, 9) (Tables 1 and 2). Furthermore, the VRC22 and VRC29 lineages had CDRH1 and/or CDRH2 indels, which are commonly found among HIV-1 antibodies (43), including many glycan-V3-reactive antibodies (8). The VRC29 insertion was surprisingly similar to the PGT128 and PGT135 families in the number of nucleotides inserted and the position of the insertion in CDR2 (Table 1). The light chain repertoire of the donor N170 MAbs was more diverse than the heavy chain repertoire and utilized Vκ1, Vκ2, Vκ3, and Vλ3 families. In further contrast to the heavy chains, most of the light chain CDRL3 lengths and somatic hypermutations were similar to the averages for healthy donor memory B cells.
TABLE 1.
Glycan-V3 antibody heavy chain genetic characteristicsa
| MAb | IGHV | IGHJ | CDRH3 length (aa) | CDRH3 amino acid sequence | V gene nt mutation frequency (%) | Indel(s) |
|---|---|---|---|---|---|---|
| N170-VRC21.01 | 3-30*03 | 4*02 | 12 | VRGDGENGGTDY | 42/288 (15) | |
| N170-VRC21.02 | 12 | VRGDGENGGTDY | 48/288 (17) | |||
| N170-VRC22.01 | 4-34*01 | 6*02 | 19 | ARAGGMSDWAYPYYYGMDV | 20/282 (7) | 3-nt CDR1 del; 15-nt CDR2 ins |
| N170-VRC28.01 | 4-34*03 | 4*02 | 26 | VRGHLSRDRLGQYAFWSGALIDYLDP | 54/284 (19) | |
| N170-VRC29.01 | 4-59*08 | 3*02 | 20 | ARHRYDRWSDSYDIRSPFDI | 35/285 (12) | 12-nt CDR2 ins |
| N170-VRC29.02 | 20 | ARHRYDVWSKGYDIRSPFDF | 45/285 (16) | 18-nt CDR2 ins | ||
| N170-VRC29.03 | 20 | ARHRYDVWSNGYDIRSPYDF | 44/285 (15) | 18-nt CDR2 ins | ||
| N170-VRC29.04 | 20 | ARHRYDPWSKGYDIRSPFDF | 46/285 (16) | 18-nt CDR2 ins | ||
| PGT121 | 4-59*01 | 6*03 | 24 | ARTLHGRRIYGIVAFNEWFTYFYMDV | 56/285 (20) | |
| PGT128 | 4-39*07 | 5*02 | 19 | ARFGGEVLRYTDWPKPAWVDL | 60/288 (21) | 18-nt CDR2 ins |
| PGT135 | 4-39*07 | 5*02 | 18 | ARHRHHDVFMLVPIAGWFDV | 56/288 (19) | 15-nt CDR1 ins |
| PCDN38A | 4-34*01 | 5*01 | 22 | ARGGRKICYDYWCGYVNNCFDT | n.a. (16) |
Indel, insertion or deletion; ins, insertion; del, deletion; nt, nucleotide; aa, amino acids; CDR, complementarity-determining region; n.a., data not published.
TABLE 2.
Glycan-V3 antibody light chain genetic characteristicsa
| MAb | IGLV | IGLJ | CDRH3 length (aa) | CDRL3 amino acid sequence | V gene nt mutation frequency (%) | Indel(s) |
|---|---|---|---|---|---|---|
| N170-VRC21.01 | λ6-57*01 | λ3*02 | 10 | QSYDSNYYWV | 24/273 (9) | |
| N170-VRC21.02 | 10 | QSYDDTYFWV | 26/273 (10) | |||
| N170-VRC22.01 | κ1-17*01 | κ1*01 | 9 | LQYNTFPRT | 16/264 (6) | |
| N170-VRC28.01 | κ2-28*01 | κ1*01 | 9 | MQGIDTPRT | 50/279 (18) | |
| N170-VRC29.01 | κ3-20*01 | κ1*01 | 9 | HQYDRSPRT | 15/267 (6) | |
| N170-VRC29.02 | 9 | QQFDFSPRT | 23/267 (9) | |||
| N170-VRC29.03 | 9 | QQYDFSPRT | 24/267 (9) | |||
| N170-VRC29.04 | 9 | EQYDVSPRT | 22/267 (8) | |||
| PGT121 | λ3-21*02 | λ3*02 | 12 | HIWDSRVPTKWV | 41/237 (17) | 21-nt FR1 del; 9-nt FR3 ins |
| PGT128 | λ3-8*01 | λ2 or 3 | 10 | GSLVGNWDVI | 20/270 (7) | 15-nt CDR1 del |
| PGT135 | λ3-15*01 | λ1*01 | 9 | QQYEEWPRT | 35/204 (17) | |
| PCDN38A | κ3-20*01 | κ1*01 | 8 | QQCGSSPT | n.a. (11) |
Indel, insertion or deletion; ins, insertion; del, deletion; nt, nucleotide; aa, amino acids; FR, framework region; CDR, complementarity-determining region; n.a., data not published.
Monoclonal antibody neutralization.
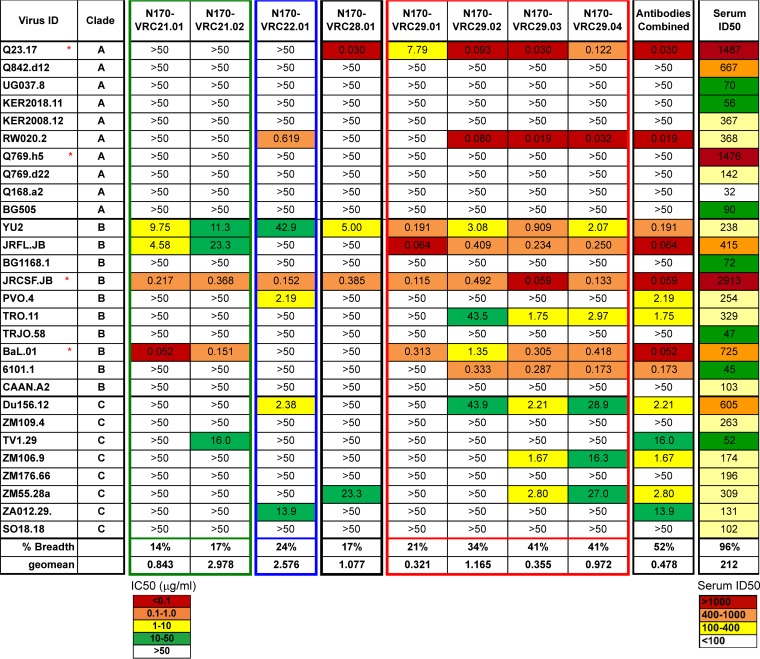
To determine the breadth and potency of the individual donor N170 MAbs, we tested their ability to neutralize a panel of 29 viral strains, including strains of clades A, B, and C. While donor N170 serum neutralized 27/28 viral strains, the four lineages in total recapitulated the neutralization of 15/28 viral strains, half of the serum breadth (Fig. 2). This suggests that there are other neutralizing antibodies in donor N170 serum that are yet to be isolated. Surprisingly, VRC28, which has the longest CDRH3 and the highest mutation frequency, was neither potent nor broad. The VRC22 and VRC29 clones were the most potent, with VRC29, the clone with the most members, demonstrating a neutralization geometric mean of 0.3 to 1.2 μg/ml and the greatest breadth (19% to 41%). Testing against a larger panel of 208 genetically diverse viruses confirmed that VRC29.03 had moderate breadth (28.8%) and potency (geometric mean titer [GMT] = 1.3 μg/ml) (Fig. 2; see also Table S1 in the supplemental material). VRC22 had a lower score on the expanded panel than on the first set of 28, in part due to the lack of neutralization of clades AE and D. Although VRC22.01 was much less broad than VRC29.03, it contributed to the total breadth of the patient's polyclonal response: 3 viruses that were resistant to VRC29.03 were neutralized by VRC22.01, and the combined breadth on the panel of 208 viruses was 30% (see Table S1 in the supplemental material).
FIG 2.
Breadth and potency of neutralization by donor N170 antibodies. Neutralization was assessed by single-round-of-infection pseudoviruses on TZMbl cells. Red stars indicate strains used to screen B-cell cultures. All Envs are considered tier 2, with the exception of Q23.17, RW020.2, and ZM109, which are considered tier 1. Values are IC50 (μg/ml) for antibodies and ID50 for serum.
Epitope mapping of donor N170 MAbs.
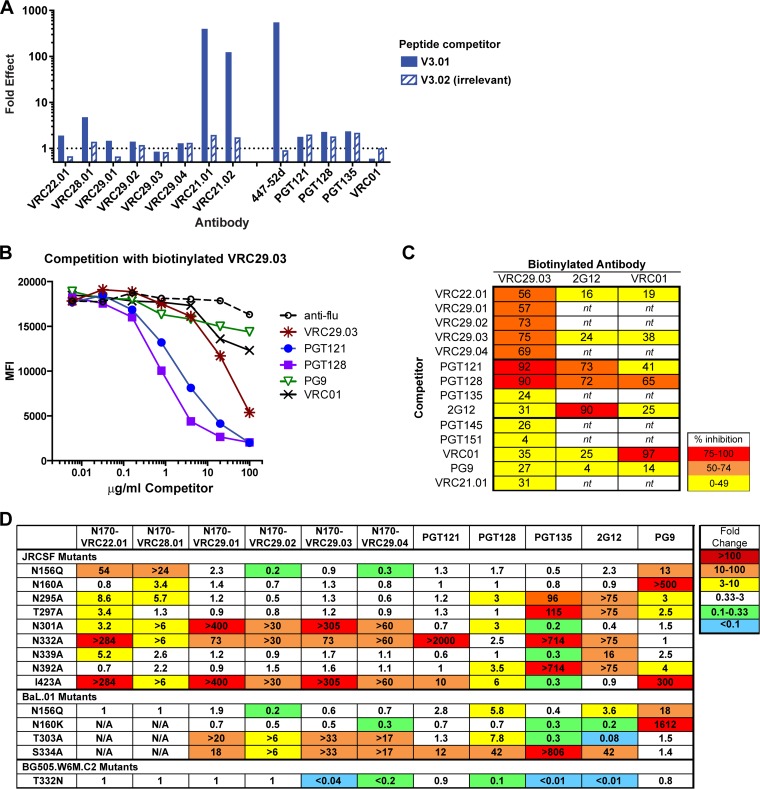
To determine the epitopes targeted by donor N170 antibodies, we first sought to exclude those MAbs that target the V3 loop. Such antibodies, which are not broad, are frequently found in the anti-HIV-1 immune response (3). We therefore performed neutralization competition assays, in which the neutralization of HIV-1 JRCSF.JB was measured in the presence of a linear V3 peptide. The activity of VRC21.01 and VRC21.02 was greatly inhibited by the V3 peptide, similar to the known V3 loop-directed antibody 447-52D (Fig. 3A). The other donor N170 antibodies were not inhibited, nor were control antibodies that target other epitopes.
FIG 3.
Donor N170 antibodies target the glycan-V3 supersite or the V3 loop. (A) Peptide competition. Shown are data for neutralization of HIV-1 JRCSF in the presence of a V3 loop peptide (V3.01) or a nonblocking, irrelevant control peptide (V3.02) compared to the no-peptide control. (B and C) Competition for binding to cell surface-expressed Env. HEK293T cells were transfected with Env JRFL.E168K gp160 dCT (deleted for the gp41 intracellular domain) and then incubated with a mix of biotinylated antibody and serially diluted unlabeled competitors, stained with streptavidin-PE, and analyzed by flow cytometry. (B) Competition with VRC29.03 (data are representative of 3 experiments). (C) Summary of competitions. Percent inhibition was calculated as the MFI of cells incubated with 100 μg/ml the competitor compared to cells with the anti-influenza virus control. Data are the averages of results from two or three experiments. nt, not tested. (D) Neutralization of viruses bearing mutations that are known to affect glycan-V3- or V1V2-directed antibodies. Neutralization activities are reported as fold changes (IC50 of the mutant/IC50 of the wild type); values of >1 indicate inhibition, and values of <1 indicate enhancement.
We next used competition assays to narrow down the site targeted by the other lineages. Binding to soluble gp120 molecules by all of them was weak (see below); however, the VRC29 antibodies bound well to cell surface-expressed JRFL E168K trimeric Env. Therefore, we assayed for the competition of biotinylated VRC29.03 binding to cell surface-expressed Env. We found that the glycan-V3-directed bNAbs PGT121 and PGT128 strongly competed with VRC29.03, consistent with the glycan-V3 supersite being the target (Fig. 3B). VRC29 clonal relatives and VRC22 also competed with VRC29.03. In this semiquantitative assay format, we saw little to no competition of VRC29.03 with PGT135 and 2G12 (Fig. 3C). Antibodies against other sites, including V1V2 (PG9 and PGT145), VRC01 (CD4 binding site), the gp120-gp41 interface (PGT151), and the V3 loop peptide (VRC21.01), did not compete with VRC29.03, nor did an anti-influenza virus control. We also tested the competition of 2G12 and VRC01 binding to transfected cells: neither antibody was competed by donor N170 antibodies; 2G12 was competed by itself, PGT121, and PGT128; and VRC01 was competed by itself and by PGT128, a previously reported effect (44) (Fig. 3C). In total, these results are consistent with VRC29 and VRC22 lineage antibody recognition of the glycan-V3 supersite.
To map the epitopes in more detail, we tested the neutralization of a series of point mutants known to affect the sensitivity to glycan-V3- and V1V2-directed antibodies (Fig. 3D). The activity of VRC22, VRC28, and VRC29 lineage antibodies was abrogated or greatly reduced against mutants of JRCSF and BaL.01 that lacked the N332 glycan (JRCSF.N332A and BaL.01.S334A), the core of the glycan-V3 supersite. Conversely, while none of the donor N170 antibodies neutralized wild-type BG505.w6M.C2 virus, VRC29.03 and VRC29.04 were able to neutralize a mutant with the N332 glycan knocked in (T332N). Removal of the N301 glycan in the JRCSF and BaL strains also abrogated sensitivity to VRC28 and the VRC29 lineage antibodies and reduced sensitivity to VRC22. Other mutations known to affect the activity of glycan-V3 antibodies were also tested. I423A conferred resistance to all donor N170 antibodies, with more modest effects on PGT121 and PGT128. This residue in the C4 domain was previously shown to be critical for some but not all members of the PGT128 family (8). Removal of glycans at positions 156, 295, and 339 had more modest effects on some of the antibodies. Notably, the patterns of sensitivity differed among the donor N170 antibodies, even within the N170-VRC29 lineage, and also differed from those of the other glycan-V3 antibodies.
Thus, the epitope mapping data illustrate that 3 of the 4 donor N170 lineages are antibodies that react with N301 and N332 as well as other glycans in the glycan-V3 supersite. The fourth lineage (VRC21) recognizes an epitope in the V3 loop and is glycan independent.
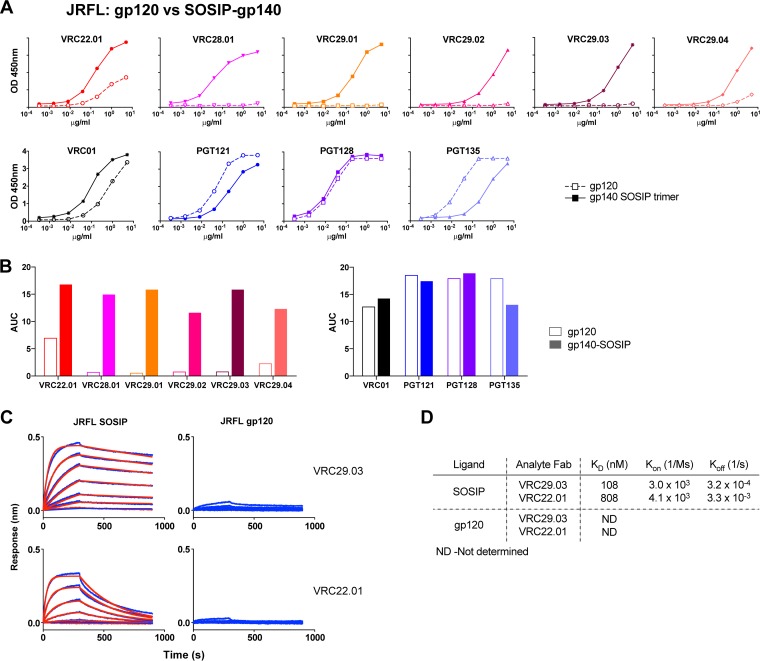
Trimer preference of donor N170 antibodies.
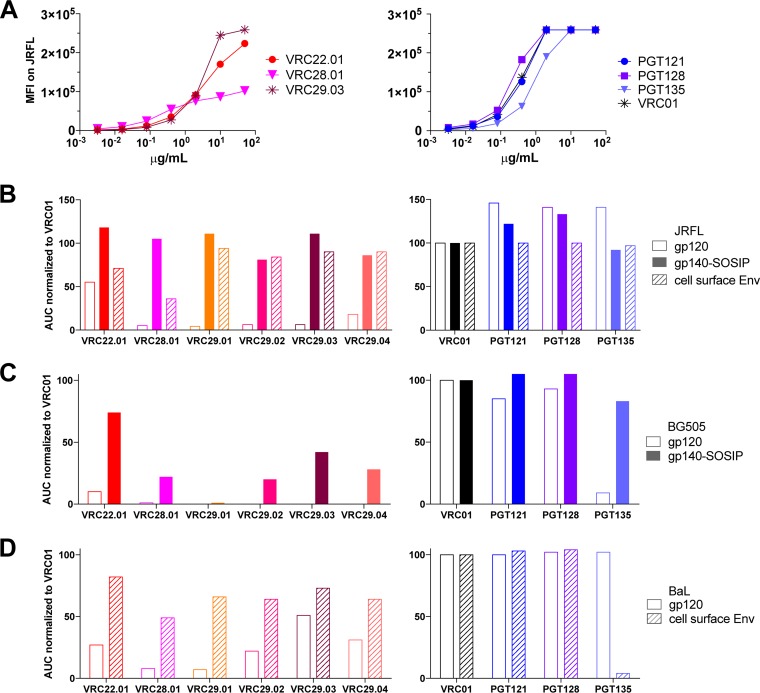
The glycan-V3-directed antibodies VRC22.01 and VRC28.01 and the VRC29 antibodies all bound poorly to gp120 derived from several HIV strains (Fig. 4 and 5). To determine whether this was due to a preference for binding to a trimer, we first assessed binding to a well-folded soluble trimer mimic, JRFL SOSIP gp140 (36), compared to a JRFL gp120 monomer under the same assay conditions. While PGT121 and PGT128 bound equally well to JRFL gp120 and JRFL SOSIP gp140 trimers by an ELISA, VRC28.01 and three of the VRC29 antibodies bound only to the trimer and not at all to gp120, while VRC22 and VRC29.04 bound much better to the trimer than to the gp120 monomer (Fig. 4A). When the ELISA data were quantified by calculating the area under the curve (AUC), striking differences between trimer and monomer binding, up to 30-fold, were noted (Fig. 4B). To confirm this observation in a different assay format, we measured the binding of Fab forms of VRC22.01 and VRC29.03 to the same proteins via biolayer light interferometry. The Fabs bound to the JRFL SOSIP trimer with equilibrium dissociation constant (KD) values of 808 and 108 nM, respectively, while binding to the monomer was too weak to quantify (Fig. 4C and D).
FIG 4.
Donor N170 antibodies bind better to JRFL SOSIP gp140 than to JRFL gp120. (A) ELISA for binding to JRFL SOSIP gp140 or gp120 of donor N170 antibodies (top graphs) and controls (bottom graphs). Graphs are representative of results from at least two independent experiments. OD, optical density. (B) Area under the curve (AUC) calculated from data shown in panel A. (C) Kinetic measurement of binding to JRFL SOSIP gp140 or gp120. Binding of Fab fragments was measured by biolayer light interferometry. (D) Kinetic parameters calculated from data shown in panel C.
FIG 5.
Donor N170 antibodies bind better to trimeric than to monomeric Envs of three HIV strains. (A) Binding of donor N170 antibodies (left) or controls (right) to cells transfected with JRFL gp160 dCT. Binding was measured by flow cytometry. Graphs are representative of results from three independent experiments. MFI, median fluorescence intensity. (B) AUC calculated from data shown in panel A or from data shown in Fig. 4A, normalized to the AUC for VRC01 measured in parallel. (C) AUC normalized to VRC01 binding for ELISAs with BG505 SOSIP gp140 or BG505 gp120. (D) AUC normalized to VRC01 binding for ELISAs with BaL gp120 or cell surface binding assays with BaL-gp160dCT.
We extended these observations by assaying a different form of JRFL trimer: cell surface-expressed envelope. The antibodies were used to stain cells transfected with JRFL gp160 dCT and measured by flow cytometry (Fig. 5A). To compare the flow cytometric data to the ELISA data, we calculated the AUC and normalized the values to those for VRC01 binding measured in parallel. VRC28.01 and VRC29.01-04 bound significantly better to cell surface JRFL Env than to gp120, while PGT121, PGT128, and PGT135 showed similar binding to all three forms of JRFL Env (Fig. 5B).
To ensure that these results were not strain specific, we compared gp120 and trimer forms of a second clade B strain, BaL, as well as the clade A strain BG505.T332N. The soluble trimer mimic BG505.T332N.SOSIP.664 (37) was bound by VRC22.01, VRC28.01, VRC29.03, and VRC29.04, while BG505.T332N gp120 was not bound by any donor N170 antibodies, except for a weak interaction with VRC22.01 (Fig. 5C). Similarly, binding to cell surface-expressed BaL Env was much stronger than binding to BaL gp120 for nearly all the donor N170 antibodies (Fig. 5D). PGT121 and PGT128 bound equally well to trimer and monomer forms of BaL and BG505, while PGT135 paradoxically showed a preference for trimeric BG505 Env but monomeric BaL Env.
Thus, unlike the quintessential glycan-V3 antibodies PGT121 and PGT128, donor N170 antibodies VRC22.01, VRC28.01, and VRC29.01 to VRC29.04 show a marked preference for binding to trimeric Env, with the VRC29 lineage being almost completely dependent on the trimeric conformation, suggesting a quaternary component to their epitopes.
DISCUSSION
In this study of a single donor, we isolated three glycan-V3-directed neutralizing antibody lineages that collectively contribute to the neutralization potency and breadth of the donor's serum against HIV-1. These lineages share many characteristics with the known glycan-V3-directed bNAbs of the PGT121, PGT128, PGT135, and PCDN families, including genetic characteristics and dependence on the glycans at N301 and N332. The members of all three donor N170 lineages were highly dependent on N301 and N332 glycans, with subtle differences providing unique fine specificities. For example, two members of the VRC29 lineage demonstrated a greater dependence on the glycan at N301 than on the glycan at N332, suggesting fine differences in the mode of recognition or angle of approach within the lineage. Unlike previously reported glycan-V3-directed bNAbs, however, the mature donor N170 antibodies preferentially bind to the native trimeric envelope rather than monomeric gp120.
A recent study of the PGT121 lineage showed that antibodies reconstructed from less-mutated sequences bound better to trimeric Env than to monomeric Env gp120, while mature isolated antibodies bound to gp120 with high affinity (25). The authors of that study concluded that as the antibodies matured, their epitope shifted, allowing binding to monomeric Env. In contrast, antibodies N170-VRC29.03 and N170-VRC29.04 are mature, as evidenced by their high levels of somatic hypermutation (15 to 16%), yet still exhibited a substantial preference for trimeric Env over monomeric gp120. To our knowledge, this is the first report of quaternary-preferring mature antibodies targeting the glycan-V3 supersite. This preference may be due to the recognition of residues or glycans on more than one Env protomer, as suggested for PG9 (45) and PGT151 (46); the angle of approach to the supersite, which is known to vary greatly among bNAbs (15, 44); or a requirement for glycans to adopt conformations specific to the trimer and not seen on gp120.
HIV-1 bNAbs often show unusual genetic characteristics (47), and the glycan-V3 antibodies have certain features in common, including CDRH3 length and mutation from the germ line. The mean CDRH3 length in healthy donor human memory B cells (29, 48–50) is 14 to 16 aa, while a length of 24 aa is 2 standard deviations above the mean (51). Strikingly, the other known glycan-V3 bNAbs have CDRH3 lengths of 18 to 24 aa, and the glycan-V3 antibodies reported here have lengths of 19, 20, or 26 aa (Table 1). Similarly, high levels of somatic hypermutation are typical for bNAbs. Mutation levels show a wide spectrum, from a mean of 4% in HIV-uninfected donor human memory B cells (21) to 6% in non-HIV-directed antibodies in HIV-infected donors (52), 9% for Env binding antibodies (52), ∼20% for glycan-V3 bNAbs (8), and extremely high levels of 30% or more for the VRC01 class of CD4bs-directed bNAbs (23, 24). The glycan-V3 antibodies from donor N170 are most similar to the other known glycan-V3 bNAbs, with 7 to 19% mutation from the heavy chain germ line. Thus, these features are common between donor N170 antibodies and the previously described glycan-V3 bNAbs. Importantly, these features are not always predictive of neutralization breadth and potency: N170-VRC28 had the highest mutation rate and the longest CDRH3 of the antibodies isolated here yet the weakest neutralization activity. Similarly, some poorly neutralizing antibodies targeting other epitopes, such as X5 and E51, have very long CDRH3 loops (53). Finally, it has been observed for multiple bNAb clonal lineages that family members with similar levels of mutation and CDRH3 lengths may have very different neutralization capacities (8, 39).
The glycan-V3-directed antibodies described here have substantial cross-clade neutralization breadth. They do not achieve the remarkable breadth shown by antibodies PG121 and PGT128; indeed, no other glycan-V3-directed lineages that match the breadth of these antibodies have as yet been reported (8, 9), and multiple clonal relatives of these antibodies that have lower breadth and potency have been isolated. We speculated that one reason for the lower breadth and potency of VRC29.03 and VRC22.01 may be a lower affinity for the trimer: previously reported data under the same assay conditions (36) showed a KD for PGT128 of 5 nM, which is 20-fold higher than that of VRC29.03 (KD of 108 nM) (Fig. 4D) and 161-fold higher than that of VRC22.01.
Despite our success in isolating multiple neutralizing antibodies from donor N170, we likely missed other neutralizing antibodies of potential interest from donor N170, since the lineages described here were unable to totally recapitulate the breadth and potency of donor N170 serum. A critical step is our dependence on serum neutralization to select an appropriate virus to screen B-cell culture supernatants; indeed, in our experience, the B-cell culture method works best when memory B cells express antibodies that are very potent neutralizers. Multiple attempts were made to screen B-cell cultures with four viruses that were sensitive to neutralization by polyclonal antibodies in the patient's serum. To our surprise, we were unable to isolate any Q769.h5-neutralizing antibodies from the B-cell culture screened with this virus, which was highly sensitive to the donor's serum. Most of the peripheral neutralizing antibodies in serum are derived from long-lived plasma cells in bone marrow, while our approach depends upon the survival of sufficient numbers of memory B cells in the peripheral circulation that are clonal relatives of the bone marrow plasma cells secreting neutralizing antibody.
In summary, this study demonstrates that the glycan-V3 supersite can be targeted by multiple lineages within a single individual. Differences in the fine specificities of the various antibodies likely contribute to the overall serum breadth and may arise in response to ongoing viral evolution during chronic infection. The quaternary specificity of some glycan-V3 antibodies implies a complex binding mode within the glycan-V3 supersite. While the developmental pathways of these antibodies, which were derived from a donor with chronic infection, is unknown, the preference of these antibodies for trimeric Env suggests that trimer mimics should be incorporated into immunogens to elicit such antibodies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marie Pancera for providing plasmids for cell surface staining and Zhongjia Yang for technical assistance. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in our neutralization panel.
Funding Statement
This work, including the efforts of Nancy S. Longo, Matthew S. Sutton, Marissa C. Jarosinski, Andrea R. Shiakolas, Ivelin S. Georgiev, Krisha McKee, Robert T. Bailer, Mark K. Louder, Sijy O’Dell, Mark Connors, John R. Mascola, and Nicole A. Doria-Rose, was funded by intramural research programs of the Vaccine Research Center, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, USA. This work, including the efforts of Javier Guenaga and Richard T. Wyatt, was funded by the International AIDS Vaccine Initiative (IAVI), made possible by generous support from many donors, including the Bill & Melinda Gates Foundation, the Ministry of Foreign Affairs of Denmark, Irish Aid, the Ministry of Finance of Japan, the Ministry of Foreign Affairs of the Netherlands, the Norwegian Agency for Development Cooperation (NORAD), the United Kingdom Department for International Development (DFID), and the United States Agency for International Development (USAID). The full list of IAVI donors is available at http://www.iavi.org.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01012-16.
REFERENCES
- 1.Overbaugh J, Morris L. 2012. The antibody response against HIV-1. Cold Spring Harb Perspect Med 2:a007039. doi: 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. 2013. Antibodies in HIV-1 vaccine development and therapy. Science 341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mascola JR, Haynes BF. 2013. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol Rev 254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. 2014. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doria-Rose NA, Klein RM, Daniels MG, O'Dell S, Nason M, Lapedes A, Bhattacharya T, Migueles SA, Wyatt RT, Korber BT, Mascola JR, Connors M. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol 84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwong PD, Mascola JR. 2012. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton DR, Hangartner L. 2016. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLeod DT, Choi NM, Briney B, Poignard P. 2016. Early antibody lineage diversification and independent limb maturation lead to broad HIV-1 neutralization targeting the Env high-mannose patch. Immunity 44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A 109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio PM, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landais E, Huang X, Havenar-Daughton C, Murrell B, Price MA, Wickramasinghe L, Ramos A, Bian CB, Simek M, Allen S, Karita E, Kilembe W, Lakhi S, Inambao M, Kamali A, Sanders EJ, Anzala O, Edward V, Bekker LG, Tang J, Gilmour J, Kosakovsky-Pond SL, Phung P, Wrin T, Crotty S, Godzik A, Poignard P. 2016. Broadly neutralizing antibody responses in a large longitudinal sub-Saharan HIV primary infection cohort. PLoS Pathog 12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pancera M, Yang Y, Louder MK, Gorman J, Lu G, McLellan JS, Stuckey J, Zhu J, Burton DR, Koff WC, Mascola JR, Kwong PD. 2013. N332-directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One 8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong CH, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien JP, Menis S, Wickramasinghe L, Seaman MS, Schief WR, Wilson IA, Poignard P, Burton DR. 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Karim QA, Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Longo NS, Satorius CL, Plebani A, Durandy A, Lipsky PE. 2008. Characterization of Ig gene somatic hypermutation in the absence of activation-induced cytidine deaminase. J Immunol 181:1299–1306. doi: 10.4049/jimmunol.181.2.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YC, Kipling D, Dunn-Walters DK. 2011. The relationship between CD27 negative and positive B cell populations in human peripheral blood. Front Immunol 2:81. doi: 10.3389/fimmu.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, NISC Comparative Sequencing Program, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. 2015. Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell 161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, Mahan A, Wang S, Kardar M, Yaari G, Walker LM, Simen BB, St John EP, Chan-Hui PY, Swiderek K, Kleinstein SH, Alter G, Seaman MS, Chakraborty AK, Koller D, Wilson IA, Church GM, Burton DR, Poignard P. 2013. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog 9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang J, Doria-Rose NA, Longo NS, Laub L, Lin CL, Turk E, Kang BH, Migueles SA, Bailer RT, Mascola JR, Connors M. 2013. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat Protoc 8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doria-Rose N, Bailer R, Louder M, Lin C-L, Turk E, Laub L, Longo N, Connors M, Mascola J. 13 September 2013. High throughput HIV-1 microneutralization assay. Protoc Exch doi: 10.1038/protex.2013.069. [DOI] [Google Scholar]
- 29.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. 2004. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol 172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 30.Brochet X, Lefranc M-P, Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russ DE, Ho KY, Longo NS. 2015. HTJoinSolver: human immunoglobulin VDJ partitioning using approximate dynamic programming constrained by conserved motifs. BMC Bioinformatics 16:170. doi: 10.1186/s12859-015-0589-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu Y, Winfrey S, Yang ZY, Xu L, Rao SS, Srivastava I, Barnett SW, Nabel GJ, Mascola JR. 2007. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine 25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montefiori DC. 2009. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 485:395–405. doi: 10.1007/978-1-59745-170-3_26. [DOI] [PubMed] [Google Scholar]
- 35.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, Bailer RT, McKee K, Nason M, O'Dell S, Ofek G, Pancera M, Srivatsan S, Shapiro L, Connors M, Migueles SA, Morris L, Nishimura Y, Martin MA, Mascola JR, Kwong PD. 2013. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 36.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. 2015. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog 11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. 2013. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pancera M, Wyatt R. 2005. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology 332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 39.Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, Staupe RP, Altae-Tran HR, Bailer RT, Crooks ET, Cupo A, Druz A, Garrett NJ, Hoi KH, Kong R, Louder MK, Longo NS, McKee K, Nonyane M, O'Dell S, Roark RS, Rudicell RS, Schmidt SD, Sheward DJ, Soto C, Wibmer CK, Yang Y, Zhang Z, Mullikin JC, Binley JM, Sanders RW, Wilson IA, Moore JP, Ward AB, Georgiou G, Williamson C, Abdool Karim SS, Morris L, Kwong PD, Shapiro L, Mascola JR. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387–400. doi: 10.1016/S0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 41.Malenbaum SE, Yang D, Cavacini L, Posner M, Robinson J, Cheng-Mayer C. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol 74:11008–11016. doi: 10.1128/JVI.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Pena AT, Derking R, Klasse PJ, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kepler TB, Liao HX, Alam SM, Bhaskarabhatla R, Zhang R, Yandava C, Stewart S, Anasti K, Kelsoe G, Parks R, Lloyd KE, Stolarchuk C, Pritchett J, Solomon E, Friberg E, Morris L, Karim SS, Cohen MS, Walter E, Moody MA, Wu X, Altae-Tran HR, Georgiev IS, Kwong PD, Boyd SD, Fire AZ, Mascola JR, Haynes BF. 2014. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe 16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, Connors M, Burton DR, Wilson IA, Klasse PJ, Ward AB, Moore JP, Sanders RW. 2015. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog 11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Pena AT, Cupo A, Julien JP, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. 2011. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One 6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpe JM, Kepler TB. 2008. Large-scale analysis of human heavy chain V(D)J recombination patterns. Immunome Res 4:3. doi: 10.1186/1745-7580-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu YC, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. 2010. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood 116:1070–1078. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian C, Luskin GK, Dischert KM, Higginbotham JN, Shepherd BE, Crowe JE Jr. 2007. Evidence for preferential Ig gene usage and differential TdT and exonuclease activities in human naive and memory B cells. Mol Immunol 44:2173–2183. doi: 10.1016/j.molimm.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briney BS, Willis JR, Crowe JE Jr. 2012. Human peripheral blood antibodies with long HCDR3s are established primarily at original recombination using a limited subset of germline genes. PLoS One 7:e36750. doi: 10.1371/journal.pone.0036750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 53.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.