ABSTRACT
The interferon-regulated Mx1 gene of the A2G mouse strain confers a high degree of resistance against influenza A and Thogoto viruses. Most other laboratory inbred mouse strains carry truncated nonfunctional Mx1 alleles and, consequently, exhibit high virus susceptibility. Interestingly, CAST/EiJ mice, derived from wild Mus musculus castaneus, possess a seemingly intact Mx1 gene but are highly susceptible to influenza A virus challenge. To determine whether the enhanced influenza virus susceptibility is due to intrinsically reduced antiviral activity of the CAST-derived Mx1 allele, we generated a congenic C57BL/6J mouse line that carries the Mx locus of CAST/EiJ mice. Adult animals of this line were almost as susceptible to influenza virus challenge as standard C57BL/6J mice lacking functional Mx1 alleles but exhibited far more pronounced resistance to Thogoto virus. Sequencing revealed that CAST-derived MX1 differs from A2G-derived MX1 by two amino acids (G83R and A222V) in the GTPase domain. Especially the A222V mutation reduced GTPase activity of purified MX1 and diminished the inhibitory effect of MX1 in influenza A virus polymerase activity assays. Further, MX1 protein was substantially less abundant in organs of interferon-treated mice carrying the CAST Mx1 allele than in those of mice carrying the A2G Mx1 allele. We found that the CAST-specific mutations reduced the metabolic stability of the MX1 protein although Mx1 mRNA levels were unchanged. Thus, the enhanced influenza virus susceptibility of CAST/EiJ mice can be explained by minor alterations in the MX1 restriction factor that negatively affect its enzymatic activity and reduce its half-life.
IMPORTANCE Although the crystal structure of the prototypic human MXA protein is known, the importance of specific protein domains for antiviral activity is still incompletely understood. Novel insights might come from studying naturally occurring MX protein variants with altered antiviral activity. Here we identified two seemingly minor amino acid changes in the GTPase domain that negatively affect the enzymatic activity and metabolic stability of murine MX1 and thus dramatically reduce the influenza virus resistance of the respective mouse inbred strain. These observations highlight our current inability to predict the biological consequences of previously uncharacterized MX mutations in mice. Since this is probably also true for naturally occurring mutations in Mx genes of humans, careful experimental analysis of any natural MXA variants for altered activity is necessary in order to assess possible consequences of such mutations on innate antiviral immunity.
INTRODUCTION
Influenza virus resistance in mice is determined largely by the Mx locus on chromosome 16, which contains two interferon (IFN)-regulated genes, designated Mx1 and Mx2 (1). MX proteins are highly conserved in vertebrates, suggesting that they play important roles in antiviral defense across species (reviewed in references 2 and 3). Mx gene induction is dependent on type I IFN (IFN-α and -β) or type III IFN (IFN-λ) signaling, resulting in instant MX protein synthesis after viral infection (4). The Mx1 allele of mouse strain A2G codes for a 72-kDa GTPase, designated MX1, that accumulates in the nuclei of IFN-stimulated cells. MX1 confers a high degree of resistance to infection with viruses of the Orthomyxoviridae family, including influenza A virus and Thogoto virus (THOV) (5, 6). Interestingly, the Mx1 gene is defective in most commonly used inbred mouse strains due to deletion of whole exons or insertion of single nonsense mutations which cause low metabolic stability of Mx1 mRNA (7). For example, C57BL/6J mice contain a large deletion of Mx1 coding exons 9 to 11, and CBA/J mice have an in-frame stop codon in exon 10. There is evidence from functional studies that wild mice may either possess or lack functional Mx1 alleles (8), but sequence information which would support this assumption is missing.
The Mx2 gene codes for a cytoplasmic protein that can inhibit some viruses with cytoplasmic replication cycles, such as vesicular stomatitis virus and hantaviruses (1, 9). In most inbred mouse strains, including C57BL/6J and A2G, the Mx2 gene is defective due to an insertion of a single nucleotide that destroys the Mx2 open reading frame (10).
The inbred mouse strain CAST/EiJ, derived from wild Mus musculus castaneus trapped in Thailand, is exceptional because it exhibits a high degree of influenza virus susceptibility although it possesses a seemingly intact Mx1 gene (11). Available sequence information indicates that MX1 from CAST/EiJ mice differs by one or two amino acids from prototypic MX1 of the A2G strain (11, 12). However, it remained unclear whether these amino acid changes account for the influenza virus susceptibility phenotype of CAST/EiJ mice or whether the antiviral activity of MX1 may simply be suppressed by some undefined regulatory factors present in Mus musculus castaneus.
To address this question, we generated a congenic C57BL/6J mouse line that carries the Mx1 allele of CAST/EiJ mice. We observed that such animals were almost as susceptible to influenza virus challenge as parental Mx1-negative C57BL/6 mice but exhibited far more pronounced resistance to Thogoto virus. CAST-derived MX1 exhibited lower GTPase activity and lower inhibitory activity in viral polymerase assays than A2G-derived MX1. MX1 also accumulated to lower levels in organs of IFN-treated mice carrying the CAST Mx1 allele than in those of animals carrying the A2G Mx1 allele due to decreased metabolic stability. These findings can explain the enhanced influenza virus susceptibility of CAST/EiJ mice.
MATERIALS AND METHODS
Infection experiments.
The following mouse strains were used for infection experiments: (i) C57BL/6J (B6) carrying a defective Mx locus (7) (purchased from Janvier Labs), (ii) congenic B6.A2G-Mx1 mice (13) carrying a functional Mx1 allele on the B6 genetic background (bred in our local specific-pathogen-free [SPF] facility), and (iii) congenic B6.CAST-Mx mice harboring the Mx locus of wild-derived inbred strain CAST/EiJ (originally purchased from Jackson Laboratories and established in our local SPF facility by 11 cycles of backcrossing to C57BL/6J mice and subsequent breeding to homozygosity). Where indicated, experiments were performed with heterozygous animals from backcross generations one to nine. Such mice, carrying one Mx allele from CAST/EiJ mice and one Mx allele from B6 mice (CAST/B6), were compared to littermates (B6/B6) carrying two defective Mx alleles derived from C57BL/6J mice. In some experiments, heterozygous mice (A2G/B6) which carried one Mx allele of A2G mice and one Mx allele of B6 mice were used.
After ketamine-xylazine anesthesia, mice were intranasally infected with a 40-μl virus inoculum containing the indicated doses of influenza A virus strain A/Seal/Massachusetts/1/1980 (H7N7) (SC35M) or A/Swan/Germany/R65/06 (H5N1) (R65). Experiments with SC35M and R65 were performed in biosafety level 2 (BSL2) and BSL3 animal facilities, respectively. Thogoto virus (THOV) infections were carried out by intraperitoneal injection of either adult mice with strain SiAr 126/FR (THOVΔML) or 3- to 5-day-old suckling mice with THOV carrying the functional IFN antagonist ML. The body weight and health conditions of infected animals were monitored daily. Animals were sacrificed in case of severe disease (ruffled fur, apathetic behavior, and limb paralysis) or else after the body weight fell below 75% of the initial value in the case of adult animals.
Ethics statement.
All mice used in the study were bred locally in our facility and handled in accordance with guidelines of the Federation for Laboratory Animal Science Associations (www.felasa.eu/recommendations) and the national animal welfare body (Gesellschaft für Versuchstierkunde; www.gv-solas.de/index.html). Animal experiments were performed in compliance with the German animal protection law (TierSchG) and approved by the local animal welfare committee of the University of Freiburg (Permit G-12/46).
Virus quantification.
Virus titers in lung homogenates (lungs homogenized in 1 ml of phosphate-buffered saline [PBS]) were quantified by plaque assays on day 4 or 5 postinfection on confluent MDCK cells by serial 10-fold dilutions in Opti-MEM (Thermo Fisher Scientific) supplemented with 0.3% bovine serum albumin (BSA). After incubating virus dilutions on MDCK cells for 1 h, virus inoculum was aspirated and cells were overlaid with 1.5% Avicel (FMC BioPolymer) in Dulbecco modified Eagle medium (DMEM) supplemented with 0.1% BSA for 72 h. To visualize and count virus plaques, cells were fixed with 4% formaldehyde and stained with 1% crystal violet solution.
Mx1 cDNA sequencing.
Total RNA was isolated from IFN-α-treated mouse embryonic fibroblasts (MEFs) using peqGOLD TriFast (Peqlab). cDNA was produced using the RevertAid H Minus first-strand cDNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The Mx1 open reading frame was amplified via PCR in four overlapping fragments. PCR fragments were gel purified with Zymoclean gel DNA recovery kit (Zymo Research) and subjected to Sanger sequencing.
Immunostaining.
Cells grown on glass coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 10 min. To reduce nonspecific antibody binding, samples were incubated with antibody diluent (PBS containing 5% normal goat serum). MX proteins were detected with the MX1-specific rabbit hyperimmune serum AP5-2771 directed against a synthetic peptide corresponding to the 16 C-terminal amino acids of MX1 (14) diluted 1:500 in antibody diluent for 1 h. After three PBS washing steps, cells were incubated with goat Alexa Fluor 488-conjugated secondary antibodies (A-11034; Thermo Fisher Scientific) diluted 1:500 for 30 min in the dark. After two washing steps, Hoechst 33258 (861405; Sigma-Aldrich) was used for nuclear counterstaining for 15 min, followed by two further washing steps.
Western blotting.
For preparation of cell lysates, MEFs grown in cell culture plates were washed with PBS, protein lysis buffer (50 mM HEPES, 1% NP-40, 1 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 100 U/ml Benzonase nuclease, 1 mM dithiothreitol [DTT], 1× cOmplete proteinase inhibitor) was added, and cells were incubated on ice for 20 min. Organ samples were homogenized in ice-cold lysis buffer with a ceramic sphere and a FastPrep-12 homogenizer (MP Biomedicals). To remove the insoluble fraction, homogenized samples were centrifuged at 4°C for 20 min. For SDS-PAGE, lysis samples were complemented with the respective amount of RunBlue 4× lithium dodecyl sulfate (LDS) sample buffer (Expedeon) containing 1/5 β-mercaptoethanol and incubated at 70°C for 10 min. Samples loaded onto 10% RunBlue SDS precast gels were run in 1× SDS running buffer (Expedeon) at 140 V. Size-separated proteins from the gel were transferred onto a methanol-activated Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore) in Towbin buffer (25 mM Tris, 192 mM glycine, 20% methanol) at 110 mA per gel for 75 min in a PerfectBlue Semi-Dry Electro Blotter device (Peqlab). The membrane was blocked with 5% SlimFast powder in PBS for 1 h, followed by incubation with primary antibody diluted in 0.1% Tween 20 and 1% SlimFast (in PBS) at room temperature for 1 h or at 4°C overnight. Primary antibodies for MX1 (a polyclonal rabbit hyperimmune serum that is cross-reactive and recognizes a large number of mammalian Mx proteins, including mouse MX2) and β-tubulin (monoclonal mouse IgG1 antibody clone TUB 2.1 [T4026; Sigma]) were used. Three washing steps with 0.1% Tween 20 in PBS were performed before secondary antibodies (diluent as for primary antibodies) were incubated with the membrane for 1 h, again followed by three washing steps. As secondary antibodies, AffiniPure horseradish peroxidase-conjugated goat anti-rabbit IgG (111-035-045; Jackson ImmunoResearch) and anti-mouse IgG (115-035-166; Jackson ImmunoResearch) were used. Primary and secondary antibodies were used at a dilution of 1:1,000. SuperSignal West Pico substrate solution served for signal detection of the membrane on an Odyssey Fc luminescence detection device (Li-Cor Biosciences).
Minireplicon assay.
For reconstitution of influenza A virus polymerase activity, HEK 293T cells were seeded in black 96-well plates. Cells were transfected at ∼30% confluence with 4 ng of pCAGGS-based expression plasmids encoding the viral polymerase subunits PB1, PB2, and PA as well as NP using FuGENE 6 transfection reagent (Promega). To generate viral minigenomes, 16 ng of plasmids encoding firefly luciferase in the negative-sense orientation flanked by the 5′ and 3′ untranslated regions (UTRs) of an influenza virus genome segment (plasmid pHW-NSluc for PR8 [15] or pPolI-FFLuc-RT [16] for KAN-1) was cotransfected together with 4 ng of pCMV-RL plasmid (Promega) encoding Renilla luciferase under the control of the constitutive cytomegalovirus (CMV) promoter. For the PR8 minireplicon, 20 ng of MX1 pCAXL expression plasmids was cotransfected. For the KAN-1 minireplicon, 5 ng of MX1 pCAXL expression plasmids was used. The calculated firefly/Renilla luciferase ratio in the presence of a transfected empty vector control was set to 100%. For each MX expression construct, triplicate wells were transfected with 10 μl of the same transfection mix. At least three independent experiments were performed. Similar MX1 expression levels were verified by Western blot analysis.
Protein purification and GTPase activity assay.
Various recombinant MX1 protein variants (designated either A2G, K49A, G83R, A222V, or CAST) were expressed from pQE9 plasmids transformed into Escherichia coli strain M15(pREP4) (Qiagen) as N-terminal His6 fusion constructs and purified as described previously (17). For GTPase activity assays, equal amounts of purified MX1 proteins were estimated by quantifying protein bands on Coomassie brilliant blue-stained SDS-polyacrylamide gels using the ImageJ software (version 1.47g). GTPase activity of recombinant MX1 was measured using the Transcreener GDP FI assay kit (BellBrook Labs) in black 96-well microtiter half-area plates (675076; Greiner). Briefly, recombinant proteins were diluted to equal MX1 concentrations, and each reaction mixture (reactions were performed in technical triplicates) was replenished with a GTP master mix to a final reaction volume of 25 μl containing 4 μM GTP and 40 nM adenosine 5′-(β,γ-imido)triphosphate lithium salt hydrate (AMP PNP). After 1 h of incubation at 37°C, the enzymatic reaction was stopped by adding 25 μl stop-and-detect buffer (20 mM HEPES [pH 7.5], 40 mM EDTA, 0.02% Brij 35, 8 nM GDP-Alexa Fluor 594 tracer, 4.56 μg/ml GDP-IRDye QC-1 antibody), and the mixture was incubated for 1 h in the dark at room temperature before detection of the fluorescent signal with a Tecan Infinite M200 plate reader (excitation, 580 nm; emission, 620 nm).
qRT-PCR.
To perform quantitative reverse transcription-PCR (qRT-PCR) of organ samples, cDNA was prepared from isolated RNA (as described in “Mx1 cDNA sequencing” above). For each qPCR sample within the 384-well plate, 50 ng of cDNA were used with a 400 nM end concentration of respective primers targeting Mx1, Mx2, Isg15, Isg56, and Hprt in 1× SensiFast SYBR Hi-Rox mix (BIO-92005; Bioline). All cDNA samples were run in technical duplicates on an ABI Prism 7900HT device (Applied Biosystems) in the ΔΔCT program mode (50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min). Data were analyzed using the RQ-Manager software version 1.2.1. Murine Hprt was used as housekeeping gene to normalize specific gene signals to the overall cDNA amount by calculating the ΔCT values (ΔCT = ΔCTgene1 − ΔCTHprt). The fold mRNA expression relative to that of Hprt was determined by calculating the 2−ΔCT values.
MX1 promoter activity assay.
Mouse 3T3 cells were seeded in black 96-well plates with clear bottoms and transfected with FuGENE 6 transfection reagent (Promega) at ∼80% confluence with 50 ng of pGL4.13[luc2/SV40]-based plasmids (Promega) carrying either the constitutive simian virus 40 (SV40) promoter or a promoter region of the A2G or CAST Mx1 allele (corresponding to A2G Mx1 nucleotide position −827 to −1). In addition, 13 ng of Renilla luciferase-encoding pRL-CMV plasmid (Promega) was cotransfected to allow normalization for transfection efficiencies (normalized luciferase activity). At 6 h posttransfection, medium supplemented with different amounts of IFN-α (0, 0.1 or 1 ng) was added to the transfected cells. The following day, luciferase activity was measured using the Dual-Glo luciferase assay system kit (Promega) with a Tecan Infinite M200 plate reader. Ratios of firefly to Renilla luciferase activity were calculated (normalized luciferase activity).
Pulse-chase.
Confluent monolayers of MEFs in 6-well plates were treated overnight with 10 ng of IFN-α per ml. The next day, cells were washed with PBS twice before newly synthesized proteins were radioactively labeled for 2 h with 4 MBq per well of 35S-labeled cysteine and methionine (EasyTag formulation [NEG772; Perkin-Elmer) in starvation medium (DMEM without methionine-cysteine and containing glutamine and 5% fetal calf serum [FCS]). The cells were then washed with PBS before addition of chase medium (DMEM containing 10% FCS, 2 mM l-methionine, and 2 mM l-cysteine). Lysates were prepared at different time points postchase by placing the plates on ice, washing cells with PBS, and lysing cells with lysis buffer (140 mM NaCl, 5 mM MgCl2, 20 mM Tris HCl [pH 7.6], 1% NP-40, cOmplete protease inhibitor) for 30 min on ice. Lysates were cleared from insoluble material by centrifugation (14,000 × g, 30 min, 4°C) and were stored at −20°C. For immunoprecipitation, the MX1-specific antibody AP5-2771 (14) was used (1:60 dilution). Briefly, lysates were incubated under constant rotation at 4°C for 1 h. Thirty microliters of wash buffer B-equilibrated protein A-Sepharose beads was added to each sample, and the samples were incubated on a rotating device at 4°C for 1 h. Beads were washed, and bound proteins were eluted by incubating in sample running buffer (80 mM Tris HCl [pH 6.8], 5 mM EDTA, 34% sucrose, bromophenol blue, 3.2% SDS, 40 mM DTT) at 95°C for 5 min and analyzed using an 8% SDS-polyacrylamide gel in running buffer (47 mM Tris, 400 mM glycine, 0.1% SDS) at 20 mA overnight. After fixing of the gel with fixation buffer (40% methanol, 10% acetic acid) for 1 to 2 h and drying the gel in a gel dryer (model 583; Bio-Rad Laboratories), radioactive MX1 band intensities were quantified on a Typhoon FLA 7000 (GE Healthcare) phosphorimager using the ImageQuant TL (GE Healthcare) software. Initial signal intensities at t = 0 h were set to 100%.
Accession number(s).
The sequences determined in this study have been submitted to GenBank under accession numbers KX774216, KX774217, KX774218, KX774219, KX774220, KX774221, KX774222, and KX774223.
RESULTS
The CAST/EiJ-derived Mx allele confers only a low degree of protection against influenza viruses.
To determine whether the reported high influenza virus susceptibility of CAST/EiJ mice is due to a functional deficiency of the Mx locus in these mice, we crossed CAST/EiJ mice with C57BL/6 mice that carry an Mx1 gene with a large deletion (7). The resulting heterozygous mice were backcrossed to C57BL/6J mice, and offspring carrying a CAST-derived Mx1 allele were identified by PCR. Backcrossing and selecting animals carrying one CAST-derived Mx1 allele were repeated 10 more times to yield the congenic mouse line B6.CAST-Mx, which contains genetic material from C57BL/6J mice except for the Mx locus and adjacent genes that originate from CAST/EiJ mice. After completion of the final backcrossing step, mice were bred to be homozygous for the CAST-derived Mx allele.
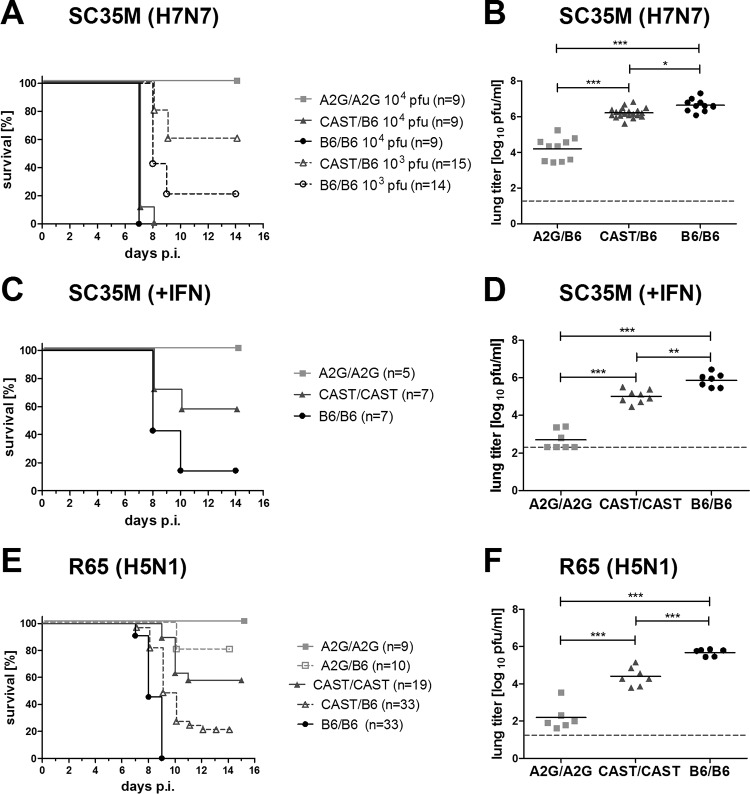
When challenged with 104 PFU of the H7N7 influenza virus strain SC35M, all mice carrying one CAST-derived Mx allele (CAST/B6) developed severe disease like Mx1-negative C57BL/6 (B6/B6) mice and had to be sacrificed for animal welfare reasons on day 7 or 8 postinfection (Fig. 1A). Under these challenge conditions, mice carrying functional A2G-derived Mx1 alleles (A2G/A2G) developed no obvious signs of disease and survived. When a 10-fold-lower dose of challenge virus was used, 40% of the CAST/B6 mice had to be euthanized because their body weight fell below the threshold of 75%, whereas the remaining 60% survived (Fig. 1A). Under identical conditions, only 20% of the Mx1-negative B6 mice survived. The survival data correlated with viral lung titers on day 4 postinfection (Fig. 1B), suggesting that the CAST-derived Mx locus confers a low but measurable degree of influenza virus resistance. To exclude the possibility that the low influenza virus resistance of CAST mice is due to poor or delayed synthesis of type I IFN after virus infection, groups of mice were treated with IFN-α prior to infection with SC35M. As expected, such treatment stimulated virus resistance to a certain extent (Fig. 1C and D) but clearly failed to confer a high degree of influenza virus resistance to mice carrying the CAST-derived Mx locus, suggesting that MX1 of CAST mice is intrinsically less potent than MX1 from A2G mice.
FIG 1.
The CAST/EiJ-derived Mx allele confers a low degree of protection against influenza viruses. Groups of homozygous (CAST/CAST) or heterozygous mice carrying one Mx allele from CAST/EiJ mice and one from C57BL/6J mice (CAST/B6) were compared to littermates carrying both Mx alleles from C57BL/6J mice (B6/B6). B6.A2G-Mx1 mice (A2G/A2G) carrying two Mx alleles from the influenza virus-resistant strain A2G served as controls. (A) Animals were infected by the intranasal route with either 103 or 104 PFU of influenza A virus strain SC35M (H7N7) as indicated. Weight loss was monitored daily, and animals were sacrificed and scored dead when body weight dropped below the 75% limit. Survival curves are shown. (B) Viral titers in the lungs of mice infected with 104 PFU of SC35M were determined at day 4 postinfection. Statistical significance was tested using analysis of variance (ANOVA) with the Bonferroni post hoc test. *, P < 0.05; ***, P < 0.001. (C) Groups of mice carrying the different Mx alleles in homozygous form were treated with 1.5 μg of IFN-α in a 40-μl volume by the intranasal route 16 h before infection with 104 PFU of SC35M. Weight loss was monitored daily, and animals were sacrificed and scored dead when body weight dropped below the 75% limit. (D) Groups of mice were pretreated with 1.5 μg of IFN-α before infection with 104 PFU of SC35M. Viral titers in the lungs were determined at day 4 postinfection. (E) Groups of mice carrying the different Mx alleles in heterozygous or homozygous form as indicated were infected with 102 PFU of the avian influenza virus strain R65 (H5N1). Weight loss was monitored daily, and animals were sacrificed and scored dead when body weight dropped below the 75% limit. Survival curves are shown. (F) Viral titers in the lungs of R65-infected mice were determined at day 4 postinfection. Statistical significance was tested using ANOVA with the Bonferroni post hoc test. ***, P < 0.001.
To confirm and extend these findings, we performed additional infection experiments with the highly pathogenic avian H5N1 influenza A virus strain R65, which is very sensitive to MX-mediated inhibition (18, 19). After infection with 100 PFU, all prototypic C57BL/6 mice had to be euthanized within 9 days postinfection, as expected, whereas all mice carrying the A2G-derived Mx1 allele in the homozygous state survived (Fig. 1E). Under these challenge conditions, we observed 58% survival of mice carrying the CAST-derived Mx allele in homozygous form (Fig. 1E). Animals carrying either the A2G- or the CAST-derived Mx allele in heterozygous form exhibited markedly reduced resistance compared to that of the corresponding homozygous controls (Fig. 1E). The results of this survival study correlated well with viral lung titers on day 4 postinfection (Fig. 1F), indicating that the beneficial effect of functional Mx1 genes results from improved inhibition of virus replication in the respiratory tract.
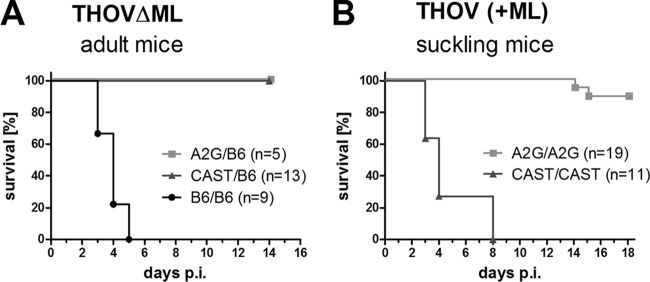
The CAST/EiJ-derived Mx allele potently protects adult but not newborn mice against THOV.
The A2G-derived Mx1 allele confers solid resistance to THOV in adult mice (6) and mediates substantial but less pronounced resistance in pups, presumably due to reduced type I IFN production in young mice (20). We observed that adult mice carrying a CAST-derived Mx allele survived intraperitoneal infection with 104 PFU of THOVΔML, whereas all Mx-negative B6 mice developed severe disease and had to be sacrificed for animal welfare reasons (Fig. 2A). These results confirmed the conclusion from the above-described experiments with H5N1 influenza virus that the CAST-derived Mx locus possesses antiviral activity. Interestingly, when suckling mice were challenged with 103 PFU of wild-type THOV, which can produce the IFN-antagonistic factor ML, the A2G- but not the CAST-derived Mx locus mediated antiviral protection (Fig. 2B), confirming the view that the Mx locus of A2G mice codes for a more powerful restriction factor than the Mx locus of CAST/EiJ mice.
FIG 2.
The CAST/EiJ-derived Mx allele potently protects adult but not newborn mice against THOV. Groups of mice carrying the different Mx alleles in heterozygous or homozygous form as indicated were infected with THOV by the intraperitoneal route. (A) Survival of adult mice infected with 104 PFU of THOV strain SiAr 126/FR. (B) Survival of 3- to 5-day-old suckling mice infected with 103 PFU of recombinant THOV expressing ML.
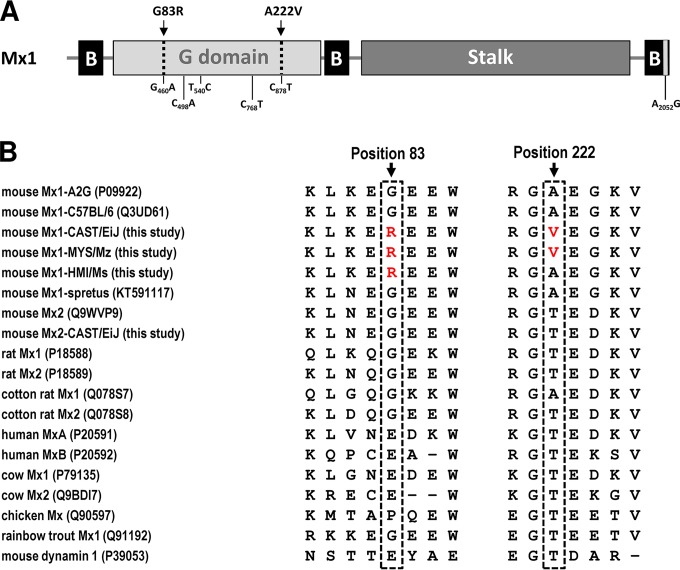
The MX1 protein of CAST/EiJ mice differs by two amino acids from MX1 of A2G mice.
cDNA sequencing of the open reading frame revealed six nucleotide differences between the A2G-derived and the CAST-derived Mx1 alleles, of which two are nonsynonymous and result in amino acid changes (Fig. 3A). Both of these changes, glycine to arginine at position 83 (G83R) and alanine to valine at position 222 (A222V), map to the G domain of MX1, the protein domain mediating GTPase activity. Glycine at position 83 shows a moderate degree of conservation among vertebrate Mx proteins. For example, it is replaced by glutamic acid in human MXA and MXB and bovine MX1 and MX2 proteins and by proline in chicken MX (Fig. 3B). In contrast, position 222 is highly conserved. Most MX proteins of mammals, including MX1 from rats, have a threonine at this position (Fig. 3B). A threonine is also present at this position in MX proteins of birds and fish as well as in mouse MX2. Even the distantly related mouse dynamin carries a threonine at the corresponding position (Fig. 3B). Remarkably, however, the MX1 proteins of all standard inbred mouse strains, which contain genetic material mostly from Mus musculus domesticus and to a lesser extent from Mus musculus musculus (21, 22), feature an alanine at position 222 (Fig. 3B). Similarly, Mus spretus has an alanine at position 222 of MX1 (17).
FIG 3.
The MX1 protein of CAST/EiJ mice differs by two amino acids from MX1 of A2G mice. (A) Schematic drawing showing the primary structure of mouse MX1 protein (NCBI accession number NM_01084). CAST-specific nucleotide changes are indicated below the graphic. Two of these nucleotide changes cause amino acid changes (G83R and A222V), as indicated above the drawing. B, tripartite bundle signaling element. (B) Alignment of MX1 sequences around amino acid positions 83 and 222 was performed using CLUSTALW with a BLOSUM cost matrix. CAST-specific amino acids at these positions are highlighted in red. Sequences from strains CAST/EiJ, HMI/Ms, and MYS/Mz originate from this study.
To determine whether the CAST/EiJ strain might mirror the situation in wild Mus musculus castaneus, we sequenced DNA samples (from the RIKEN Bioresource Center DNA bank) of two additional inbred mouse strains, HMI/Ms and MYS/Mz, that were both derived from wild Mus musculus castaneus. We found that these two strains have an arginine at position 83 like CAST/EiJ mice but differ at position 222. HMI/Ms mice have an alanine at position 222 like A2G and most other inbred mouse strains, whereas MYS/Mz mice have a valine at this position like CAST/EiJ mice (Fig. 3B). Further, HMI/Ms and MYS/Mz mice both show the synonymous C498A mutation found in CAST/EiJ mice. Thus, the various Mus musculus castaneus inbred strains are not uniform with regard to the primary structure of restriction factor MX1.
Subcellular localization and antiviral activity of CAST-derived MX1 protein.
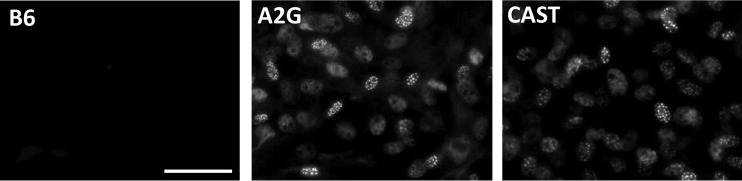
Immunofluorescence studies using MEFs derived from congenic C57BL/6J mice carrying either the A2G- or the CAST-derived Mx locus revealed no obvious differences in the intracellular distribution of MX1. Both MX1 variants were found predominantly in the nuclei of IFN-α-treated cells, where they seemed to accumulate in distinct structures or aggregates (Fig. 4).
FIG 4.
Normal intracellular distribution of CAST-derived MX1 protein. MEFs from C57BL/6J mice lacking functional Mx1 alleles (B6) or carrying the Mx1 allele of strain CAST/EiJ (CAST) or A2G (A2G) were treated with 10 ng IFN-α per ml for 16 h before cells were fixed and stained for MX1 protein using the MX1-specific rabbit antiserum AP5.
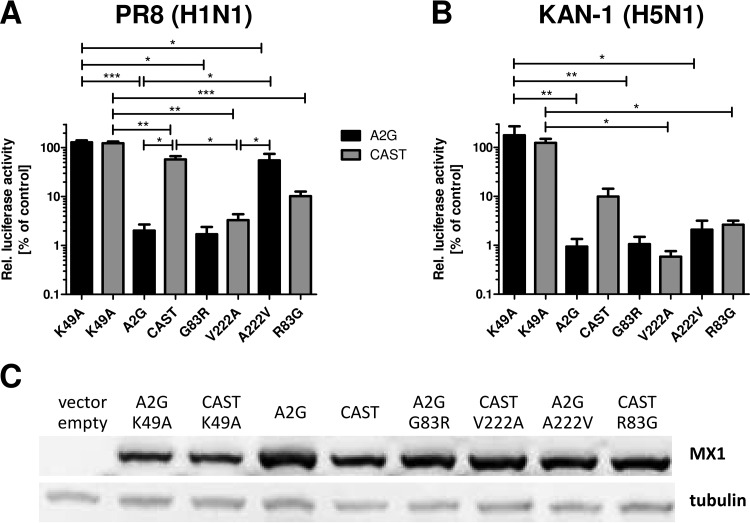
To determine whether the G83R and A222V mutations might influence the antiviral activity of MX1, we performed influenza virus minireplicon assays. In an H1N1-based (PR8) minireplicon assay system, the A2G variant of MX1 reduced viral polymerase activity to about 2%, whereas the GTPase-deficient K49A mutant was inactive, as expected (Fig. 5A). Under these experimental conditions, the CAST variant of MX1 carrying the G83R and A222V mutations exhibited only minimal inhibitory activity (Fig. 5A). A construct of A2G MX1 carrying only the G83R mutation exhibited potent antiviral activity, whereas a construct carrying only the A222V mutation exhibited poor antiviral activity, similar to the case for the CAST MX1 variant (Fig. 5A). To confirm and extend these findings, we performed additional experiments using an H5N1-based (KAN-1) minireplicon assay system that is substantially more sensitive to inhibition by MX proteins than the PR8-based system (23, 24). CAST-derived MX1 reduced the polymerase activity of KAN-1 to about 10% (Fig. 5B). The G83R mutant showed full antiviral activity in this assay, whereas the A222V mutant showed slightly reduced activity, but this difference was not statistically significant (Fig. 5B). Taken together, these results showed that CAST-derived MX1 is less active in influenza virus minireplicon assays than A2G-derived MX1, although it shows normal nuclear localization and the expression levels of the different MX1 variants were comparable in this assay (Fig. 5C). Our results further showed that position 222 plays a more important role in antiviral activity of MX1 than position 83.
FIG 5.
CAST-derived MX1 exhibits low inhibitory activity in influenza virus polymerase reconstitution assays. 293T cells were cotransfected with plasmids encoding NP, the three viral polymerase subunits PB1, PB2, and PA, a polymerase I-driven construct for an influenza virus minigenome encoding firefly luciferase, an expression plasmid encoding Renilla luciferase, and expression constructs for the indicated MX1 variants, including the GTPase-deficient K49A mutant protein (K49A). Firefly luciferase activity was normalized to Renilla luciferase activity, and values in the absence of coexpressed MX1 (control) were set to 100%. (A) A minireplicon system based on polymerase subunits derived from the influenza virus strain A/Puerto Rico/8/34 (PR8) was employed, and 20 ng of the indicated MX1 plasmids was cotransfected. Normalized mean luciferase values ± standard errors of the means (SEM) from five independent experiments are indicated, except for the G83R construct, which was included in two experiments only. (B) A minireplicon system based on polymerase subunits from the H5N1 influenza virus strain A/Thailand/1/2004 (KAN-1) was employed, and 5 ng of the indicated MX1 plasmids was cotransfected. Normalized mean luciferase values ± SEM from three independent experiments are shown. The K49A construct was used in two of three experiments. Statistical significance was tested using ANOVA with the Bonferroni post hoc test. *, P < 0.05; ***, P < 0.001. (C) Western blot analysis of representative samples used for panels A and B, demonstrating that the various MX1 variants were expressed at comparable levels.
CAST-derived MX1 exhibits slightly lower GTPase activity than A2G-derived MX1.
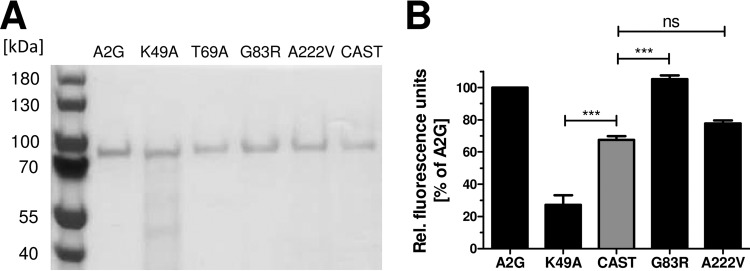
To determine whether the poor antiviral activity of CAST-derived MX1 might be explained by reduced GTP hydrolysis activity, we purified N-terminal histidine-tagged recombinant protein from E. coli (Fig. 6A) and measured GTPase activity using a fluorescence-based assay system which detects GDP resulting from hydrolysis of GTP. A2G-derived MX1 carrying either lysine (A2G) or alanine (K49A, GTPase deficient) at position 49 served as positive and negative controls, respectively. The GTPase activity of CAST-derived MX1 was clearly reduced compared to that of A2G-derived MX1, but it was evidently higher than the background nuclease activity of this assay system (Fig. 6B). The G83R mutation in A2G MX1 did not affect GTPase activity, whereas the A222V mutation did (Fig. 6B), indicating that amino acid position 222 modulates the enzymatic activity of MX1.
FIG 6.
CAST MX1 shows slightly reduced GTPase activity compared to A2G MX1. N-terminally His6-tagged MX1 proteins were expressed in E. coli strain M15(pREP4) from pQE-9 expression vectors and purified via Ni-nitrilotriacetic acid (NTA) affinity chromatography. (A) SDS-PAGE analysis and Coomassie brilliant blue staining was used to verify that the samples used for the GTPase assays contained similar amounts of the various MX1 variants. (B) GTPase activities of the different MX1 protein variants were determined using the Transcreener GDP FI assay kit (BellBrook Labs), in which newly synthesized GDP is monitored by fluorescence. To compensate for variations in fluorescence between individual experiments, values were normalized to the A2G MX1 signal, which was set to 100%. Normalized fluorescence values ± SEM from nine independent experiments are shown. A2G MX1 and CAST MX1 were present in all nine experiments, the K49A and A222V variants in eight of the nine experiments, and the G83R variant in five of the nine experiments. Statistical significance was tested using ANOVA with the Bonferroni post hoc test. ns, not significant (P > 0.05); ***, P < 0.001.
CAST-derived MX1 has lower metabolic stability than A2G-derived MX1.
MEFs from B6 mice carrying the CAST Mx locus contained only low levels of MX1 protein after treatment with a low dose (0.1 ng per ml) of IFN-α. Even when treated with a 10-fold-higher dose of IFN-α for 16 h, the levels of MX1 in cells carrying the CAST Mx locus were substantially lower than those in cells carrying the A2G-derived Mx locus (Fig. 7A). The former cells accumulated high levels of MX2 (Fig. 7A), which is not present in cells carrying the A2G Mx locus due to a genetic defect (1, 25).
FIG 7.
CAST MX1 exhibits lower metabolic stability than A2G MX1. (A) MEFs from C57BL/6J mice carrying either no functional Mx alleles (B6), two CAST-type Mx alleles (CAST), or two A2G-type Mx alleles (A2G) were stimulated with IFN-α for 16 h, and MX protein levels were measured by Western blot using an antibody that can detect MX1 and MX2. (B) Groups of adult B6, CAST, or A2G mice (n = 3) were either stimulated with 1 μg IFN-α by the intraperitoneal route for 14 h or were left untreated. Lungs were harvested, and MX protein levels were detected by Western blotting using an antibody that can detect MX1 and MX2. (C) RNA was isolated from the lung samples described for panel B, followed by cDNA synthesis. Products were subjected to quantitative SYBR green-based real-time PCR, and transcript levels of Mx1, Mx2, Isg15, and Isg56 were determined. The fold expression of the indicated genes relative to Hprt is represented by the mean 2−ΔCT values ± SEM among three mice. Statistical significance was tested using ANOVA with the Bonferroni post hoc test. ns, not significant (P > 0.05); ***, P < 0.001. (D) Mouse 3T3 cells were transfected with firefly pGL4.13 reporter plasmids carrying either a constitutive promoter (SV40) or the Mx1 promoter region from A2G or CAST mice, followed by overnight treatment with different doses of IFN-α. Activity of the firefly luciferase was normalized to Renilla luciferase activity encoded by a cotransfected control plasmid. Statistical significance was tested using one-way ANOVA with the Bonferroni post hoc test. ns, not significant (P > 0.05); **, 0.01 ≥ P > 0.001. (E) MEFs from C57BL/6J mice carrying either no functional Mx alleles (B6), two CAST-type Mx alleles (CAST), or two A2G-type Mx alleles (A2G) were treated with 10 ng IFN-α per ml (+) for 12 h or were left untreated (−) before newly synthesized proteins were metabolically labeled with 35S-labeled l-methionine-l-cysteine for 2 h. Cells were then maintained in label-free medium for the indicated times before cells were lysed. MX1 in the lysates was immunoprecipitated, antibody-bound material was analyzed by SDS-PAGE, and radioactivity in the MX1 bands (arrow) was quantified. (F) Graphic representation of radioactivity detected in the MX1 bands at various time points after starting the pulse-chase experiments. Data points are from three independent experiments, except for the 3-h time point, which is derived from a single experiment. Means ± SEM are shown. The correlation coefficients (r2) of the regression curves indicate a first-order exponential decay.
To determine MX1 levels in vivo, we injected 1 μg of IFN-α into the peritoneums of three mice per group and measured MX protein levels in the lungs by Western blot analysis at 14 h posttreatment. We observed substantially higher levels of MX1 in the lungs of C57BL/6J mice carrying the A2G Mx locus than in those of congenic mice carrying the CAST Mx locus (Fig. 7B, bottom panel). Interestingly, lungs of untreated control mice also contained low levels of MX1 when the A2G locus was present but not when the CAST Mx locus was present (Fig. 7B, top panel).
To determine whether these differences in MX protein levels were due to different mRNA levels, we first quantified mRNAs encoding MX1 and other IFN-stimulated proteins in the lungs of IFN-treated mice by qRT-PCR. We used the same samples for RNA extraction in which we had observed the differences in MX1 protein levels (Fig. 7B). Interestingly, we observed no significant differences between the Mx1 mRNA levels in lungs of mice carrying the A2G or the CAST Mx locus (Fig. 7C). In particular, Mx1 mRNA levels in the lungs of untreated mice carrying the A2G-derived Mx locus were not higher than those in lungs of untreated mice carrying the CAST-derived Mx locus, although the former contained substantially higher levels of MX1 protein, indicating differences in the metabolic stabilities of the two MX1 protein variants. As expected, Mx2 mRNA was detected exclusively in mice carrying the CAST Mx locus, whereas the products of other IFN-stimulated genes, such as Isg15 and Isg56, were present at comparable levels irrespective of the Mx allele (Fig. 7C). We next sequenced the promoter region of the Mx1 gene. There were only six nucleotide differences (including an insertion of four nucleotides at position −380) between mice carrying the A2G or the CAST Mx allele in an 827-nucleotide-long fragment from the region located immediately upstream of the transcription initiation site of the Mx1 gene (data not shown).
To functionally test the promoter activities of these DNA fragments, they were cloned upstream of a firefly luciferase reporter gene and the resulting constructs were tested for IFN-mediated activation of luciferase activity in transiently transfected mouse 3T3 cells. Both promoters were strongly activated by IFN treatment, but no significant differences in activity were observed between promoter fragments originating from the A2G or the CAST Mx locus (Fig. 7D). Thus, it seemed unlikely that differences in promoter activity or mRNA stability can explain the observed decreased MX1 protein levels in cells and organs of mice carrying the CAST Mx locus.
To determine whether the CAST-specific mutations might influence MX1 protein stability, we measured the metabolic half-life of MX1 in MEFs from mice carrying either the A2G or the CAST Mx locus. To do this, cells were treated with 10 ng per ml of IFN-α for 12 h before newly synthesized proteins were metabolically labeled with 35S-labeled l-methionine–l-cysteine for 2 h. The cells were then maintained in medium containing unlabeled amino acids for 3, 6, 22, or 47 h before cell lysates were prepared. MX1 in cell lysates was immunoprecipitated using an MX1-specific antibody, bound material was analyzed by SDS-PAGE, and the MX1 signal was visualized (Fig. 7E). Under these conditions, labeling of MX1 was consistently more efficient in cells carrying the A2G Mx locus, and the signal became fainter with reduced kinetics. Quantification revealed that the metabolic half-life of MX1 encoded by the A2G Mx allele was about 16 h, whereas the half-life of MX1 encoded by the CAST Mx allele was only about 7 h (Fig. 7F).
DISCUSSION
High influenza virus susceptibility of inbred mouse strain CAST/EiJ that correlates with amino acid changes in restriction factor MX1 has been documented (11, 12). However, because genetic factors that negatively act on MX1 have recently been detected in the mouse strain DBA/2 (26), it remained unclear whether the minor alterations in MX1 reduce restriction factor activity in CAST/EiJ mice or whether the genetic background of CAST/EiJ mice has a negative effect. We show here that a congenic C57BL/6J mouse line carrying the Mx locus of CAST/EiJ remained susceptible to influenza virus. We found that one seemingly minor amino acid change at position 222 in the G domain of MX1 negatively influences GTPase activity and that this alteration reduces the ability of MX1 to inhibit the activity of the influenza virus polymerase. We further found that the metabolic stability of CAST-derived MX1 is strongly reduced. Taken together, our findings strongly suggest that distinct mutations in restriction factor MX1 rather than unknown modulatory factors are responsible for the high influenza virus susceptibility phenotype of CAST/EiJ mice.
Unexpectedly, our study demonstrated that the CAST Mx locus harbors diminished but not completely abrogated restriction factor activity. The residual antiviral activity of the CAST Mx locus became apparent in our experiments only when using challenge viruses that are highly sensitive to MX-mediated restriction. In the case of influenza A virus, residual antiviral activity was detected only when the animals were infected with an H5N1 avian influenza virus (Fig. 1E and F) that is known to exhibit high MX sensitivity (18, 19). This differential behavior was accurately mirrored in influenza virus minireplicon assays in which CAST-derived MX1 showed significant activity only against the highly MX-sensitive polymerase complex of the H5N1 virus and not against the polymerase complex of a less MX-sensitive H1N1 virus. The residual antiviral activity of the CAST Mx locus manifested itself most prominently when adult animals were challenged with THOVΔML (Fig. 2), a virus of the Orthomyxoviridae family that is highly susceptible to MX-mediated inhibition (3). Adult mice carrying the CAST Mx locus controlled THOVΔML very well (Fig. 2A). However, suckling mice, in which ML-expressing THOV is much more virulent due to the immature IFN system (20), readily succumbed to infection with THOV if they carried the CAST- but not the A2G-derived Mx locus (Fig. 2B). Thus, CAST/EiJ mice carry a functional Mx locus that confers far less pronounced resistance to orthomyxoviruses than the Mx locus of A2G mice.
Our functional studies revealed that the A222V exchange in CAST MX1 plays a more important role with regard to GTPase activity and ability to inhibit influenza A virus polymerase activity than the G83R exchange, as expected from the fact that position 222 shows a higher degree of evolutionary conservation (Fig. 3B). Interestingly, MX proteins of most other vertebrate species, including rats, have a threonine rather than an alanine at the corresponding position. Remarkably, mouse MX2 also has a threonine at this position, suggesting that unknown evolutionary pressure has shaped the Mx1 allele in mice. Curiously, human MXA with either T256A or T256V exchanges (position 256 corresponds to position 222 in mouse MX1) exhibited normal antiviral activity in influenza virus and Thogoto virus minireplicon assays (unpublished data), further suggesting that position 222 serves a unique role in mouse MX1. Since position 222 maps to the well-defined GTPase domain dimerization interface (16), it is reasonable to assume that the CAST-specific A222V mutation accounts for the reduced GTPase activity of this variant MX1 protein by altering intermolecular contacts of biologically active MX multimers. Since GTPase activity is required for antiviral activity of MX1, reduced enzymatic activity can account for the observed reduced inhibitory activity of CAST-derived MX1 in viral polymerase assays.
We currently do not know whether the A222V mutation is also the main cause for metabolic instability of CAST-derived MX1. MX proteins are believed to function as oligomers. These oligomeric structures are stabilized by intermolecular bonds via stalk/stalk and stalk/tripartite bundle signaling element (BSE) interactions (27). Further, two separate MX oligomers can transiently dimerize via their G domains in a GTP-dependent manner, which stimulates GTPase activity (16, 28). The latter dimerization event is central to catalytic activity, as it leads to a mechanical power stroke mediated by GTP hydrolysis which is transmitted to the stalk via conformational changes of the BSE (28). Thus, the CAST-specific sequence alterations may change the dimerization interface in more than one way; they could lead to reduced enzymatic activity of MX1 and/or affect MX oligomer stability.
Assessing the role of MX1 in influenza virus resistance of CAST/EiJ mice is complicated by the fact that the Mx locus of CAST mice contains a second functional Mx gene, designated Mx2. Available data indicate that MX2 plays no decisive role in resistance toward either influenza or Thogoto virus. It is well documented that the MX2 protein of mice can inhibit the replication of certain RNA viruses with cytoplasmic replication cycles, such as vesicular stomatitis virus (1), but there is no evidence that MX2 can influence the growth of viruses from the Orthomyxovirus family. In line with this view, our attempts to demonstrate antiviral activity of MX2 against influenza or Thogoto virus in viral minireplicon assays yielded only negative results (unpublished data). Nevertheless, we cannot exclude the remote possibility that MX2 can influence the metabolic stability of MX1 by direct or indirect interactions. Due to the tight genetic linkage of the Mx1 and Mx2 genes (10), it is difficult to breed animals carrying the Mx1 allele of CAST/EiJ mice and the defective Mx2 allele of B6 or A2G mice in a homozygous form. However, since heterozygous CAST/A2G hybrid mice carrying one functional Mx2 allele were as resistant to influenza virus challenge as B6/A2G hybrid mice lacking a functional Mx2 gene (unpublished data), it is rather unlikely that MX2 exhibits regulatory activity on MX1. Similarly, it remains possible that some other genes tightly linked to Mx1, such as Tmprss2, influenced the influenza virus susceptibility phenotype of our congenic mice. A role of this gene is unlikely because the amino acid sequences of TMPRSS2 from B6 and CAST/EiJ mice are identical (data not shown).
When measuring the metabolic half-lives of MX1, we noted reduced stability of radiolabeled MX1 in IFN-treated cells from mice carrying the CAST Mx locus compared to that in cells from mice carrying the A2G Mx locus (Fig. 7E and F). Interestingly, we further noted reduced initial levels of radiolabeled CAST MX1 in all three experiments performed. Since mRNA levels did not differ significantly, it is tempting to speculate that ribosomes might translate mRNAs encoding CAST MX1 at reduced rates compared to those for mRNAs encoding A2G MX1. Altered codon usage may dictate the rhythm of elongation, which in turn may influence cotranslational protein folding and even metabolic stability of the product (29–31). The open reading frame of CAST-derived Mx1 mRNA contains six nucleotide changes which all affect codon usage, and the changes at nucleotide positions 460, 498, 768, and 878 create codons which are less frequently used in Mus musculus (32). Of course, the alternative interpretation is that the intrinsically low stability of CAST MX1 is due to the A222V mutation and that the altered ability of this mutant protein to participate in G domain interactions can explain our observations. Since we measured the metabolic stabilities of the MX1 variants in IFN-treated cells from congenic mice, final proof that G83 and A222 determine MX1 stability is still lacking. We did not manage to generate transfected cell lines expressing the various MX1 variants in an inducible manner that might ideally be used for such experiments.
Our results indicate that the CAST/EiJ inbred mouse strain does not accurately mirror the genetic situation in wild populations of Mus musculus castaneus. If this were the case, the MX1 proteins of all Mus musculus castaneus inbred strains should be identical. However, strain HMI/Ms has an alanine at position 222, whereas strain MYS/Mz has a valine like in strain CAST/EiJ (Fig. 3B). All three strains have an arginine at position 83 and carry a synonymous nucleotide change at position 498, indicating that this might represent a Mus musculus castaneus signature. Interestingly, sequence information from 10 individual wild mice trapped in northwest India (33) predicts that the MX1 protein of wild Mus musculus castaneus has a glycine at position 83 and an alanine at position 222 (D. Tautz, personal communication), like in Mus musculus domesticus. We currently do not understand this unexpected discrepancy. We cannot exclude the possibility that the observed MX1 mutations were acquired during the inbreeding process. However, we favor the alternative possibility that Mus musculus castaneus animals from different parts of the world differ with regard to MX1. Indeed, the founders of inbred strains CAST/EiJ and HMI/Ms originated from Thailand (www.jax.org) and Taiwan (http://dna.brc.riken.jp/en/BRCmgenome2en.html), respectively, whereas the wild Mus musculus castaneus animals mentioned above were trapped in northwest India. Unfortunately, no information is available on the origin of the inbred strain MYS/Mz. Since mice do not appear to represent natural hosts of influenza A viruses, it is tempting to speculate that some other evolutionary pressure is at work that selects for an Mx locus with reduced anti-influenza virus activity that can still confer sufficient protection against tick-borne thogotoviruses, which are believed to represent bona fide pathogens of rodents. It is conceivable that such natural selection pressure by thogotoviruses is not uniform around the globe, which in turn may favor the existence of distinct Mx1 alleles in various parts of Southeast Asia where Mus musculus castaneus is the dominant Mus species.
Taken together, irrespective of whether CAST/EiJ mice carry a rare or a frequent natural allelic variant of the Mx1 gene, our study revealed that seemingly minor genetic changes in MX restriction factors can have dramatic consequences for host resistance toward pathogen challenge. In the case of CAST/EiJ mice, enhanced influenza virus susceptibility is mainly, if not entirely, due to a single alanine-to-valine change in a region of the GTPase domain of MX1 that has not previously been predicted to play a central role in antiviral activity. These findings highlight our current inability to accurately predict the biological consequences of distinct genetic changes in MX restriction factors. Based on our results in the mouse model, it seems likely that similar difficulties are experienced when we try to assess the role of genetic variations in the human MXA and MXB restriction factors, which are involved in innate immune control of influenza virus and HIV, respectively (3). Thus, it may be necessary to perform extensive functional studies to identify physiologically important activity changes in naturally occurring MX variants of humans.
ACKNOWLEDGMENTS
We thank Otto Haller for helpful discussions, Katrin Friedrich and Annette Ohnemus for technical assistance, Diethard Tautz (MPI, Plön) for providing access to the Mx1 sequence tracks in the USCS Genome Browser, and Anne Halenius and Cosima Zimmermann for providing reagents and technical help for the pulse-chase experiments. Genomic DNAs of mouse strains MYS/Mz and HMI/Ms were kindly provided by Riken BRC through the National Bio-Resource Project of the MEXT, Japan.
REFERENCES
- 1.Zürcher T, Pavlovic J, Staeheli P. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187:796–800. doi: 10.1016/0042-6822(92)90481-4. [DOI] [PubMed] [Google Scholar]
- 2.Verhelst J, Hulpiau P, Saelens X. 2013. Mx proteins: antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 77:551–566. doi: 10.1128/MMBR.00024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Holzinger D, Jorns C, Stertz S, Boisson-Dupuis S, Thimme R, Weidmann M, Casanova JL, Haller O, Kochs G. 2007. Induction of MxA gene expression by influenza A virus requires type I or type III interferon signaling. J Virol 81:7776–7785. doi: 10.1128/JVI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenmann J, Lane CA, Hobson D. 1963. The resistance of A2g mice to myxoviruses. J Immunol 90:942–951. [PubMed] [Google Scholar]
- 6.Haller O, Frese M, Rost D, Nuttall PA, Kochs G. 1995. Tick-borne Thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J Virol 69:2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol 8:4518–4523. doi: 10.1128/MCB.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haller O, Acklin M, Staeheli P. 1987. Influenza virus resistance of wild mice: wild-type and mutant Mx alleles occur at comparable frequencies. J Interferon Res 7:647–656. doi: 10.1089/jir.1987.7.647. [DOI] [PubMed] [Google Scholar]
- 9.Jin HK, Yoshimatsu K, Takada A, Ogino M, Asano A, Arikawa J, Watanabe T. 2001. Mouse Mx2 protein inhibits hantavirus but not influenza virus replication. Arch Virol 146:41–49. doi: 10.1007/s007050170189. [DOI] [PubMed] [Google Scholar]
- 10.Staeheli P, Sutcliffe JG. 1988. Identification of a second interferon-regulated murine Mx gene. Mol Cell Biol 8:4524–4528. doi: 10.1128/MCB.8.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, Haagmans BL, McMillan L, Miller DR, Rosenzweig E, Valdar W, Wang J, Churchill GA, Threadgill DW, McWeeney SK, Katze MG, Pardo-Manuel de Villena F, Baric RS, Heise MT. 2013. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog 9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leist SR, Pilzner C, van den Brand JM, Dengler L, Geffers R, Kuiken T, Balling R, Kollmus H, Schughart K. 2016. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genomics 17:143. doi: 10.1186/s12864-016-2483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horisberger MA, Staeheli P, Haller O. 1983. Interferon induces a unique protein in mouse cells bearing a gene for resistance to influenza virus. Proc Natl Acad Sci U S A 80:1910–1914. doi: 10.1073/pnas.80.7.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier E, Fah J, Grob MS, End R, Staeheli P, Haller O. 1988. A family of interferon-induced Mx-related mRNAs encodes cytoplasmic and nuclear proteins in rat cells. J Virol 62:2386–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhelst J, Parthoens E, Schepens B, Fiers W, Saelens X. 2012. Interferon-inducible protein Mx1 inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J Virol 86:13445–13455. doi: 10.1128/JVI.01682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dick A, Graf L, Olal D, von der Malsburg A, Gao S, Kochs G, Daumke O. 2015. Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity. J Biol Chem 290:12779–12792. doi: 10.1074/jbc.M115.650325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhelst J, Spitaels J, Nurnberger C, De Vlieger D, Ysenbaert T, Staeheli P, Fiers W, Saelens X. 2015. Functional comparison of Mx1 from two different mouse species reveals the involvement of loop L4 in the antiviral activity against influenza A viruses. J Virol 89:10879–10890. doi: 10.1128/JVI.01744-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol 81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomon R, Staeheli P, Kochs G, Yen HL, Franks J, Rehg JE, Webster RG, Hoffmann E. 2007. Mx1 gene protects mice against the highly lethal human H5N1 influenza virus. Cell Cycle 6:2417–2421. doi: 10.4161/cc.6.19.4779. [DOI] [PubMed] [Google Scholar]
- 20.Pichlmair A, Buse J, Jennings S, Haller O, Kochs G, Staeheli P. 2004. Thogoto virus lacking interferon-antagonistic protein ML is strongly attenuated in newborn Mx1-positive but not Mx1-negative mice. J Virol 78:11422–11424. doi: 10.1128/JVI.78.20.11422-11424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Hunter KW, Gandolph M, Rowe WL, Finney RP, Kelley JM, Edmonson M, Buetow KH. 2005. A high-resolution multistrain haplotype analysis of laboratory mouse genome reveals three distinctive genetic variation patterns. Genome Res 15:241–249. doi: 10.1101/gr.2901705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade CM, Kulbokas EJ 3rd, Kirby AW, Zody MC, Mullikin JC, Lander ES, Lindblad-Toh K, Daly MJ. 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J Virol 85:8133–8140. doi: 10.1128/JVI.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mänz B, Dornfeld D, Gotz V, Zell R, Zimmermann P, Haller O, Kochs G, Schwemmle M. 2013. Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein. PLoS Pathog 9:e1003279. doi: 10.1371/journal.ppat.1003279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin HK, Takada A, Kon Y, Haller O, Watanabe T. 1999. Identification of the murine Mx2 gene: interferon-induced expression of the Mx2 protein from the feral mouse gene confers resistance to vesicular stomatitis virus. J Virol 73:4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin DL, Hatesuer B, Bergmann S, Nedelko T, Schughart K. 2015. Protection from severe influenza virus infections in mice carrying the Mx1 influenza virus resistance gene strongly depends on genetic background. J Virol 89:9998–10009. doi: 10.1128/JVI.01305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. 2011. Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity 35:514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Rennie ML, McKelvie SA, Bulloch EM, Kingston RL. 2014. Transient dimerization of human MxA promotes GTP hydrolysis, resulting in a mechanical power stroke. Structure 22:1433–1445. doi: 10.1016/j.str.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59:744–754. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders R, Deane CM. 2010. Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res 38:6719–6728. doi: 10.1093/nar/gkq495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supek F. 2016. The code of silence: widespread associations between synonymous codon biases and gene function. J Mol Evol 82:65–73. doi: 10.1007/s00239-015-9714-8. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y. 2007. Codon usage database. http://www.kazusa.or.jp/codon/.
- 33.Harr B, Karokac E, Neme R, Teschke M, Pfeifle C, Pezer Z, Babiker H, Linnenbrink M, Montero I, Scavetta R, Abai MR, Molins MP, Schlegel M, Ulrich RG, Altmüller J, Franitza M, Büntge A, Künzel S, Tautz D. 2016. Genomic resources for wild populations of the house mouse, Mus musculus and its close relative Mus spretus. Scientific Data 3:160075. doi: 10.1038/sdata.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]