ABSTRACT
Accumulating evidence suggests that viruses hijack cellular proteins to circumvent the host immune system. Ubiquitination and SUMOylation are extensively studied posttranslational modifications (PTMs) that play critical roles in diverse biological processes. Cross talk between ubiquitination and SUMOylation of both host and viral proteins has been reported to result in distinct functional consequences. Enterovirus 71 (EV71), an RNA virus belonging to the family Picornaviridae, is a common cause of hand, foot, and mouth disease. Little is known concerning how host PTM systems interact with enteroviruses. Here, we demonstrate that the 3D protein, an RNA-dependent RNA polymerase (RdRp) of EV71, is modified by small ubiquitin-like modifier 1 (SUMO-1) both during infection and in vitro. Residues K159 and L150/D151/L152 were responsible for 3D SUMOylation as determined by bioinformatics prediction combined with site-directed mutagenesis. Also, primer-dependent polymerase assays indicated that mutation of SUMOylation sites impaired 3D polymerase activity and virus replication. Moreover, 3D is ubiquitinated in a SUMO-dependent manner, and SUMOylation is crucial for 3D stability, which may be due to the interplay between the two PTMs. Importantly, increasing the level of SUMO-1 in EV71-infected cells augmented the SUMOylation and ubiquitination levels of 3D, leading to enhanced replication of EV71. These results together suggested that SUMO and ubiquitin cooperatively regulated EV71 infection, either by SUMO-ubiquitin hybrid chains or by ubiquitin conjugating to the exposed lysine residue through SUMOylation. Our study provides new insight into how a virus utilizes cellular pathways to facilitate its replication.
IMPORTANCE Infection with enterovirus 71 (EV71) often causes neurological diseases in children, and EV71 is responsible for the majority of fatalities. Based on a better understanding of interplay between virus and host cell, antiviral drugs against enteroviruses may be developed. As a dynamic cellular process of posttranslational modification, SUMOylation regulates global cellular protein localization, interaction, stability, and enzymatic activity. However, little is known concerning how SUMOylation directly influences virus replication by targeting viral polymerase. Here, we found that EV71 polymerase 3D was SUMOylated during EV71 infection and in vitro. Moreover, the SUMOylation sites were determined, and in vitro polymerase assays indicated that mutations at SUMOylation sites could impair polymerase synthesis. Importantly, 3D is ubiquitinated in a SUMOylation-dependent manner that enhances the stability of the viral polymerase. Our findings indicate that the two modifications likely cooperatively enhance virus replication. Our study may offer a new therapeutic strategy against virus replication.
INTRODUCTION
Small ubiquitin-like modifier (SUMO) modification, a member of the group of posttranslational modifications (PTMs), is important for the regulation of many cellular proteins and pathways (1). SUMO is a novel protein modifier similar to the well-studied ubiquitin (Ub). Structural analysis revealed high similarity between the two PTMs despite limited (18%) sequence identity (2). Four SUMO isoforms exist in vertebrates, each having an approximate molecular mass of 12 kDa. SUMO-2 and SUMO-3 can form polymeric SUMO chains via a single conserved lysine residue (3, 4). Without such a site, SUMO-1 cannot act as a link to elongate chains in vivo but as a chain terminator on poly-SUMO-2 and -3 chains (5–7).
Similar to ubiquitination, SUMO proteins are initially synthesized as inactive precursors and cleaved by a specific protease belonging to the family of sentrin-specific proteases (SENPs) to expose the COOH-terminal diglycine motif (8). The carboxyl terminus of mature SUMO peptide is linked to a cysteine residue in E1-activating enzyme (SUMO-activating enzymes 1 and 2 [SAE1/SAE2]) via a thioester in an ATP-dependent manner. The activated SUMO is subsequently transferred to a cysteine residue of the SUMO-conjugating enzyme Ubc9. Although Ubc9 can transfer SUMO to the target protein, mutual action of Ubc9 and E3 SUMO ligases is required for efficient modification (9–11). An isopeptide bond is formed between the lysine residue of the target protein and SUMO. The modification is dynamically reversible. In addition to a role in SUMO precursor maturation, SENPs also act as SUMO deconjugation enzymes to complete the SUMO modification (SUMOylation) cycle (8).
Typically, the lysine in the amino acid consensus motif ψ-K-X-D/E or the inverted motif E/D-X-K-ψ (where ψ represents a large hydrophobic residue) of a protein is recognized as the SUMOylation site (6, 12, 13). Apart from covalent modification, target proteins may be modified noncovalently via SUMO-interacting motifs (SIMs) (6, 12). In general, the SIMs contain a hydrophobic core (V/I-X-V/I-V/I or V/I-V/I-X-V/I/L) flanked by acidic residues either upstream or downstream (14–17). In addition to being structurally related, ubiquitin and SUMO modifications of substrates occur preferentially on lysine residues, and ubiquitin can be hybridized with the SUMO moiety by anchoring to the lysine on poly-SUMO (18, 19). Cross talk between the two PTMs may be involved in genome stability and cell growth, which are associated with pathogen resistance and cancer treatment (20). Studies indicate that SUMOylation can antagonize ubiquitin/proteasome-mediated degradation by competing the same lysine to stabilize the target protein or cooperatively degrade the target protein by sharing the same lysine with ubiquitin (20, 21).
Viruses, as obligate pathogenic organisms, must usurp the host proteins and cellular pathways throughout their life cycles to evade antiviral defenses by creating an environment that facilitates their replication (7). Many viral proteins have been identified as substrates for SUMOylation. Viruses can interact with and exploit the enzymes of the SUMO pathways to promote their assembly or replication or to evade the host immune system. Viruses could also be constrained by the host SUMO systems, and vice versa (22). Some viruses possess more than one SUMOylated protein, indicating the complexity of virus-host interactions. For instance, SUMOylation of human cytomegalovirus (HCMV) DNA polymerase subunit UL44 and immediate-early 1 and 2 (IE1 and IE2) proteins is required for efficient viral replication (23–26). At least five influenza virus proteins are SUMO targets, and infection with influenza virus leads to a global increase in cellular SUMOylation (27). SUMOylation of nonstructural protein 1 (NS1) accelerates virus growth, M1 protein SUMOylation facilitates viral ribonucleoprotein export and the assembly of virus particles, and NP SUMOylation regulates the intracellular trafficking of NP and efficient virus production (28–30). The early lytic gene product (K-bZIP), the major transcriptional factor (K-Rta), and latency-associated nuclear antigen 2 of Kaposi's sarcoma-associated herpesvirus are SUMOylated to achieve efficient viral replication (31–33). SUMOylation of the p6 domain of the Gag polyprotein and integrase of human immunodeficiency virus type 1 is important for virus replication (34, 35). Nevertheless, SUMOylated viral proteins from other viruses remain to be discovered. A global study of viruses and host SUMOylation will enhance our understanding of the interactions between viruses and their hosts.
Enterovirus 71 (EV71) belongs to the genus Enterovirus (human enterovirus A) of the family Picornaviridae, whose members cause severe neurological diseases in young children under 5 years old, known as hand, foot, and mouth disease (36). Similar to other human enteroviruses, including poliovirus, coxsackie virus, and echovirus, EV71 is a single-stranded RNA virus with a single open reading frame encoding a precursor protein. After infection, the precursor is cleaved into four structural (VP1, VP2, VP3, and VP4) and seven nonstructural (2A, 2B, 2C, 3A, 3B, 3C, and 3D) proteins (37). The 3D protein is an RNA-dependent RNA polymerase (3Dpol) and is responsible for the process of RNA replication. 3C protease is SUMOylated at lysine residue 52 (K52) by SUMO-1. SUMOylation of 3C increases ubiquitination, which leads to the degradation of 3C and eventually causes the reduction of viral replication (38). Our bioinformatics analysis suggested that EV71 3D polymerase is likely to be SUMOylated and may contain both covalent and noncovalent motifs. However, whether 3D is indeed SUMOylated and the role of such modification during infection remain to be addressed.
In the present study, we demonstrate that EV71 3D is modified by SUMO-1 both in vivo and in vitro. We further identify K159 and 150-152 SIM as the sites for SUMOylation. Mutations at both SUMOylation sites impaired polymerase synthesis in in vitro polymerase assays. In addition, we found that the K63-linked ubiquitin modification of 3D is SUMOylation dependent, and a combination of the two modifications resulted in higher 3D stability and enhanced viral replication. Although SUMOylation of 3C and 3D resulted in opposite effects on virus reproduction, enhanced EV71 replication in cells with elevated SUMO-1 indicates that 3D SUMOylation plays a predominant role. Our findings together imply that EV71 exploits host cellular modification for effective replication, revealing a potential target for antiviral therapy.
MATERIALS AND METHODS
Cell culture and virus manipulation.
Human embryonic kidney cells (HEK 293T; China Center for Type Culture Collection [CCTCC]) and African green monkey kidney epithelial cells (Vero; CCL-81; American Type Culture Collection) were cultured in Dulbecco's modified Eagle medium. Human rhabdomyosarcoma cells (RD; CCL-136; American Type Culture Collection) were grown in minimal essential medium. All the cells were maintained in medium containing 10% fetal bovine serum (Life Technology, Australia) in 5% CO2 at 37°C.
EV71 strain BrCr was obtained from the Institute of Medical Biology, Chinese Academy of Medical Science (39). Virus titers were measured by 50% tissue culture infectious dose (TCID50) in RD cells using the Reed-Muench formula (40). One-step growth curves of viruses were determined with parental EV71 at a desired multiplicity of infection (MOI) (41). The EV71 infectious clone was kindly provided by Minetaro Arita (National Institute of Infectious Diseases, Tokyo, Japan) (42). The EV71 infectious clone and its mutant recombinant RNAs were in vitro transcribed with a RiboMax large-scale RNA production system T7 kit (Promega) (43). The RNAs were transfected into Vero cells to rescue EV71 wild-type (WT) and mutant viruses, and these viral strains were amplified in RD cells (39).
Antibodies and reagents.
EV71 3D, 3C, and VP1 polyclonal antibodies were prepared by immunizing rabbits with His tag-3D, -3C, and -VP1 fusion proteins, respectively. Rabbit anti-hemagglutinin (HA) and mouse anti-V5 antibodies were purchased from CoWin Biotech (Beijing, China). Mouse anti-β-actin antibody was acquired from Proteintech (Wuhan, China). Rabbit anti-SUMO-1, -2, and -3 antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Mouse monoclonal anti-Flag M2 and mouse anti-HA antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Mouse anti-myc, mouse monoclonal anti-Ub, and mouse control IgG were obtained from Abmart (Shanghai, China), Santa Cruz (CA, USA), and Boster (Wuhan, China), respectively. MG132, a proteasome inhibitor, and complete protease inhibitor cocktail were purchased from Calbiochem (San Diego, CA, USA) and Roche (Indianapolis, IN), respectively. N-Ethylmaleimide (NEM) and cycloheximide (CHX) were obtained from Sigma-Aldrich. Western and immunoprecipitation (IP) lysis buffer and radioimmunoprecipitation assay (RIPA) lysis buffer were purchased from Beyotime (Jiangsu, China).
Plasmid construction.
Flag-3D and HA-3D were generated by inserting the EV71 3D coding sequence into pCAGGS as previously described (44, 45). All the mutations of 3D (K159R, L150A/D151A, L150A/D151A/L152A, K159R/L150A/D151A/L152A, V263A/S264A/L265A, and I317A/D318A/L319A) were created by overlapping extension PCR. Recombinant EV71 mutants harboring 3D mutations were constructed by overlapping extension PCR using the infectious clone as a template at the indicated sites. Verification of the mutants was performed by RNA extraction, reverse transcription, and PCR (39).
pcDNA3-HA-SUMO-1 (Addgene plasmid 21154) was obtained from Junying Yuan (46). SRa-HA-SUMO-2, pcDNA3/HA-SUMO-3, and Flag-SENP-1 (Addgene plasmids 17360, 17361, and 17357) were gifts from Edward Yeh (47, 48). pCMV hUbc9 wt HA (Addgene plasmid 14438) was a gift from Peter Howley (49). pcDEF-HA-SUMO-1, pcDEF-Myc-Ubc9, and pcDEF-Flag-SUMO-2 were generously provided by Hong Tang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) (50). pRK-Flag-SUMO-2 was kindly provided by Yanyi Wang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) (51). Myc–SUMO-1 and V5–SENP-1 were constructed by inserting the SUMO-1 and SENP-1 coding sequences, respectively, into pCAGGS. Flag–SUMO-1, -2, and -3 were created by amplifying and inserting their coding sequences into pCAGGS. HA-K48-Ub and HA-K63-Ub were mutants of HA-Ub with all the lysine residues except K48 and K63 mutated into arginines (52). HA-UB-KO (knockout) was a mutant of ubiquitin created by the replacement of lysine residue 63 (Lys63) of HA-K63-Ub with arginine.
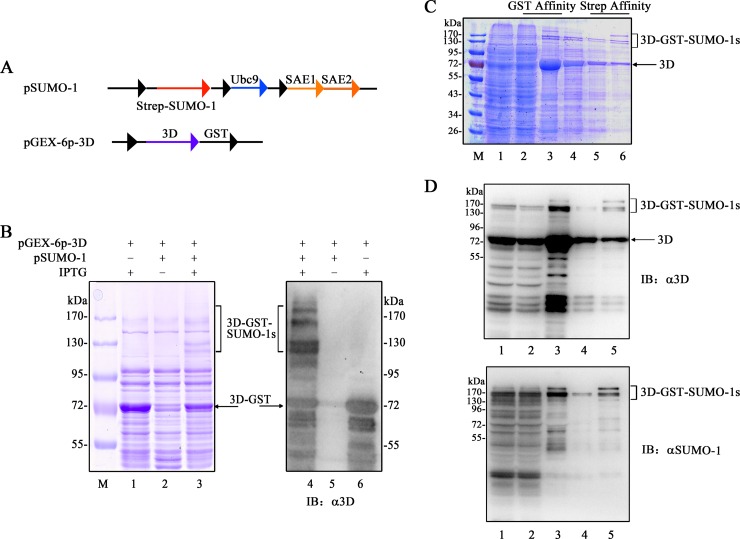
The plasmid expressing WT 3D was generated by cloning the 3D gene into a pET26-Ub vector (53, 54), and the 3D K159R, K159A, and L150A/D151A mutants were further constructed using the WT plasmid as the template. 3D was cloned into pGEX-6p-1 (GE Healthcare) to produce glutathione S-transferase (GST) tag-3D. pSUMO-1 (Addgene plasmid 52258) containing SAE1 and SAE2 was from Primo Schär (55). The His tag in pSUMO-1 was replaced with Strep-tag II (an eight-residue minimal peptide sequence [Trp-Ser-His-Pro-Gln-Phe-Glu-Lys] that exhibits intrinsic affinity toward streptavidin columns) to create pSUMO-1-Strep.
Protein preparation and in vitro polymerase assays.
The plasmids for the expression of WT 3D and its variants were transformed into Escherichia coli strain BL21(DE3)(pCG1) (kindly supplied by Craig Cameron) for expression (54). The bacteria were cultured and purified as described previously, except that the induction condition before harvesting was 11 h at 25°C (56). The preparation of the 33-nucleotide RNA template (T33) was as described previously (57, 58). The 10-nucleotide RNA primer (P10) was purchased from Integrated DNA Technologies. The T33/P10 construct was annealed at 45°C for 3 min at a molar ratio of 1:0.9 in an RNA annealing buffer (RAB) (50 mM NaCl, 5 mM Tris [pH 7.5], 5 mM MgCl2). A typical 20-μl reaction mixture containing 6 μM 3D or its mutants, 50 mM Tris (pH 7.0), 20 mM NaCl, 55 mM KCl, 5 mM MgCl2, 4 mM TCEP [tris(2-carboxyethyl)phosphine], 300 μM ATP, 4 μM (T33/P10) construct, and 300 μM GTP was incubated at 22.5°C for 15, 30, or 60 min before being quenched with an equal volume of stop solution (95% [vol/vol] formamide, 20 mM EDTA [pH 8.0], 0.02% [wt/vol] bromphenol blue, and 0.02% [wt/vol] xylene cyanol). The quenched samples were heated at 100°C for 3 min. The RNA species in the reaction mixture were resolved by 20% polyacrylamide-7 M urea gel electrophoresis before staining with Stains-All (Sigma-Aldrich). Color images obtained by scanning the stained gels were converted to grayscale images prior to quantification by Image J (http://imageJ.nih.gov/ij/). The Stains-All-based quantification method was used previously to assess the amounts of RNAs with similar lengths (58).
In vitro SUMOylation assay.
The SUMOylated 3D proteins were produced by introducing pGEX-6p-3D into E. coli BL21(DE3) harboring pSUMO-1-Strep by electroporation. The strains were grown under selective pressure at 50 mg/liter ampicillin and 25 mg/liter streptomycin for coexpressing the two plasmids. The bacterial strains were cultured, and the protein was purified as previously described in the purification of 3D (53, 59), except that the cell lysates were first loaded on a GSTrap HP column (GE Healthcare, Waukesha, WI, USA). Subsequently, the eluted fractions were passed through a StrepTrap HP column (GE Healthcare, Waukesha, WI, USA). After the two steps of enrichment, the extractions were separated by anion exchange and size exclusion chromatography for further purification (55). The collected fractions were electrophoresed on SDS-PAGE and detected by Coomassie blue staining or immunoblotting using a Bio-Rad imaging system. The immunoblot analysis was performed as previously described (45).
SUMOylation and ubiquitination assays.
SUMOylation and ubiquitination assays were performed with Dynabeads protein G (Life Technology, Grand Island, NY) for IP. 293T cells were cotransfected with the indicated plasmids using the calcium phosphate reagent ProFection (Promega, Southampton, United Kingdom). For assay of ubiquitination, 30 h posttransfection, cells were lysed with Western and IP lysis buffer plus complete protease inhibitors according to the manufacturer's instructions. For the SUMOylation assay, after 30 h of transfection, cells were harvested and lysed with Western and IP lysis buffer plus complete protease inhibitors and 20 mM NEM. The lysates were centrifuged at 16,000 × g at 4°C for 10 min. According to the protocol, 10 μg of corresponding antibody was incubated with 50 μl of protein G Dynabeads for 20 min before incubation with the supernatants from the cell lysis for 25 min. Following several washes with PBST (PBS with 0.02% Tween 20), the complexes were boiled with electrophoresis sample buffer, followed by immunoblot (IB) analysis with the indicated antibodies as previously described (45, 52).
In assays involving EV71 infection, cells were cultured on 100-mm dishes. At 24 h posttransfection, 293T cells were infected with EV71 at an MOI of 10 for 18 h before harvesting. For the infection of RD cells, the cells were lysed with RIPA lysis buffer after infection with EV71 at an MOI of 10 for 8 h. The supernatants were collected for IP as described above.
CHX chase analysis.
The CHX chase experiment was performed as described previously (60). Cells were seeded on 35-mm culture dishes at a density of 6 × 106 cells. The cells were transfected with Flag-3D and its mutant Flag-K159R/L150A/D151A/L152A. After 24 h, the cells were treated with 100 μg/ml of CHX dissolved in dimethyl sulfoxide (DMSO) in the presence or absence of 20 μM MG132. Cells were then collected at different time points and subjected to immunoblotting. The quantification of protein levels was analyzed with Image J software.
Statistical analysis.
Each of the experiments was repeated at least three times. Data from the viral titer, one-step growth curve, primer-dependent polymerase assays, and CHX chase are presented as means ± standard deviations. Student's t test was used to determine the statistical significance between different tests, with significance defined as a P value of <0.05 or <0.01.
RESULTS
3D polymerase is SUMOylated during EV71 infection.
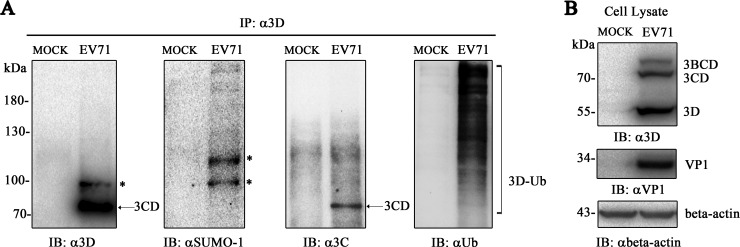
SUMOylation is one of the PTMs that play critical roles in diverse biological processes. However, little is known concerning how host PTM systems interact with enterovirus 71. Online prediction suggested that EV71 3D polymerase may bear SUMOylation sites. Here, we performed experiments to investigate the SUMOylation of 3D during EV71 infection. RD cells infected with EV71 were harvested at 8 h postinfection before immunoprecipitation with an anti-3D antibody. Structural proteins VP1 and 3D polymerase of EV71 were detected in cell lysates, and the anti-3D antibody was able to detect 3D and its precursors, 3BCD and 3CD (Fig. 1B). SUMOylation and ubiquitination of 3D were detected by anti-SUMO-1 antibody and anti-Ub antibody (Fig. 1A). Notably, IP with anti-3D antibody could obtain 3D, 3CD, and 3BCD complexes, whereas 3C was proven to be SUMOylated, and bands detected by anti-SUMO-1 antibody might be a mixture of 3D SUMOylation, 3CD SUMOylation, and 3BCD SUMOylation. Anti-3C antibody detected only 3CD protein (∼73 kDa), anti-3D antibody detected both 3CD and SUMOylated 3D, and anti-SUMO-1 antibody detected SUMOylated bands (Fig. 1A). The bands detected by anti-3C antibody were different from those detected by anti-SUMO-1 antibody or by anti-3D antibody, excluding the influence of 3CD and 3BCD SUMOylation (Fig. 1A). The SUMOylated 3Dpol (asterisks) was detected by IP with anti-3D antibody, followed by Western blotting with anti-SUMO-1 antibody or anti-3D antibody. The result of anti-SUMO-1 antibody treatment showed one more SUMOylated band than that with anti-3D antibody, which might be due to the antigenic epitope limitation of anti-3D antibody. Altogether, these results confirmed that EV71 3D could be SUMOylated during infection.
FIG 1.
EV71 3D is SUMOylated during infection. RD cells (1 × 107) were infected with EV71 (MOI = 10) and harvested at 8 h postinfection before immunoprecipitation with an anti-3D antibody. IP and IB analyses were performed with the indicated antibodies in the presence of NEM. (A) Immunoblot detection by anti-SUMO-1, anti-Ub, anti-3C, and anti-3D antibodies during EV71 infection after immunoprecipitation by the anti-3D antibody. (B) Immunoblot analysis of cell lysis during infection. Anti-VP1, anti-3D, and anti-β-actin antibodies were used to detect the expression of VP1, 3D, and β-actin during infection. Lysis of RD cells without infection was set as a mock control. SUMO-1-modified 3D, ubiquitin-modified 3D, and 3CD are indicated by asterisks, brackets, and arrows, respectively.
EV71 3D is modified by SUMO-1 and -3.
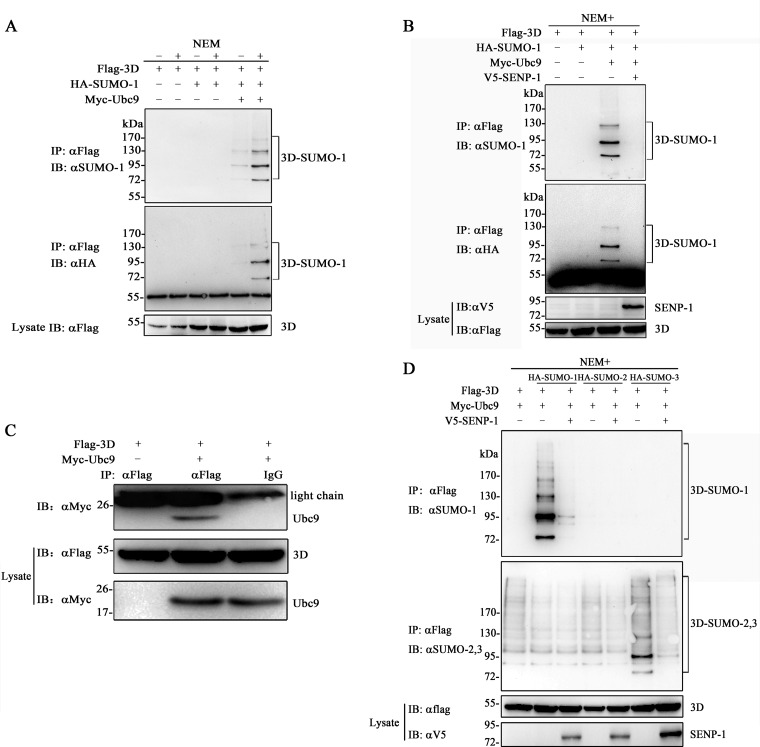
We then performed experiments to confirm the SUMOylation of EV71 3D. SUMOylation is reversible by SENPs, which could be irreversibly inhibited by NEM; hence, the successful detection of this modification relies on the addition of NEM to the lysis buffer. We designed two experiments (Fig. 2A and B). Flag-3D, HA–SUMO-1, and Myc-Ubc9 (SUMO E2-conjugating enzyme) were overexpressed in 293T cells. At 30 h posttransfection, the cells were subjected to immunoprecipitation with anti-Flag antibody. Higher-molecular-mass bands above unmodified 3D (∼52 kDa) ranging between 80 and 130 kDa were detected by an anti-SUMO-1 or anti-HA antibody in the presence of NEM (Fig. 2A). The basal SUMO-1 moiety of 3D was removed when coexpressed with SENP-1 (Fig. 2B). For further verification, we performed coimmunoprecipitation experiments to study the relationship between 3D and Ubc9 with an anti-Flag antibody. The result showed that the SUMO-conjugating enzyme Ubc9 was present in the immunoprecipitated complex (Fig. 2C), indicating that 3D could interact with Ubc9. Furthermore, when 3D was coexpressed with SUMO-1, -2, and -3 in 293T cells, 3D was preferentially modified by SUMO-1 and -3 in a poly-SUMO chain conjugation, which could be de-SUMOylated by SENP-1 (Fig. 2D).
FIG 2.
EV71 3D is modified by SUMO-1 and -3. (A) SUMOylation of EV71 3D in the presence of NEM. 293T cells were transfected with Flag-3D, Myc-Ubc9, and HA–SUMO-1. The cells were lysed with or without the addition of NEM, which was added to a final concentration of 20 mM prepared in ethanol. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies for the SUMOylation assay. (B) De-SUMOylation of 3D by SENP-1. 293T cells were transfected with Flag-3D, Myc-Ubc9, HA–SUMO-1, and V5–SENP-1. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies in the presence of NEM. (C) 3D interacts with Ubc9. 293T cells were transfected with Flag-3D, Myc-Ubc9, or empty vector. Coimmunoprecipitation was performed with anti-Flag or control mouse IgG, and the immunoblots were probed with the corresponding antibodies. (D) 3D was modified by SUMO-1 and -3. Flag-3D, Myc-Ubc9, HA–SUMO-1, HA–SUMO-2, and HA–SUMO-3 were overexpressed in 293T cells. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies in the presence of NEM. Molecular mass markers are shown. SUMO-1-modified Flag-3Ds are indicated by brackets.
K159 and L150/D151/L152 are responsible for the SUMOylation of EV71 3D.
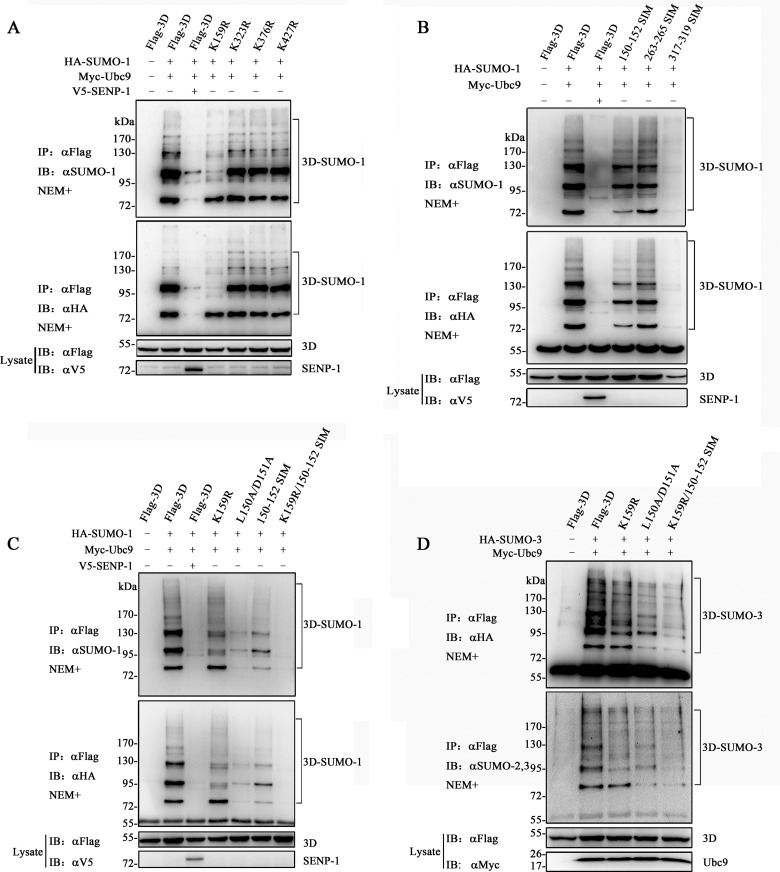
SUMO-1 conjugation has been reported to bind virtually to target proteins to regulate physiological processes, whereas SUMO-2 and -3 are more widely expressed as free, unconjugated forms, which are available for stress responses (22, 61). The unconjugated pool of SUMO-1 is smaller than that of SUMO-2 and -3 (62); therefore, our study focused on SUMO-1 modification. Bioinformatics analysis with the SUMOplot analysis program predicted that several lysine residues in 3D may be modified by SUMO. K159, K376, and K427 of 3D following the consensus motif ψ-K-X-D/E are the top three lysine residues on the list (see Fig. 4A). K323 fits the inverted SUMOylation consensus motif E/D-X-K-ψ (see Fig. 4A). The four lysine residues were mutated to arginines to create K159R, K376R, K427R, and K323R mutants. 3D and its mutants were coexpressed with SUMO-1 and Ubc9 in 293T cells to perform SUMOylation assays. Immunoprecipitation experiments indicated that the K323R, K376R, and K427R mutants had no obvious effect on SUMO-1 reduction, whereas K159R largely abolished the larger migration bands above 90 kDa (Fig. 3A). These results also indicated that, besides K159, there is likely to be more than one site that could be modified.
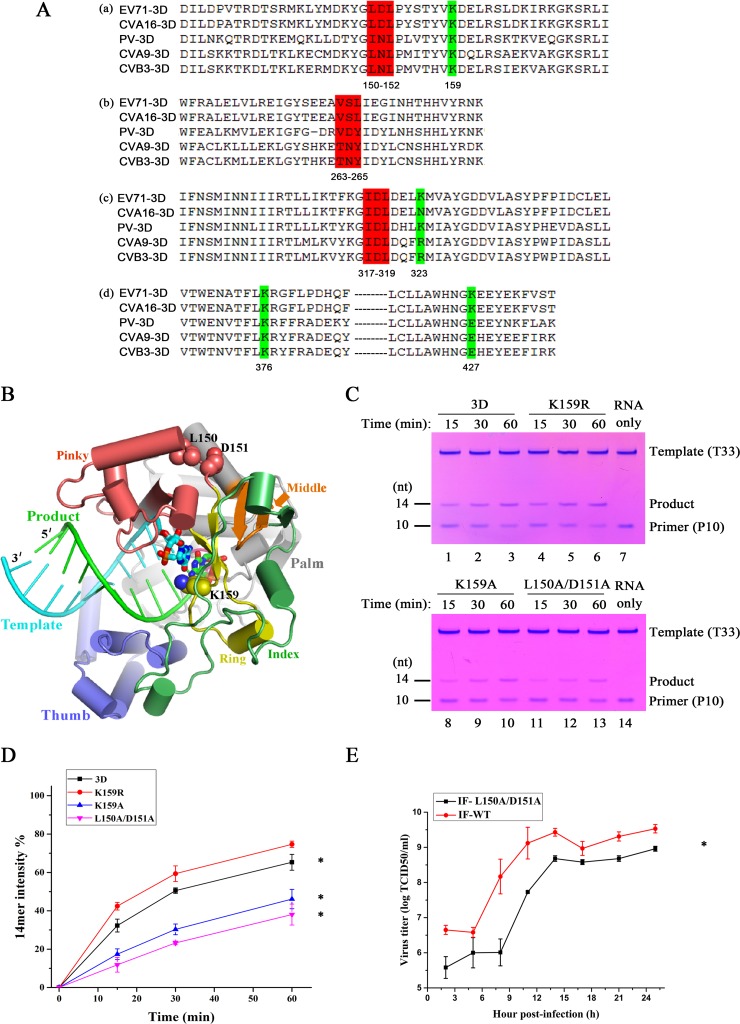
FIG 4.
Assessments of the relevance of the 3D SUMOylation sites to polymerase activity and virus replication. (A) Schematic representation of the potential lysine residues and SIMs in enterovirus polymerase by sequence alignment. Polyprotein sequences of enterovirus 71 (GenBank accession no. AB204852), coxsackievirus A16 (GenBank accession no. AY790926), poliovirus (GenBank accession no. KT353719), coxsackievirus A9 (GenBank accession no. D00627), and coxsackievirus B3 (GenBank accession no. M16572) were used in the alignment. (a) Putative sites of 150-152 SIM and K159. (b) Putative sites of 263-265 SIM. (c) Putative sites of 317-319 SIM and K323. (d) Putative sites of K376 and K427. The SIMs are shaded in red and the lysine residues in green. (B) EV71 3D elongation complex crystal structure with a closed conformation active site (Protein Data Bank [PDB] entry 5F8J) depicting structural features of the SUMOylation sites. The polymerase adopts a circular human right hand architecture with thumb (in slate), palm (in gray), and finger domains surrounding the active site. The finger domain is further divided into index (in green), middle (in orange), ring (in yellow), and pinky (in pink) finger subdomains. The template (in cyan) and product (in green) RNA duplex are shown in a ladder format with the active-site nucleotide pair shown as thick sticks. K159 (side chain in spheres) is within the ring finger subdomain and is an active-site residue interacting with the nascent base pair, while L150 and D151 (side chains in spheres) are on the polymerase surface, forming part of the pinky finger. (C and D) In vitro polymerase activity of 3D and its K159R, K159A, and L150A/D151A mutants. The time-dependent conversion of the primer P10 into the 14-mer product was compared among the WT and three mutant enzymes. The values and error bars (means ± standard deviations) in panel C were taken from the results of three independent experiments with representative gels shown in panel B (Student's t test; *, P < 0.05). nt, nucleotides. (E) Growth curves of EV71 and its IF-L150A/D151A mutant in RD cells. RD cells (5 × 105) were infected with wild-type EV71 or its IF-150A/D151A variant (MOI = 10) and harvested at the indicated times postinfection. Viral growth curves were generated by plotting the virus titer against time. The experiments were carried out in triplicate. Significant differences were determined using the Student t test (*, P < 0.01).
FIG 3.
Identification of Lys159 and Leu150/Asp151/Leu152 as major SUMOylation sites. (A) SUMOylation assay of 3D and its single-site mutants based on the canonical consensus motif. The lysine residues at positions 159, 323, 376, and 427 were replaced by arginines to create K159R, K323R, K376R, and K427R. (B) SUMOylation assay of 3D and its SIM mutants. Leu150/Asp151/Leu152, Val263/Ser264/Leu265, and Ile317/Asp318/Leu319 were mutated into three consecutive alanine residues, namely, 150-152 SIM, 263-265 SIM, and 317-319 SIM. (C) SUMOylation assay by SUMO-1 of 3D and its selected mutants. K159R/150-152 SIM was generated by mutating Lys159 into Arg159 and Leu150/Asp151/Leu152 into Ala150/Ala151/Ala152. L150A/D151A was created by mutating Leu150/Asp151 into Ala150/Ala151. (D) SUMOylation assay by SUMO-3 of 3D and its selected mutants. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies in the presence of NEM. Molecular mass markers are shown. SUMO-1-modified Flag-3Ds are indicated by brackets.
As described above, SIMs feature hydrophobic amino acid cores flanked by acidic residues (V/I)(V/I)(D/E)(V/I/L)(T/D/E) and (V/I)(V/I)(V/I/L)(V/I/L)(D/E) (14, 17). We created three mutants of 3D, namely, 150-152 SIM and 317-319 SIM, which were created by inspecting the 3D amino acid sequence, and 263-265 SIM, which was predicted by a SUMO-binding-motif online prediction website (GPS-SBM) (Fig. 4A) (63). Three alanines were introduced to replace the LDL (150 to 152), VSL (263 to 265), and IDL (317 to 319) of the SIMs to create the mutants. Figure 3B shows that the band modified by SUMO-1 (∼80 kDa) was considerably decreased in 150-152 SIM, whereas 263-265 SIM maintained almost the same bands as the wild-type 3D. 317-319 SIM completely attenuated the modified bands, which was likely due to protein reduction in lysis. Considering the characteristics of the mutants, we constructed a mutant, K159R/150-152 SIM, by combining the K159R mutation and 150-152 SIM. This mutant had remarkably decreased SUMO-1 modification in comparison with 3D or K159R and with 150-152 SIM (Fig. 3C). In the assay of SUMO-3, K159R lost the bands above 90 kDa, which were similar to those in SUMO-1 (Fig. 3D), and K159R/150-152 SIM almost totally lost SUMOylation. These results indicated that SUMO-3 modification of 3D shares the same sites with SUMO-1. Furthermore, the single mutation of K159 resulted in the reduction of higher bands but not the band around 80 kDa in the assay with both SUMO-1 and SUMO-3 (Fig. 3C and D), indicating that position 159 was first modified by SUMO-3 and subsequently by SUMO-1 into a poly-SUMOylated chain. These results were observed because SUMO-1 could not be elongated and always acts as a terminator of poly-SUMO chains (6, 7). In conclusion, K159 and L150/D151/L152 may be responsible for the SUMOylation of EV71 3D.
Mutations of SUMOylation sites impair 3D polymerase activity and virus replication.
Of the two SUMOylation sites in 3D, K159 is a highly conserved active-site residue in RNA-dependent RNA polymerase (RdRp) motif F and within hydrogen-bonding distance from the edge of the nascent base pair of the RdRp elongation complex, while the 150-to-152 site is on the surface of 3D and does not directly participate in RNA binding, NTP entry, and catalysis (Fig. 4B) (56). With an aim of assessing the relevance of both SUMOylation sites to the polymerase function, we conducted primer-dependent polymerase assays (58, 64) for WT 3D and the K159R, K159A, and L150A/D151A mutants. With the T33/P10 RNA construct and GTP/ATP as the only substrates, the 10-nucleotide primer (P10) was expected to be elongated by 4 nucleotides, producing a 14-nucleotide product (58). For the WT enzyme, the majority of the primer was converted to the product within a 60-min incubation time (Fig. 4C and D). The K159R mutant enzyme exhibited a similarly high conversion yield, while the K159A and L150A/D151A mutants had obviously lower yields, with only 40% of the primer converted, as judged by the band intensity (Fig. 4C and D). These data, in general, suggest that both SUMOylation sites in 3D may modulate EV71 polymerase activity, despite their differences in structural properties based on available crystallographic data (56, 64).
To assess the effect of 3D SUMOylation sites on EV71 replication, we constructed the IF-K159R and IF-K159R/150-152 SIM recombinant viruses. Unfortunately, both recombinant viruses were unable to be rescued. However, transfection of WT 3D protein and K159R recombinant RNA resulted in the rescue of wild-type virus. A L150A/D151A mutant was constructed to map the critical region. Since the L150A/D151A mutant can greatly decrease the SUMO-1 modification (Fig. 3C), we also tested the IF-L150A/D151A recombinant virus. As shown in Fig. 4E, the growth curve of this recombinant virus was significantly lower than that of wild-type virus. Therefore, the lower titer of IF-L150A/D151A was likely due to the decreased enzyme activity and/or lack of SUMOylation.
3D protein is SUMOylated in vitro.
SUMOylation is a process of enzyme-catalyzed reactions. Two-component vector systems carrying SAE1, SAE2, Ubc9, SUMO-1, and the target protein in E. coli were used to generate SUMOylated proteins in vitro (55). In the present study, we used the method by transforming E. coli with pSUMO-1-Strep and pGEX-6p-3D to produce SUMOylated proteins (Fig. 5A). SUMO-1-modified proteins were confirmed by SDS-PAGE and immunoblotting. Under IPTG (isopropyl-β-d-thiogalactopyranoside) induction, the results of Coomassie blue staining and immunoblotting showed that BL21(pSUMO-1-Strep)(pGEX-6p-3D) strains not only expressed 3D-GST, but also 3D–GST–SUMO-1s (Fig. 5B, lanes 3 and 4). In contrast, BL21(pGEX-6p-3D) produced only 3D-GST (Fig. 5B, lanes 1 and 6). After induction, the cell lysates of BL21(pSUMO-1-Strep)(pGEX-6p-3D) strains were subjected to subsequent GST and Strep affinity chromatography. The collected protein samples were resolved by SDS-PAGE and visualized by Coomassie blue staining (Fig. 5C). SUMOylated 3D was verified by immunoblotting with the anti-3D or anti-SUMO-1 antibody (Fig. 5D). These results indicate that EV71 3D can be modified by SUMO-1 in vitro.
FIG 5.
In vitro SUMO-1 modification of 3D. (A) Schemes of the gene arrangement of pSUMO-1-Strep and pGEX-6p-3D. pSUMO-1-Strep carrying Ubc9 (SUMO-conjugating enzyme E2), SAE1 and SAE2, and SUMO-1 (tagged at the N terminus with Strep tag) was used for the expression of the enzymes required for SUMOylation. pGEX-6p-1 carrying 3D was used for the expression of GST-tagged 3D. (B) E. coli BL21(pGEX-6p-3D) and BL21(pSUMO-1-Strep)(pGEX-6p-1) were used to produce GST-3D and GST–3D–SUMO-1. The supernatants of the two strains were analyzed by Coomassie blue staining (lanes 1 to 3) and immunoblotting (lanes 4 to 6) with the anti-3D antibody under the induction of IPTG (500 μM) at 25°C for 10 h. BL21(pSUMO-1-Strep)(pGEX-6p-1) without IPTG induction was used as the control (lanes 2 and 5). (C and D) Cell lysates of E. coli BL21(pSUMO-1-Strep)(pGEX-6p-1) were subjected to subsequent GST and Strep affinity purification to separate SUMOylated proteins. Fractions during purification of SUMOylated proteins were analyzed by SDS-PAGE and subsequent Coomassie blue staining (C) and immunoblotting (D) with anti-3D and anti-SUMO-1 antibodies. M, marker. (C) Lanes: 1, crude lysate supernatants; 2, GST column flowthrough; 3, GST column eluted fraction; 4, dialyzed elution fractions; 5, Strep column flowthrough; 6, Strep column eluted fraction. (D) Lanes: 1, crude lysate supernatants; 2, GST column flowthrough; 3, GST column eluted fraction; 4, Strep column flowthrough; 5, Strep column eluted fraction. Molecular mass markers are shown. SUMO-1-modified and unmodified GST-3Ds are indicated by brackets and arrows, respectively.
3D is ubiquitinated in a SUMOylation-dependent manner.
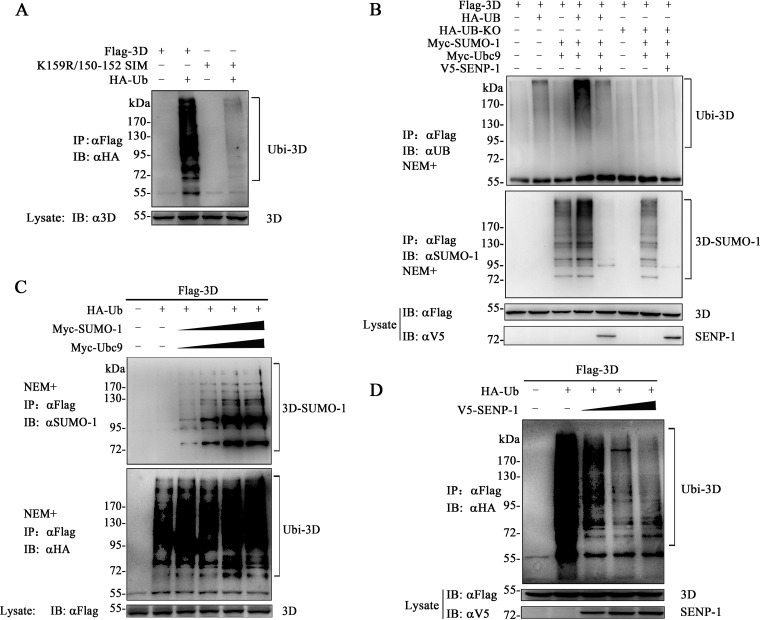
The ubiquitin-proteasome system (UPS) is required for the effective replication of both coxsackievirus B3 (CVB3) (65) and EV71 (66). Since SUMOylation can interact with ubiquitination in many viral proteins, we determined whether EV71 3D is ubiquitinated. Flag-3D and K159R/150-152 SIM were coexpressed with HA-Ub in 293T cells for the ubiquitination assay (Fig. 6A). The cell lysates were subjected to immunoprecipitation with the anti-Flag antibody. Immunoblotting by the anti-HA antibody showed that 3D could be polyubiquitinated (poly-Ub), whereas the mutant K159R/150-152 SIM almost lost this modification.
FIG 6.
Conjugation of ubiquitin to 3D based on its SUMOylation levels. (A) Ubiquitination assay of 3D and K159R/150-152 SIM. 293T cells were transfected with the indicated constructs. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies for the ubiquitination assay. (B) Ubiquitination and SUMOylation assay of 3D. 293T cells were transfected with the indicated constructs. In particular, HA-UB-KO was coexpressed to remove endogenous ubiquitin, while V5–SENP-1 was coexpressed for de-SUMOylation. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies. (C and D) SUMOylation affected the ubiquitination of 3D in a dose-dependent manner. 293T cells were transfected with 2 μg of Flag-3D and 2 μg of HA-Ub or Myc-Ubc9 (0.5, 1, 1.5, and 2 μg), Myc–SUMO-1 (0.5, 1, 1.5, or 2 μg), or V5–SENP-1 (0.5, 1, 1.5, and 2 μg) in 60-mm dishes. The empty vector was used to adjust the total DNA. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies. Molecular mass markers are shown. SUMO-1- and ubiquitin-modified 3Ds are indicated by brackets.
The above-described experiments implied that ubiquitination and SUMOylation of 3D may share the same lysine residues and that 3D ubiquitination was SUMOylation related. To determine the association between the two modifications, SENP-1 and lysineless ubiquitin (Ub-KO) were introduced into 3D SUMOylation and ubiquitination experiments. Ub-KO terminates the ubiquitin chain, and it was created by replacing all lysines with arginines. Ub-KO had a dose-dependent effect on the ubiquitin system when Ub-KO was overexpressed to compete with the endogenous ubiquitin (67). In our study, 3D ubiquitination was increased when Ub was coexpressed with SUMO and Ubc9 (Fig. 6B). Coexpression of SENP-1 decreased both 3D SUMOylation and ubiquitination, which was in accordance with the results for the mutant K159R/150-152 SIM. The coexpression of Ub-KO resulted in the reduction of 3D ubiquitination, but it had hardly any effect on 3D.
We further investigated whether the 3D ubiquitination level could be altered according to the gradient expression of SENP-1 or SUMO-1 in 293T cells. The ubiquitination level was enhanced when the SUMOylation of 3D was increased (Fig. 6C). On the other hand, the increased expression of SENP-1 gradually reduced the amount of ubiquitin conjugated on 3D (Fig. 6D). Taken together, these data imply that the two modifications occur sequentially, with ubiquitination being highly SUMOylation dependent.
SUMO-1 modification enhances the stability of 3D.
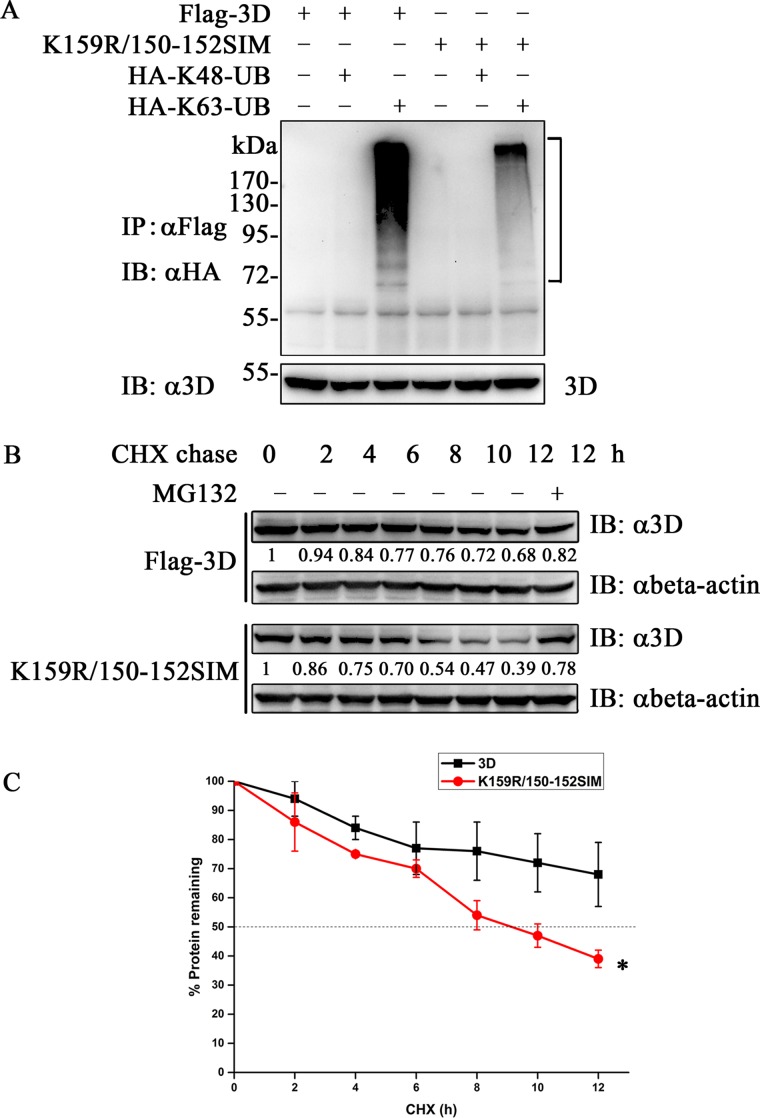
Normally, the poly-Ubs are primarily linked through K48 or K63 to play diverse roles in regulating cellular activities, which account for 52% and 38% of all ubiquitination events, respectively (68). K48-linked ubiquitination chains are involved in proteasomal degradation, whereas K63-linked ubiquitination is a docking site for mediating protein-protein interactions or conformational changes (69). To explore which kind of poly-Ub 3D is linked and whether 3D is degraded, we transfected 3D and K159R/150-152 SIM, together with HA-K48-Ub or HA-K63-Ub, to perform the ubiquitination assay. 3D was specifically modified by K63-linked ubiquitin, whereas the K159R/150-152 SIM significantly reduced the conjugation (Fig. 7A). Notably, no degradation of 3D was observed in the assay (Fig. 7A).
FIG 7.
SUMOylation of 3D increased protein stability. (A) 3D was modified by K63-linked ubiquitin. Flag-3D or K159R/150-152 SIM and HA-K48-Ub or HA-K63-Ub were transfected into 293T cells for the ubiquitination assay. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies. The ubiquitin-modified 3Ds are indicated by brackets. (B and C) Cycloheximide chase assay of 3D and K159R/150-152 SIM. 293T cells (1 × 105) were transfected with 3D or K159R/150-152 SIM. After 24 h, the cells were treated with 100 μg/ml of CHX for the indicated periods in the presence or absence of 20 μM MG132. The cell lysates were analyzed by immunoblotting with the anti-Flag antibody. 3D and its mutant bands were quantitated by Image J software. The amounts of 3D and its mutant relative to levels in untreated cells are indicated below the bands. The plot shows the half-lives of 3D and K159R/150-152 SIM from three independent experiments. Significant differences were determined using the Student t test (*, P < 0.01). Means ± standard deviations are shown.
CHX chase experiments were then carried out to determine the stability difference between 3D and K159R/150-152 SIM. 3D exhibited a longer half-life than K159R/150-152 SIM, and the MG132 treatment attenuated K159R/150-152 SIM degradation (Fig. 7B and C). These results imply that SUMOylation-related K63-linked ubiquitination may stabilize 3D.
An elevated SUMO-1 expression level increases EV71 replication.
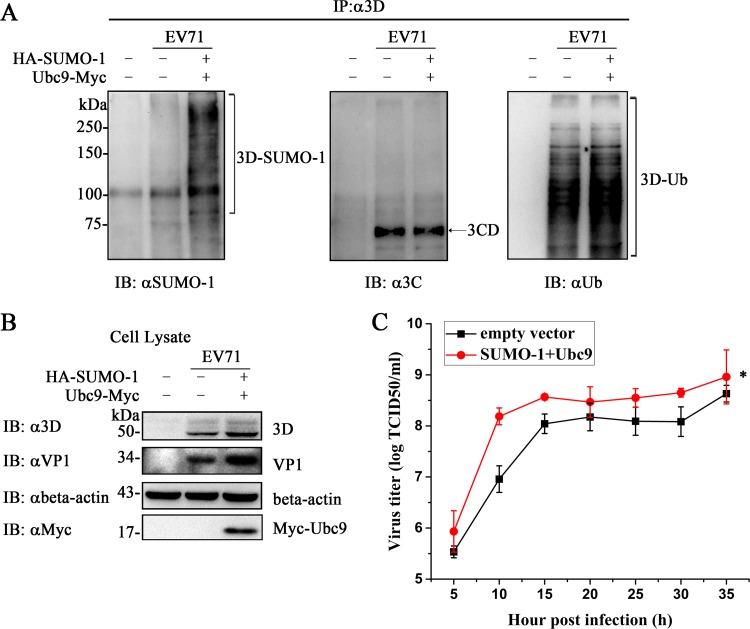
To analyze the effects of SUMO-1 overexpression on virus production, SUMO-1 and Ubc9 were transiently transfected into 293T cells. The cells were subsequently infected with EV71 for 18 h. The SUMOylation and ubiquitination levels of viral 3D increased under the condition of SUMO-1 overexpression (Fig. 8A). 3CD protein was detected by anti-3C antibody. These results implied that IP by an anti-3D antibody could coimmunoprecipitate 3D, 3CD, and 3BCD, while data on the anti-SUMO-1 antibody indicated that 3D was indeed modified by SUMO-1. The expression levels of viral 3D and VP1 were increased when the SUMOylation level was enhanced (Fig. 8B). The titer of EV71 produced from 293T cells with increased SUMOylation was higher than that from 293T cells transfected with the empty vector (Fig. 8C). Collectively, these results indicate that elevation of the global cell SUMOylation increased both SUMOylation and ubiquitination levels of 3D, which augmented 3D expression and consequently enhanced viral replication.
FIG 8.
SUMO-1 modification promoted EV71 replication. 293T cells (7 × 106) were transfected with HA–SUMO-1, Myc-Ubc9, or empty vector in 100-mm dishes. At 24 h posttransfection, the cells were infected with EV71 (MOI = 10) for 18 h. One of the dishes with no transfection was used as the control. Immunoprecipitation and immunoblot analyses were performed with the indicated antibodies. (A) SUMOylation and ubiquitination assay by immunoprecipitation of 3D in cells with elevated SUMO-1 after EV71 infection. (B) Immunoblot analysis of cell lysis during infection. Anti-VP1, anti-3D, and anti-β-actin antibodies were used to detect the expression of VP1, 3D, and β-actin during infection. Anti-myc antibody was used to detect the expression of Ubc9. SUMO-1- and ubiquitin-modified 3Ds are indicated by brackets and 3CD by an arrow. (C) Growth curves of EV71 produced from transfected 293T cells in RD cells. 293T cells were transfected with HA–SUMO-1, Myc-Ubc9, or empty vector for 24 h, followed by infection with EV71 at an MOI of 10 for 18 h. The cells were harvested at the indicated times postinfection to plot the growth curves. The data are presented as means ± standard deviations obtained from the results of three independent experiments. Significant differences were determined using the Student t test (*, P < 0.01).
DISCUSSION
A number of viral proteins have been reported to be modified by SUMO moieties (22, 62). However, knowledge concerning SUMOylation in enteroviruses is limited. Only EV71 3C protease is SUMOylated, while coxsackievirus B5 is involved in the host cell SUMOylation system in the family Picornaviridae (38, 62, 70). Current understanding of viral polymerase SUMOylation is limited to RdRp of Turnip mosaic virus (71), DNA polymerase subunit UL44 of HCMV (25), nonstructural protein 5 (NS5) of dengue virus (72), and polymerase basic protein 1 (PB1) of influenza virus (30). In the currently study, we reveal that the 3D protein of EV71, an RNA-dependent RNA polymerase, is SUMOylated. We found that EV71 3D polymerase is modified by SUMO-1 both in vivo and in vitro and that 3D is stabilized by SUMOylation-related ubiquitination. K159 and L150/D151/L152 are responsible for 3D SUMOylation. Mutations of SUMOylation sites impaired 3D polymerase activity and virus replication. Moreover, elevated SUMOylation levels during EV71 infection resulted in promotion of 3D stability, which enhanced viral replication. We propose that SUMOylation at specific 3D sites helps to maintain the cellular level of 3D protein and that the corresponding de-SUMOylation may be necessary for 3D to resume its polymerase activity. The 3D polymerase function may be required only when it is responsible for RNA genome replication, which is SUMOylation free. These findings imply that EV71 exploits host proteins for effective infection and that the SUMO system is likely to be a putative antiviral target.
Online prediction indicated that 3D bears both covalent and noncovalent sites for SUMOylation. According to the predictions, K159, K427, and K376 are the top three lysine residues following the pattern of the ψ-K-X-D/E motif (http://www.abgent.com/sumoplot.html), while V263/S264/L265 is the putative SIM (63). In our study, K159 and L150/D151/L152 as the SIM were proven to be the SUMOylation sites, while K159 follows the classic consensus motif ψ-K-X-D/E and is the major site for SUMO-1 modification (Fig. 3A). However, although L150/D151/L152 is not within the predicted SUMOylation sites of 3D, they have been proven to be a SIM in our study. Additionally, the inverted consensus motif E/D-X-K-ψ worked for EV71 3C, but not for 3D (38). In fact, the bioinformatics prediction software is based on the calculation of the reported SUMOylation site, and some unknown limitations may exist. Therefore, there may be SUMOylation sites that cannot be predicted by bioinformatics software. For instance, mutation of the predicted SUMOylation sites in the human cytomegalovirus DNA polymerase processivity factor UL44 hardly attenuates the modification (25). Here, our finding of L150/D151/L152 as a SIM site may expand the data pool for developing a higher-confidence algorithm.
Normally, the interplay between SUMOylation and ubiquitination often involves the stability of the target protein (73). SUMOylation and K48-linked ubiquitination of IRF3 share the same lysine residues, and the two processes are competitive, indicating that SUMOylation of IRF3 stabilizes the protein by antagonizing ubiquitination (51). SUMOylation of EV71 3C protease shares the same K52 with its ubiquitination and consequently enhances ubiquitination, which leads to the degradation of 3C (38). Another study showed how the two PTMs cooperatively stabilized target proteins in various ways. For instance, the E3 protein of vaccinia virus is modified by both SUMOylation and ubiquitination. Wild-type E3 exhibited a longer half-life than its SIM mutant. It was speculated that the different stabilities observed between wild-type E3 and the SIM mutant were due to the longer ubiquitin chains conjugated on the SIM mutant (74). SUMOylation stabilizes dengue virus NS5 against proteasome degradation, which supports virus replication (72). SUMO-1 stabilizes phosducin, and the authors inferred that SUMO moieties conjugated on phosducin may mask the region recognized by the ubiquitin-proteasome pathway (75). Following gradient overexpression of SUMO-1 and Ubc9 or SENP-1 with 3D, the results showed that the level of ubiquitination was highly dependent on SUMOylation. Moreover, when Ub was replaced with Ub-KO, 3D ubiquitination was reduced, although it had hardly any effect on 3D SUMOylation. Taken together, these results implied that the poly-SUMO chain likely acts as a signal for 3D ubiquitination. In such a case, ubiquitin might be hybridized with poly-SUMO chains or conjugated on the exposed internal residues of 3D due to the conformational change induced by SUMOylation. The importance of UPS during replication of both CVB3 and EV71 may rely on 3D SUMOylation (65, 66). We found that the degradation of 3D by K63-linked ubiquitination was not observed and that wild-type 3D showed a longer half-life than the SUMOylation-deficient mutant K159R/150-152 SIM. K63-linked polyubiquitination generally has nonproteolytic cellular functions, including DNA damage repair, stress responses, and inflammatory pathways (76). Taken together, SUMO moieties conjugated on 3D act as a signal for K63-linked ubiquitination, which cooperatively stabilizes 3D.
K159 is within RdRp catalytic motif F and is highly conserved in positive-strand RNA viruses (77). Structurally, this residue is located in the polymerase active site and within hydrogen-bonding distance from the nascent base pair (Fig. 4B) (56, 64, 78). Therefore, it is not surprising that the K159A mutation reduces in vitro polymerase activity (Fig. 4C and D). The K159R mutation, however, retained a WT level of polymerase activity in our test, but the corresponding recombinant virus was not viable. Despite being an amino acid with a basic side chain, arginine may be not ideal as a replacement for the native lysine at this critical position. Due to its critical roles in polymerase catalysis, we tend to believe that the defects in polymerase catalysis (although not clarified in our in vitro assays) rather than the loss of the SUMOylation property account for the nonviability of the recombinant IF-K159R virus. Compared to K159, the 150-152 SIM is moderately conserved only in enteroviruses and is located near the polymerase surface (Fig. 4B), and therefore, it is unlikely to play a major role in polymerase catalysis. Although the in vitro enzyme activity of L150A/D151A was lower than that of 3D, the recombinant virus IF-L150A/D151A was successfully rescued. The fact that the titer of IF-L150A/D151A recombinant virus was lower than that of WT EV71 was likely due to the reduced enzyme activity and/or the impairment of SUMOylation. To reinforce this deduction, we adopted a method, by overexpressing SUMO-1, to determine how the SUMOylation system could affect EV71 infection. EV71 infection in 293T cells with elevated SUMOylation showed that more SUMO-1 led to higher ubiquitination of 3D, which increased virus production compared with the cells transfected with empty vector. It has been reported that EV71 3C K52R SUMOylation-deficient virus showed elevated virus titers compared to wild-type virus (38). The increased titer of EV71 from SUMO-1-overexpressing cells was likely due to higher SUMOylation and ubiquitination on 3D, which may eventually stabilize 3D and the augmented polymerases, promoting EV71 replication. In addition to the results from EV71 3C SUMOylation, the increased global SUMOylation level is beneficial for EV71 replication. Although 3C and 3D SUMOylation have opposite consequences during EV71 replication, the influence of 3D SUMOylation is greater than that of 3C SUMOylation. As a polymerase, any modification of 3D may affect virus replication, and in the case of SUMOylation, the impact is favorable for virus replication. These results suggested that EV71 may exploit the SUMOylation-related ubiquitination to facilitate its replication by protecting the 3D polymerase.
Timely turning on and off of SUMO signaling on viral proteins is important for virus replication (32). Given that 3D is an RdRp, the maintenance of an appropriate level of RdRp in infected cells is crucial for viral growth, as a smaller amount of RdRp may be beneficial for the virus to escape host immune systems (79, 80). It is worth noting that the SUMOylation of 3D and RNA synthesis by 3D may well be two relatively independent processes. Based on the chemical nature of polymerase catalysis and the SUMOylation process, the covalent SUMOylation at K159, at least, is not compatible with polymerase catalysis. We propose that SUMOylation at specific 3D sites helps to maintain the cellular level of 3D protein and that the corresponding de-SUMOylation may be necessary for 3D to resume its polymerase activity, in particular for the critical site of K159. In summary, our findings showed that ubiquitination of EV71 3D is SUMOylation dependent. EV71 may exploit the cross talk of SUMOylation and ubiquitination to stabilize the 3D polymerase and enhance viral replication, which may provide new insights into antivirus drug development by targeting the SUMOylation of EV71 3D.
ACKNOWLEDGMENTS
We thank Hsiu-Ming Shih (Institute of Biomedical Sciences, Academia Sinica, Taiwan, Republic of China) for his helpful suggestions on conducting the experiments. We thank Hong Tang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) for providing plasmids pcDEF-HA-SUMO-1, pcDEF-Myc-Ubc9, and pcDEF-Flag-SUMO-2. We thank Yanyi Wang (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) for providing plasmid pRK-Flag-SUMO-2. We thank Huimin Yan (Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China) for providing anti-3D antibody, and we thank Minetaro Arita (National Institute of Infectious Diseases, Tokyo, Japan) for providing the infectious clone of EV71.
This work was supported by the National Natural Science Foundation of China (grant 31300152 and grant 81371811). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Meulmeester E, Melchior F. 2008. Cell biology: SUMO. Nature 452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 2.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 3.Tatham MH, Jaffray E, Vaughan OA, Desterro JMP, Botting CH, Naismith JH, Hay RT. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 4.Bylebyl GR, Belichenko I, Johnson ES. 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- 5.Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal ACO. 2008. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics 7:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang YG, Dasso M. 2009. SUMOylation and deSUMOylation at a glance. J Cell Sci 122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett RD, Boutell C, Hale BG. 2013. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol 11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- 8.Hay RT. 2007. SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17:370–376. doi: 10.1016/j.tcb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Johnson ES, Blobel G. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem 272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi T, Seki M, Maeda D, Wang WS, Kawabe Y, Seki T, Saitoh H, Fukagawa T, Yagi H, Enomoto T. 2002. Ubc9 is essential for viability of higher eukaryotic cells. Exp Cell Res 280:212–221. doi: 10.1006/excr.2002.5634. [DOI] [PubMed] [Google Scholar]
- 11.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345–356. doi: 10.1016/S0092-8674(02)00630-X. [DOI] [PubMed] [Google Scholar]
- 12.Gareau JR, Lima CD. 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay RT. 2005. SUMO: a history of modification. Mol Cell 18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 14.Merrill JC, Melhuish TA, Kagey MH, Yang SH, Sharrocks AD, Wotton D. 2010. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One 5:e8794. doi: 10.1371/journal.pone.0008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO modified proteins. Proc Natl Acad Sci U S A 101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song J, Zhang ZM, Hu WD, Chen Y. 2005. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem 280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 17.Sun HY, Hunter T. 2012. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem 287:42071–42083. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kravtsova-Ivantsiv Y, Ciechanover A. 2012. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci 125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 19.Guzzo CM, Matunis MJ. 2013. Expanding SUMO and ubiquitin-mediated signaling through hybrid SUMO-ubiquitin chains and their receptors. Cell Cycle 12:1015–1017. doi: 10.4161/cc.24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Praefcke GJK, Hofmann K, Dohmen J. 2012. SUMO playing tag with ubiquitin. Trends Biochem Sci 37:23–31. doi: 10.1016/j.tibs.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Denuc A, Marfany G. 2010. SUMO and ubiquitin paths converge. Biochem Soc Trans 38:34–39. doi: 10.1042/BST0380034. [DOI] [PubMed] [Google Scholar]
- 22.Mattoscio D, Segre CV, Chiocca S. 2013. Viral manipulation of cellular protein conjugation pathways: the SUMO lesson. World J Virol 2:79–90. doi: 10.5501/wjv.v2.i2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nevels M, Brune W, Shenk T. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J Virol 78:7803–7812. doi: 10.1128/JVI.78.14.7803-7812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann H, Floss S, Stamminger T. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J Virol 74:2510–2524. doi: 10.1128/JVI.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinigalia E, Alvisi G, Segre CV, Mercorelli B, Muratore G, Winkler M, Hsiao HH, Urlaub H, Ripalti A, Chiocca S, Palu G, Loregian A. 2012. The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner. PLoS One 7:e49630. doi: 10.1371/journal.pone.0049630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim ET, Kim YE, Huh YH, Ahn JH. 2010. Role of noncovalent SUMO binding by the human cytomegalovirus IE2 transactivator in lytic growth. J Virol 84:8111–8123. doi: 10.1128/JVI.00459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal S, Santos A, Rosas JM, Ortiz-Guzman J, Rosas-Acosta G. 2011. Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res 158:12–27. doi: 10.1016/j.virusres.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Xu K, Klenk C, Liu B, Keiner B, Cheng J, Zheng BJ, Li L, Han Q, Wang C, Li T, Chen Z, Shu Y, Liu J, Klenk HD, Sun B. 2011. Modification of nonstructural protein 1 of influenza A virus by SUMO1. J Virol 85:1086–1098. doi: 10.1128/JVI.00877-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CY, Jeng KS, Lai MM. 2011. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J Virol 85:6618–6628. doi: 10.1128/JVI.02401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han QL, Chang C, Li L, Klenk C, Cheng JK, Chen YX, Xia NS, Shu YL, Chen Z, Gabriel G, Sun B, Xu K. 2014. Sumoylation of influenza A virus nucleoprotein is essential for intracellular trafficking and virus growth. J Virol 88:9379–9390. doi: 10.1128/JVI.00509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumiya Y, Ellison TJ, Yeh ET, Jung JU, Luciw PA, Kung HJ. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J Virol 79:9912–9925. doi: 10.1128/JVI.79.15.9912-9925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izumiya Y, Kobayashi K, Kim KY, Pochampalli M, Izumiya C, Shevchenko B, Wang DH, Huerta SB, Martinez A, Campbell M, Kung HJ. 2013. Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog 9:e1003506. doi: 10.1371/journal.ppat.1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcos-Villar L, Campagna M, Lopitz-Otsoa F, Gallego P, Gonzalez-Santamaria J, Gonzalez D, Rodriguez MS, Rivas C. 2011. Covalent modification by SUMO is required for efficient disruption of PML oncogenic domains by Kaposi's sarcoma-associated herpesvirus latent protein LANA2. J Gen Virol 92:188–194. doi: 10.1099/vir.0.024984-0. [DOI] [PubMed] [Google Scholar]
- 34.Gurer C, Berthoux L, Luban J. 2005. Covalent modification of human immunodeficiency virus type 1 p6 by SUMO-1. J Virol 79:910–917. doi: 10.1128/JVI.79.2.910-917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamborlini A, Coiffic A, Beauclair G, Delelis O, Paris J, Koh Y, Magne F, Giron ML, Tobaly-Tapiero J, Deprez E, Emiliani S, Engelman A, de The H, Saib A. 2011. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J Biol Chem 286:21013–21022. doi: 10.1074/jbc.M110.189274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AbuBakar S, Chee HY, Al-Kobaisi MF, Xiaoshan J, Chua KB, Lam SK. 1999. Identification of enterovirus 71 isolates from an outbreak of hand, foot and mouth disease (HFMD) with fatal cases of encephalomyelitis in Malaysia. Virus Res 61:1–9. doi: 10.1016/S0168-1702(99)00019-2. [DOI] [PubMed] [Google Scholar]
- 37.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol Rev 26:91–107. doi: 10.1111/j.1574-6976.2002.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen SC, Chang LY, Wang YW, Chen YC, Weng KF, Shih SR, Shih HM. 2011. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J Biol Chem 286:31373–31384. doi: 10.1074/jbc.M111.254896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Z, Ke X, Wang M, He S, Li Q, Zheng C, Zhang Z, Liu Y, Wang H. 2013. Human microRNA hsa-miR-296-5p suppresses enterovirus 71 replication by targeting the viral genome. J Virol 87:5645–5656. doi: 10.1128/JVI.02655-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pizzi M. 1950. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum Biol 22:151–190. [PubMed] [Google Scholar]
- 41.Gong Y, Trowbridge R, MacNaughton TB, Westaway EG, Shannon AD, Gowans EJ. 1996. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J Gen Virol 77:2729–2736. doi: 10.1099/0022-1317-77-11-2729. [DOI] [PubMed] [Google Scholar]
- 42.Arita M, Shimizu H, Nagata N, Ami Y, Suzaki Y, Sata T, Iwasaki T, Miyamura T. 2005. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J Gen Virol 86:1391–1401. doi: 10.1099/vir.0.80784-0. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Cao RY, Chen R, Mo L, Han JF, Wang X, Xu X, Jiang T, Deng YQ, Lyu K, Zhu SY, Qin ED, Tang R, Qin CF. 2013. Rational design of thermostable vaccines by engineered peptide-induced virus self-biomineralization under physiological conditions. Proc Natl Acad Sci U S A 110:7619–7624. doi: 10.1073/pnas.1300233110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Z, Sahashi Y, Matsuo K, Asanuma H, Takahashi H, Iwasaki T, Suzuki Y, Aizawa C, Kurata T, Tamura S. 1998. Comparison of the ability of viral protein-expressing plasmid DNAs to protect against influenza. Vaccine 16:1544–1549. doi: 10.1016/S0264-410X(98)00043-7. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Z, Li H, Zhang Z, Meng J, Mao D, Bai B, Lu B, Mao P, Hu Q, Wang H. 2011. Enterovirus 71 2C protein inhibits TNF-alpha-mediated activation of NF-kappaB by suppressing IkappaB kinase beta phosphorylation. J Immunol 187:2202–2212. doi: 10.4049/jimmunol.1100285. [DOI] [PubMed] [Google Scholar]
- 46.Terui Y, Saad N, Jia S, McKeon F, Yuan J. 2004. Dual role of sumoylation in the nuclear localization and transcriptional activation of NFAT1. J Biol Chem 279:28257–28265. doi: 10.1074/jbc.M403153200. [DOI] [PubMed] [Google Scholar]
- 47.Kamitani T, Nguyen HP, Kito K, Fukuda-Kamitani T, Yeh ET. 1998. Covalent modification of PML by the sentrin family of ubiquitin-like proteins. J Biol Chem 273:3117–3120. doi: 10.1074/jbc.273.6.3117. [DOI] [PubMed] [Google Scholar]
- 48.Cheng J, Kang XL, Zhang S, Yeh ETH. 2007. SUMO-specific protease 1 is essential for stabilization of HIF1 alpha during hypoxia. Cell 131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasugi T, Howley PM. 1996. Identification of the structural and functional human homolog of the yeast ubiquitin conjugating enzyme UBC9. Nucleic Acids Res 24:2005–2010. doi: 10.1093/nar/24.11.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mi Z, Fu J, Xiong Y, Tang H. 2010. SUMOylation of RIG-I positively regulates the type I interferon signaling. Protein Cell 1:275–283. doi: 10.1007/s13238-010-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ran Y, Liu TT, Zhou Q, Li S, Mao AP, Li Y, Liu LJ, Cheng JK, Shu HB. 2011. SENP2 negatively regulates cellular antiviral response by deSUMOylating IRF3 and conditioning it for ubiquitination and degradation. J Mol Cell Biol 3:283–292. doi: 10.1093/jmcb/mjr020. [DOI] [PubMed] [Google Scholar]
- 52.Luo H, Zhang Z, Zheng Z, Ke X, Zhang X, Li Q, Liu Y, Bai B, Mao P, Hu Q, Wang H. 2013. Human bocavirus VP2 upregulates IFN-beta pathway by inhibiting ring finger protein 125-mediated ubiquitination of retinoic acid-inducible gene-I. J Immunol 191:660–669. doi: 10.4049/jimmunol.1202933. [DOI] [PubMed] [Google Scholar]
- 53.Gong P, Campagnola G, Peersen OB. 2009. A quantitative stopped-flow fluorescence assay for measuring polymerase elongation rates. Anal Biochem 391:45–55. doi: 10.1016/j.ab.2009.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gohara DW, Ha CS, Kumar S, Ghosh B, Arnold JJ, Wisniewski TJ, Cameron CE. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr Purif 17:128–138. doi: 10.1006/prep.1999.1100. [DOI] [PubMed] [Google Scholar]
- 55.Weber AR, Schuermann D, Schar P. 2014. Versatile recombinant SUMOylation system for the production of SUMO-modified protein. PLoS One 9:e102157. doi: 10.1371/journal.pone.0102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu B, Gong P. 2016. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc Natl Acad Sci U S A 113:E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Batey RT, Kieft JS. 2007. Improved native affinity purification of RNA. RNA 13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu J, Lu G, Zhang B, Gong P. 2015. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J Virol 89:249–261. doi: 10.1128/JVI.02085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campagnola G, Weygandt M, Scoggin K, Peersen O. 2008. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J Virol 82:9458–9464. doi: 10.1128/JVI.00647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kao S, Wang W, Chen C, Chang Y, Wu Y, Wang Y, Wang S, Nesvizhskii AI, Chen Y, Hong T, Yang P. 2015. Analysis of protein stability by the cycloheximide chase assay. Bio Protoc 5:e1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 62.Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J Virol 86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42:W325–W330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong P, Peersen OB. 2010. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 107:22505–22510. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Si XN, Gao G, Wong J, Wang YH, Zhang JC, Luo HL. 2008. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One 3:e2585. doi: 10.1371/journal.pone.0002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin L, Qin Y, Wu H, Chen Y, Wu S, Si X, Wang H, Wang T, Zhong X, Zhai X, Tong L, Pan B, Zhang F, Zhong Z, Wang Y, Zhao W. 2015. Pyrrolidine dithiocarbamate inhibits enterovirus 71 replication by down-regulating ubiquitin-proteasome system. Virus Res 195:207–216. doi: 10.1016/j.virusres.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 67.Bloom J, Pagano M. 2005. Experimental tests to definitively determine ubiquitylation of a substrate. Methods Enzymol 399:249–266. doi: 10.1016/S0076-6879(05)99017-4. [DOI] [PubMed] [Google Scholar]
- 68.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. 2011. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem 286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mallette FA, Richard S. 2012. K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res 22:1221–1223. doi: 10.1038/cr.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomes R, Guerra-Sa R, Arruda E. 2010. Coxsackievirus B5 induced apoptosis of HeLa cells: effects on p53 and SUMO. Virology 396:256–263. doi: 10.1016/j.virol.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Xiong RY, Wang AM. 2013. SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of turnip mosaic virus, is required for viral infection. J Virol 87:4704–4715. doi: 10.1128/JVI.02828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Su CI, Tseng CH, Yu CY, Lai MM. 2016. SUMO modification stabilizes dengue virus nonstructural protein 5 to support virus replication. J Virol 90:4308–4319. doi: 10.1128/JVI.00223-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miteva M, Keusekotten K, Hofmann K, Praefcke GJK, Dohmen RJ. 2010. SUMOylation as a signal for polyubiquitylation and proteasomal degradation, p 195–215. In Groettrup M. (ed), Conjugation and deconjugation of ubiquitin family modifiers. Landes Bioscience, Austin, TX. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Santamaria J, Campagna M, Garcia MA, Marcos-Villar L, Gonzalez D, Gallego P, Lopitz-Otsoa F, Guerra S, Rodriguez MS, Esteban M, Rivas C. 2011. Regulation of vaccinia virus E3 protein by small ubiquitin-like modifier proteins. J Virol 85:12890–12900. doi: 10.1128/JVI.05628-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klenk C, Humrich J, Quitterer U, Lohse MJ. 2006. SUMO-1 controls the protein stability and the biological function of phosducin. J Biol Chem 281:8357–8364. doi: 10.1074/jbc.M513703200. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Xu M, Scotti E, Chen ZJJ, Tontonoz P. 2013. Both K63 and K48 ubiquitin linkages signal lysosomal degradation of the LDL receptor. J Lipid Res 54:1410–1420. doi: 10.1194/jlr.M035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruenn JA. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res 31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Lou Z, Miao Y, Yu Y, Dong H, Peng W, Bartlam M, Li X, Rao Z. 2010. Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell 1:491–500. doi: 10.1007/s13238-010-0061-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kerkvliet J, Papke L, Rodriguez M. 2011. Antiviral effects of a transgenic RNA-dependent RNA polymerase. J Virol 85:621–625. doi: 10.1128/JVI.01626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi AG, Wong J, Marchant D, Luo H. 2013. The ubiquitin-proteasome system in positive-strand RNA virus infection. Rev Med Virol 23:85–96. doi: 10.1002/rmv.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]