Abstract
On reaching maturity, animal organs cease to increase in size because of inhibition of cell replication activities. It follows that maintenance of optimal organ function depends on the elimination of oxidatively damaged cells and their replacement with new cells. To examine the effects of oxidative stress and apoptosis on the accumulation of oxidized proteins, we exposed acute promyelocytic leukemia cells to arsenic trioxide (As2O3) in the presence and absence of a general caspase inhibitor (benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone), which is known to inhibit caspase-induced apoptosis. We confirm that treatment of cells with As2O3 induces apoptosis and leads to the accumulation of oxidized proteins. Furthermore, inhibition of caspase activities prevented As2O3-induced apoptosis and led to a substantial increase in accumulation of oxidized proteins. Moreover, inhibition of caspase activity in the absence of As2O3 led to elevated levels of the LMP2 immunoproteasome protein. We also show that caspase inhibition leads to increases in the levels of oxidized proteins obtained by treatments with hydrogen peroxide plus ferrous iron. Collectively, these results suggest the possibility that an age-related loss in capacity to carry out apoptosis might contribute to the observed accumulation of oxidized proteins during aging and in age-related diseases.
Keywords: arsenic trioxide, programmed cell death, protein carbonyl NB4 cells, H2O2
Proteins, lipids, and nucleic acids are subject to oxidation by reactive oxygen species (ROS) generated during normal metabolism and even more so under conditions of oxidative stress. The intracellular levels of oxidized proteins have been shown to increase during aging and in the development of many age-related diseases, including Alzheimer's disease, rheumatoid arthritis, atherosclerosis, and Parkinson's disease (1–6). Moreover, an increase in intracellular ROS leads to initiation of various types of cell death (7). In vitro investigations with acute promyelocytic leukemia (APL)-derived NB4 cells indicated that clinically achievable concentrations of arsenic trioxide (As2O3) (1–2 μM) induce apoptosis by means of an ROS-dependent pathway (8, 9). Intracellular production of ROS causes disruption of the mitochondrial membrane potential and consequently to release of cytochrome c into the cytosol and thereby to activation of the caspase cascade, leading to programmed cell death through apoptosis (9). ROS play an important role in triggering cell death and in determining whether cells die by apoptosis or oncosis (10). Apoptosis is the main mechanism by which multicellular organisms eliminate damaged, nonfunctional cells to ensure proper development and maintenance of cellular homeostasis (11). The present study was undertaken to test the hypothesis that a loss in the ability to eliminate oxidatively damaged cells by apoptosis would favor accumulation of oxidized proteins, as has been observed during aging and in age-related diseases (1). To this end, cultured APL-derived NB4 cells (referred to hereafter as APL cells) were exposed to As2O3, which is known to induce oxidative stress and apoptosis (12), in the presence and absence of a tripeptide derivative, benzyloxycarbonyl-Val-Ala-Asp-f luoromethyl ketone (z-VAD-fmk), which is a general caspase inhibitor (GCI) (13). Direct measurements confirm that GCI inhibits intracellular caspase activity and also the ability of As2O3 to induce apoptosis and concomitantly leads to the accumulation of oxidized protein (i.e., protein carbonyl derivatives). Furthermore, it was found that in the absence of As2O3, treatment with GCI leads to accumulation of the LMP2 and LMP7 immunoproteasome proteins and to a significant decrease in 20S proteasome activity.
Materials and Methods
Materials. As2O3 was purchased from Sigma. A 100 μmol/liter stock solution of As2O3 was obtained by dissolving As2O3 in 5 M NaOH and by diluting in H2O, followed by adjustment of the pH to 7.0. RPMI medium 1640 and FBS were from Cellgro (Herndon, VA). Primary Ab against procaspase-3 was from Gene Therapy Systems (San Diego). Poly(ADP-ribose) polymerase (PARP) was from Cell Signaling Technology (Beverly, MA). Abs against the immunoproteasome proteins LMP2 and LMP7 were from Affinity BioReagents (Golden, CO). Goat anti-mouse and goat anti-rabbit IgG conjugated with alkaline phosphatase secondary Abs (Santa Cruz Biotechnology) were used for Western blot. Rabbit anti-2,4-dinitrophenyl Ab was obtained from DAKO. Caspase-3 substrate Ac-Asp-Glu-Val-Asp (DEVD)-7-amino-4-methylcoumarin (AMC) (Ac-DEVD-AMC) and the z-VAD-fmk (referred to here as GCI) were purchased from Pharmingen. The enhanced chemiluminescence immunoblot detection system was purchased from Applied Biosystems.
Cell Culture. APL cells (DSMZ Braunschweig, Braunschweig, Germany) were cultured in suspension and were routinely passaged to maintain an optimal density of 0.3–1.5 × 106 cells per ml. Cells were split every 4–6 days by centrifugation followed by aspiration of the culture medium. The cells were cultured in RPMI medium 1640 supplemented with 10% FBS and antibiotics (50 units/ml penicillin and 50 mg/ml streptomycin) in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Immediately before the various treatments described herein, the cells were centrifuged and resuspended in fresh medium and the cell density adjusted to 1 × 106 cells per ml.
Preparation of Cell Extracts and Protein Samples. After 24 h of treatment, cells in 5 ml of each culture were harvested by centrifugation, and the sedimented pellets were lysed by addition of 100 μl of lysis buffer, prepared by dissolving one tablet of EDTA-free protease inhibitor (Roche Diagnostics) in 10 ml of PBS (pH 7.2) containing 0.1% vol/vol Triton X-100. After lysis, the suspensions were sonicated, the soluble cell proteins were separated from insoluble debris by centrifuging at 14,000 rpm for 3 min, and the supernatant solutions were frozen at –20°C until used. Protein concentration was determined by using bicinchoninic acid reagents with BSA as standard.
Measurement of Protein Carbonyl by Spectrophotometry. Protein carbonyl content was measured by using 2,4-dinitrophenyl-hydrazine (DNPH), as described in refs. 14 and 15. Five hundred microliters of 10 mM DNPH dissolved in 2 M HCl was added to 100 μl of soluble protein fraction. In a parallel, a blank was prepared by addition of 500 μl of 2 M HCl containing no DNPH. After incubation at 20°C for 1 h in the dark, 500 μl of 20% trichloroacetic acid was added to the sample and was allowed to stand for 15 min at 4°C. The precipitated protein was collected by centrifugation at 11,000 rpm for 3 min, and the supernatant was discarded. The pellet was washed three times with 1 ml of a 1:1 (vol/vol) mixture of ethanol and ethyl acetate and centrifuged at 11,000 rpm for 3 min to remove free reagent. The final precipitated protein was redissolved in 200 μl of 6 M guanidine hydrochloride solution. After incubation for 30 min at 37°C, the amount of protein dinitrophenylhydrozone derivative was quantified by measuring the absorbance at 370 nm (SpectraMax Plus384, Molecular Devices) and converted to nanomoles of hydrazone by using a molar absorption coefficient of 22,000 M–1·cm–1. Carbonyl content was expressed as nanomoles of DNPH incorporated per milligram of protein.
Immunoblot Analysis. Assays were performed as described in refs. 14 and 16, with slight modifications. Equal amounts (25 μg) of the soluble protein in the cell extracts were derivatized by adding 15 μl of DNPH reagent to 14 μl of sample in lysis buffer containing 6% SDS (pH 7.2). The reaction was allowed to continue at room temperature for 10 min. The derivatization reaction was stopped by adding 12 μl of neutralizing solution containing 2 M Tris base, 30% glycerol (final concentration 0.52 M), and 1.3 μl of 2-mercaptoethanol (pH 6.9), followed by vortexing. Derivatized protein samples were then loaded onto a 12% Nu-PAGE gel, and electrophoresis was run at 150 V for 75 min. Subsequently, the proteins were transferred onto a nitrocellulose membrane. The membrane was blocked by 5% of dry milk before immunostaining for the detection of DNPH-bound protein by using anti-2,4-dinitrophenyl IgG as a primary Ab (1:2,000 dilution) in PBS-Tween 20 and alkaline phosphatase-conjugated anti-rabbit IgG as secondary Ab (1:5,000 dilution).
Immunoblot Analyses of Caspase-3, PARP, and the Immunoproteasomes LMP2 and LMP7. Extracted proteins were detected with anti-caspase-3, anti-PARP, anti-LMP2, and anti-LMP7 primary Abs (dilution 1:2,000), respectively. The membranes were incubated with alkaline phosphatase-conjugated goat anti-mouse or goat anti-rabbit secondary Abs for 30 min (dilution 1:5,000). Specific proteins were detected with the blotting reagent (disodium 3-(4-methoxyspiro(1,2-dioxetane-3,2′-(5′-chloro)tricyclo-[3.3.1.13.7]decan}-4-yl)phenyl phosphate containing nitroblock, and chemiluminescence was detected by exposure of membranes to Kodak X-Omat films.
Caspase-3 Activity. Caspase-3 enzymatic activity was determined by using the CaspACE Assay System Kit (Promega). Briefly, a 100-μl reaction mixture containing an equal amount (25 μg) of clear supernatant of cell extract and 32 μl of caspase assay buffer, 2 μl of DMSO, and 10 μl of 100 mM DTT (pH 7.2) was prepared on a 96-well plate. Two microliters of 2.5 mM synthetic caspase-3 substrate (Ac-DEVD-AMC) was added just before the reaction. During the assay, the levels of released AMC were monitored every 10 min at 37°C by using a CytoFluor 4000 Fluorescence Multiwell Plate Reader (PerSeptive Biosystems, Framingham, MA) with excitation and emission wavelengths at 360 and 460 nm, respectively. To determine whether caspase is involved in As2O3-induced cell death, the caspase inhibition assay was performed with 100 μM of the GCI, z-VAD-fmk (Biomol, Plymouth Meeting, PA), solubilized in DMSO as described by the manufacturer.
Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL)-Peroxidase Staining. For TUNEL-peroxidase staining, we used the In Situ Cell Death kit (Roche Diagnostics). The treated cells were attached on poly(l-lysine)-coated glass-bottomed dishes and fixed for 30 min in 4% paraformaldehyde in PBS (pH 7.4). After the cells were washed three times with PBS, the endogenous peroxidase was inactivated by incubation with 3% hydrogen peroxide (H2O2) in methanol for 30 min at room temperature. The cells were then washed with PBS and subsequently permeabilized for 2 min on ice with 0.1% sodium-citrate solution containing 0.1% Triton X-100. The cells were then washed twice with PBS, stained with the TUNEL reaction mixture for 60 min at 37°C, washed twice with PBS, and labeled with peroxidase-conjugated goat Ab for 30 min at 37°C. DNA fragmentation was detected by staining with diaminobenzidine and observed under a microscope (Spot Insight with digital camera, Diagnostic Instruments, Sterling Heights, MI).
Measurement of Cell Death. Cytotoxicity was measured by using the Cell Counting Assay Kit-8 (Dojindo Molecular Technologies, Gaithersburg, MD) according to the manufacturer's protocol. One hundred microliters of APL cells were plated on 96-well plates at a density of 1 × 106 cells per ml followed by preincubation with 100 μM GCI for 1 h, then treated with 2 μM As2O3 for 24 h. Ten microliters of Cell Counting Assay Kit-8 solution was added to each well, the cells were incubated for another 1 h, and the absorbance at 450 nm was measured by using a microplate reader (SpectraMax Plus384, Molecular Devices). The amount of the formazan dye, generated by the activities of dehydrogenases in cells, is directly proportional to the number of living cells.
Assessment of DNA Fragmentation. Total genomic DNA was isolated from As2O3-treated APL cells in the absence and presence of GCI by using an Apoptotic DNA Ladder kit (Roche) according to manufacturer's protocol. Briefly, the cells were collected by centrifugation and washed twice with cold PBS. The cell pellet was suspended in 400 μl of lysis buffer (pH 4.4) at room temperature for 15 min, and DNA-bound to glass fibers in the presence of chaotropic salts was eluted in 200 μl of prewarmed (70°C) elution buffer (pH 8.5). An equal amount of DNA sample was then electrophoresed on a 1% agarose gel containing 0.1 μg/ml ethidium bromide. The agarose gel was run at 50 V for 90 min in a TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) and evaluated and photographed under UV illumination.
20S Proteasome Activity. The Proteasome Activity kit (Chemicon) was used to measure the 20S proteasome activity. An equal amount (20 μg) of clear whole-cell extract was incubated with fluorogenic proteasome substrate, Suc-LLVY-AMC, in 100 μl of reaction mixture for 1 h at 37°C, followed by the detection of fluorophore AMC. The free AMC fluorescence was quantified by using the CytoFluor multiwell plate reader (Series 4000, PerSeptive Biosystems) with excitation and emission wavelengths at 380 and 460 nm, respectively.
Results
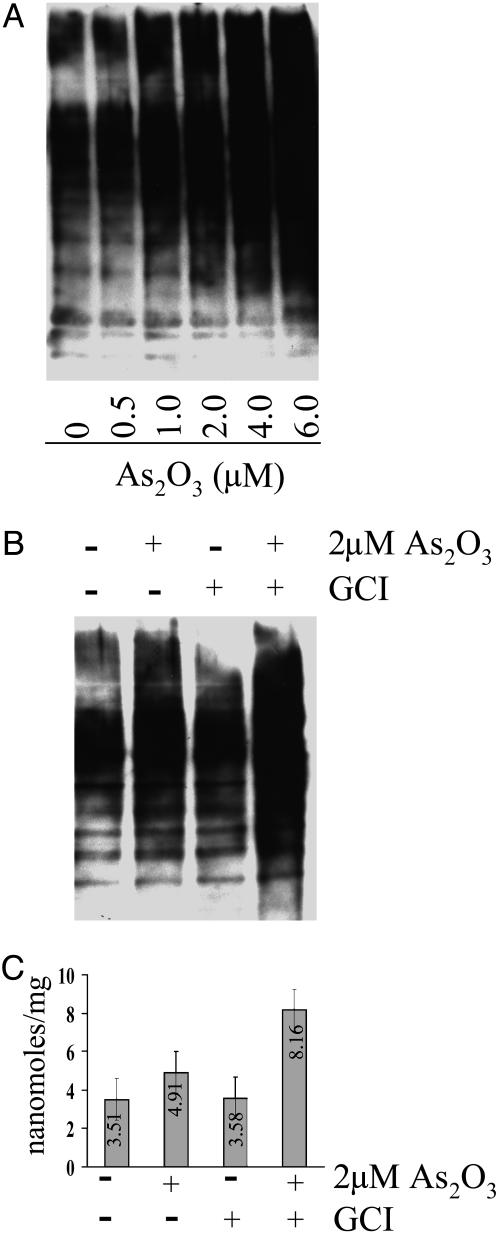
Inhibition of Caspase Activity Leads to Loss of Apoptotic Capacity and Enhanced Accumulation of Oxidized Proteins. To investigate the possibility that apoptosis might play an important role in determining the intracellular level of oxidized protein, APL cells were treated with As2O3 (a known inducer of apoptosis) at concentrations ranging from 0 to 6 μM for 24 h. After incubation, the proteins in cell extracts were derivatized with DNPH and run on Nu-PAGE, followed by Western blotting with anti-2,4-dinitrophenyl Ab for the detection of DNPH-modified protein carbonyl groups. As shown in Fig. 1A, the protein carbonyl content of proteins increased progressively with rising concentration of As2O3. Because 6 μM As2O3 is toxic, as determined by actin degradation (data not shown), we elected to use 2-μM concentrations in all subsequent studies. To determine whether caspase inhibition has any effect on protein carbonyl formation, we treated APL cells with the GCI for 1 h before treatment with 2 μMAs2O3 for 24 h. The protein carbonyl content of cell extracts proteins was then analyzed by using both immunochemical and spectrophotometric methodologies. The immunochemical analysis (Fig. 1B) confirmed that treatment with As2O3 alone leads to a substantial increase in the protein carbonyl content and that treatment with both GCI and As2O3 leads to an even greater increase in protein carbonyl content. Similar results were obtained by using the spectrophotometric procedure (Fig. 1C). Treatment with As2O3 alone led to a 40% increase in carbonyl content compared with the control value of 3.51 nmol/mg, whereas treatment with both GCI and As2O3 led to a 128% increase over the control value of 3.58 nmol/mg.
Fig. 1.
Immunochemical and spectrophotometric analyses of protein carbonyl formation in APL cells. (A) Immunoblot analysis of protein carbonyl formation in cells after incubation for 24 h in the absence (lane 1) and presence of various concentrations (0.5–6 μM) of As2O3 (lanes 2–6), as indicated. (B) Immunoblot analysis of protein carbonyl formation in cells after 24-h treatments with and without 2 μM As2O3 and/or 100 μM GCI supplements indicated as follows: lane 1, no supplement (control); lane 2, plus As2O3; lane 3, plus GCI; lane 4, plus both As2O3 and GCI. (C) Spectrophotometric analysis of protein carbonyl content of the same samples as in B. Results shown are the means of three independent experiments (bars, ±SD).
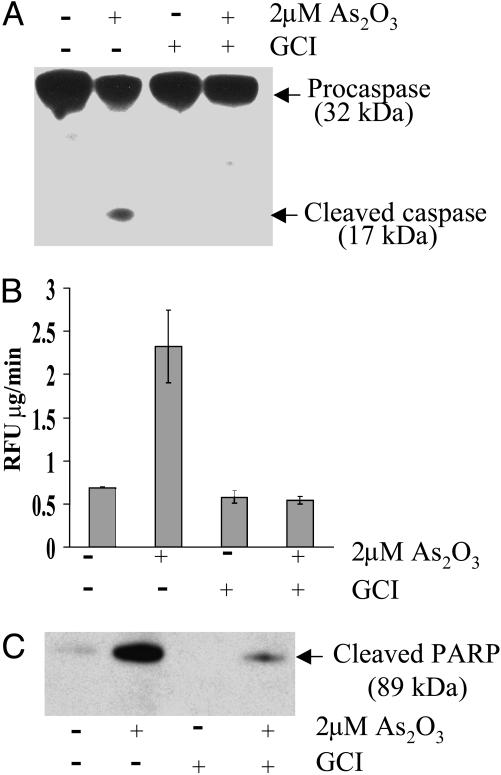
Effect of As2O3 on Caspase Activity. Because conversion of the 32-kDa procaspase-3 to the active 17-kDa caspase-3 triggers apoptosis, we examined the possibility that treatment of APL cells with As2O3 facilitates cleavage of procaspase-3 to the 17-kDa peptide. As shown in Fig. 2A, As2O3 stimulates cleavage of procaspase-3 to the 17-kDa peptide derivative, as measured by Western blot assays. Results summarized in Fig. 2A also indicate that the As2O3-induced cleavage reaction is inhibited by GCI. Upon activation, caspase-3 is known to catalyze cleavage of the model substrate Ac-DEVD-AMC to form the fluorescent product AMC and also cleavage of PARP to form an 89-kDa derivative. Results of studies summarized in Fig. 2B indicate that treatment of APL cells with As2O3 led to a 3-fold increase in the cleavage of the Ac-DEVD-AMC substrate, measured fluorometrically, and also promoted cleavage of PARP, as determined by immunochemical analysis of the 89-kDa peptide formed (Fig. 2C). These results indicate that under our experimental conditions, As2O3 induces conversion of procaspase-3 to its catalytically active caspase-3 peptide derivative. In parallel experiments, it was found that both As2O3-induced cleavage reactions are inhibited by the GCI, providing further proof that caspases are involved in the cleavage reactions (Fig. 2C).
Fig. 2.
Effect of As2O3 in presence and absence of GCI on caspase activation in APL-derived NB4 cells. The cells were treated with 2 μM As2O3 for 24 h in both the presence and absence of 100 μM GCI. (A) Immunoblot analyses of the cleavage of procaspase-3 (32 kDa) to the active caspase fragment (17 kDa). Conditions: lane 1, no supplements (control); lane 2, plus As2O3; lane 3, plus GCI; lane 4, plus both As2O3 and GCI. (B) Samples were as in A, but caspase-3 activity was measured fluorometrically by using Ac-DEVD-AMC as a substrate (see Materials and Methods). (C) Samples were as in A, but cleavage of full-length PARP into 89-kDa fragment was measured by the immunoblot procedure described in Materials and Methods. Results are the means of three independent experiments (bars, ±SD).
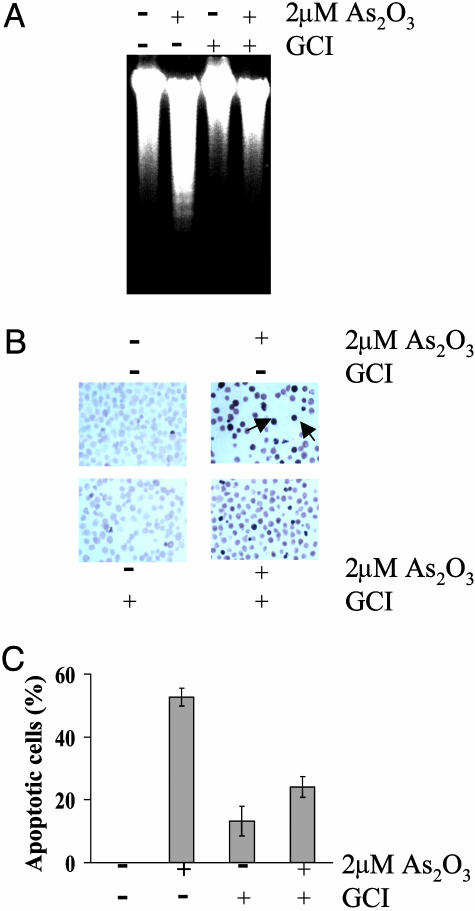
Detection of Apoptosis. Evidence that As2O3 induces apoptosis in APL cells was obtained by showing that As2O3 treatment led to DNA laddering, nuclear TUNELing, and cytotoxicity (Fig. 3). As2O3 treatment led to the generation of low-molecular-weight DNA fragments (≈180–200 bp), but this fragmentation was prevented by pretreatment of the cells with GCI (Fig. 3A). In agreement with caspase-3 and PARP data, we found that the population of TUNEL-positive nuclei was increased in response to the As2O3 treatment, and that the TUNEL-positive nuclei were remarkably decreased in the presence of GCI (Fig. 3B). Cytotoxicity measurements obtained by using the Cell Counting Assay Kit-8 assay technique (see Materials and Methods) indicated that after As2O3 treatment of APL cells for 24 h in the absence of GCI, 52.7% of the cell population was dead cells, whereas the fraction of dead cells present after treatment in the presence of GCI was 24.1%, and there were no dead cells in the untreated cell population (Fig. 3C).
Fig. 3.
Effect of caspase inhibition on As2O3-induced apoptosis in APL-derived NB4 cells. (A) Induction of internucleosomal DNA fragmentation ladder. Conditions: lane 1, no supplements (control); lane 2, plus 2 μMAs2O3; lane 3, plus 100 μM GCI; lane 4, plus 2 μMAs2O3 and 100 μM GCI. (B) TUNEL assay. TUNEL-positive nuclei were identified by dark brown bodies (see Materials and Methods). Arrows, TUNEL-positive nuclei. (C) Cytotoxic assay. Conditions: lane 1, no supplements (control); lane 2, plus 2 μMAs2O3; lane 3, plus 100 μM GCI; lane 4; plus 2 μMAs2O3 and 100 μM GCI. Results are means of three independent experiments (bars, ±SD).
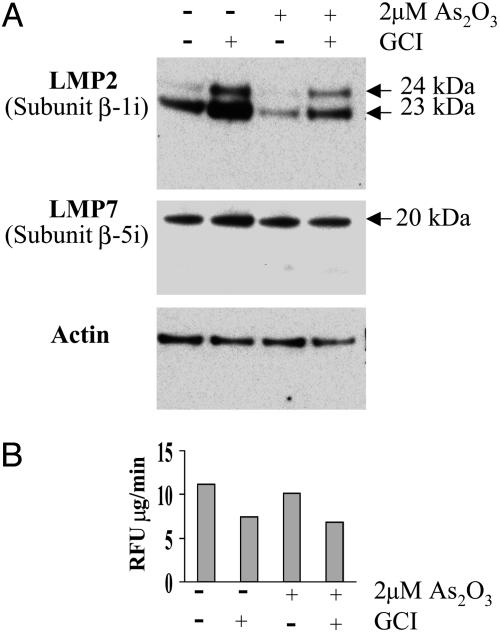
Effect of GCI on Immunoproteasome Levels. Unexpectedly, it was observed that treatment of APL cells with GCI in the absence of AS2O3 led to a substantial increase in levels of the 20-kDa LMP7 and 23-kDa LMP2 immunoproteasome proteins and the 24-kDa precursor of the LMP2 protein (Fig. 4A). Curiously, treatment of the cells with As2O3, both in the presence and absence of GCI, led to a significant suppression of LMP2 protein accumulation. Even more puzzling was the finding that the GCI-provoked increase in immunoproteasome levels was accompanied by a 30% decrease in the 20S proteasome catalytic activity as measured by the increase in fluorescence associated with proteolytic cleavage of the suc-LLVY-AMC substrate to the free AMC derivative (Fig. 4B). Validity of the fluorescence technique as a measure of 20S proteasome activity is supported by the demonstration that the observed changes in fluorescence are almost completely prevented by the general proteasome inhibitor lactacystin (data not shown).
Fig. 4.
Effects of GCI and As2O3 on production of immunoproteasome proteins in APL-derived NB4 cells. The cells were treated with and without As2O3 and/or GCI as described in Fig. 2. (A) Immunoblot of the 23- and 24-kDa LMP2 proteins (Top) and the LMP7 protein (Middle). Conditions: lane 1, no supplements (control); lane 2, plus 100 μM GCI; lane 3, plus 2 μMAs2O3; lane 4, plus 2 μMAs2O3 and 100 μM GCI. (B) 20S proteasome activity was measured by fluorometric analysis of the cleavage of the suc-LLVY-AMC peptide as described in Materials and Methods. The same samples as in A were used.
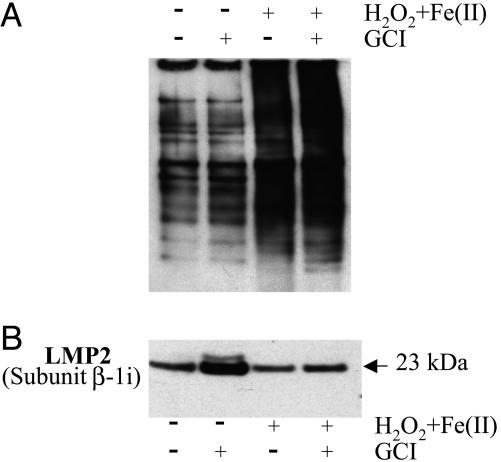
Effect of H2O2 Plus Iron on Protein Carbonyl and LMP2 Levels. Because treatment of cells with As2O3 leads to up-regulation of ROS formation and because, as is well known, exposure of proteins to a mixture of H2O2 and ferrous iron leads to the generation of protein carbonyl derivatives (16), we examined the possibility that exposure of APL cells to a mixture of H2O2 and Fe(II) might mimic the effects of arsenite on these cells. As shown in Fig. 5A, treatment with the H2O2 system led, as expected, to the oxidation of cellular proteins to carbonyl derivatives, and this oxidation was somewhat greater if GCI was also present. Other data in Fig. 5 confirm that exposure of cells to GCI leads to an increase in the level of the 23-kDa LMP2 immunoproteasome, and (as with As2O3) this increase is lowered by the presence of H2O2 (Fig. 5B).
Fig. 5.
Effect of caspase inhibition on H2O2 and ferrous iron treatment on formation of protein carbonyl derivatives and immunoproteasome proteins formation in APL-derived NB4 cells. Cells were incubated for 24 h in the presence and absence of 500 μMH2O2 plus 500 μM Fe(II) and 100 μM GCI. (A) Immunoblot analysis of protein carbonyl formation in cell lysates (see Materials and Methods). Lane 1, no additions (control); lane 2, plus 100 μM GCI; lane 3, plus 500 μMH2O2 and 500 μM Fe (II); lane 4, plus 500 μMH2O2 and 500 μM Fe(II) plus 100 μM GCI. (B) Immunoblot analysis of immunoproteasome LMP2 protein. The same samples as in A were used.
Discussion
The demonstration that treatment of cultured APL-derived NB4 cells with increasing concentrations (0.5–6 μM) of As2O3 leads to progressive increases in oxidatively modified proteins (protein carbonyl derivatives) and to apoptosis (Figs. 1A and 3) confirms results of earlier studies indicating that As2O3 induces generation of H2O2 and other forms of ROS that are involved in oxidation of DNA, lipids, and proteins, and in the initiation of apoptosis. APL cells have relatively low levels of glutathione peroxidase and catalase and have a constitutively higher H2O2 content than other leukemia cells (17). As2O3 inhibits glutathione peroxidase activity and increases cellular H2O2 content in APL cells (17). At a concentration of 6 μM, As2O3 seemed to be toxic as judged by the promotion of actin degradation (data not shown). This concentration is much higher than the concentrations (1–2 μM) previously shown to be of therapeutic value in the treatment of patients with APL (17, 18). Therefore, we have used a concentration of 2 μM for other studies reported here. The observation that the GCI inhibits As2O3-induced apoptosis (Figs. 2 and 3) is also understandable. It is well established that conversion of procaspases to their catalytically active forms is involved in the initiation of apoptosis and that GCI inhibits this conversion (17). We confirm that GCI inhibits conversion of the 32-kDa procaspase-3 to its active 17-kDa derivative, and that this reaction inhibits the ability of caspase-3 to catalyze the cleavage of its model substrate, Ac-DEVD-AMC, and also cleavage of PARP (Fig. 2). The most intriguing result of this study is the demonstration that GCI inhibition of apoptosis of cultured APL cells is associated with an extensive increase in the level of oxidized proteins (Fig. 1 B and C). This finding supports the proposition (19) that loss of apoptotic capacity may contribute to the accumulation of oxidized proteins as occurs during aging and in the development of some age-related diseases (1–6). This proposition is based on the consideration that when animals reach maturity, many of their organs become fixed in size. Therefore, maintenance of optimal organ function depends on the ability to get rid of oxidatively damaged cells and replace them with good cells. This observation raises an important question: Why are highly toxic ROS (H2O2, O2·–) used to activate cell-signaling pathways that lead to apoptosis on the one hand and to cell replication on the other (20–24). It was proposed that these signaling pathways may serve to monitor the intracellular concentrations of ROS so that if the steady-state level exceeds a normal, tolerable value, the cells are primed to set in place the ability to eliminate oxidatively damaged cells and replace them with good cells (19). Failure to mediate apoptosis and cell replication under conditions of oxidative stress would lead to loss of organ function. We assume that the increase in levels of oxidized protein that are observed when APL cells are treated with GCI is due to the inhibition of caspase activities leading to apoptosis, but other possibilities are not excluded. The possibility that GCI is involved in the inhibition of proteases that play a role in the degradation of oxidized proteins should be examined. Assuming that the average molecular weight of cellular protein subunits is ≈50 kDa (25) and that only one carbonyl group is incorporated into a given protein molecule, it follows that a value of 2 nmol of DNPH-reactive protein carbonyl per mg corresponds to modification of ≈10% of the cellular protein. Accordingly, the levels of protein carbonyls observed in extracts of APL cells after treatment with As2O3 in the absence and presence of the GCI would correspond to 29.5% and 41%, respectively, of the protein (Fig. 1C). Also of particular interest is the finding that treatment of APL cells with GCI alone leads to a substantial increase in the accumulation of the 23-kDa LMP2 immunoproteasome protein (Fig. 4). This protein is able to replace some subunits of the 20S proteasome and to facilitate antigen processing (26). Curiously, the increases in levels of LMP2 and LMP7 proteins were accompanied by a 30% decrease in 20S proteasome activity (Fig. 4B). It remains to be determined whether, under the experimental conditions used in this study, these immunoproteasomes are incorporated into the 20S proteasome. The mechanisms that underlie the ability of As2O3 to induce apoptosis and stimulate the generation of oxidized proteins are still poorly understood. It is known that As2O3 treatment leads to generation of H2O2 and to a decrease in the GSH level (11, 17, 27, 28). As2O3 also reacts with cysteine sulfhydryl groups of proteins and is known to inhibit glutathione peroxidase (28). The possibility that it may compromise the activities of other antioxidant proteins, such as thioredoxin, thioredoxin reductase, peroxiredoxins, and glutaredoxin reductase, all of which have functional cysteine residues (29–33), deserves consideration. Such interactions may play an important role in the ability of As2O3 to induce protein oxidation and apoptosis. It is noteworthy that solutions of As2O3 used in these studies were prepared, as described in refs. 34 and 35, by dissolving the As2O3 in 5 M NaOH and then diluting with deionized water and adjusting the pH to 7.0. It is stated (36) that, upon treatment with alkali, As2O3 is converted to a mixture of [AsO(OH)2]–, [AsO2(OH)]2–, and [AsO3]2–. If so, it remains to be established how concentrations of these species vary with subsequent adjustment of the pH to 7.0 and which, if any, of these species are implicated in the modification of proteins.
Acknowledgments
We thank Dr. Mohammed Akbar (National Institute of Alcohol Abuse and Alcoholism) for his technical assistance with the TUNEL assay.
Abbreviations: Ac-DEVD-AMC, Ac-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin; APL, acute promyelocytic leukemia; As2O3, arsenic trioxide; DNPH, 2,4-dinitrophenyl hydrazine; PARP, poly(ADP-ribose) polymerase; ROS, reactive oxygen species; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; z-VAD-fmk, benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone.
References
- 1.Stadtman, E. R. (2001) Ann. N.Y. Acad. Sci. 928, 22–38. [DOI] [PubMed] [Google Scholar]
- 2.Carrard, G., Bulteau, A., Petropoulos, I. & Friguet, B. (2002) Int. J. Biochem. Cell Biol. 34, 1461–1474. [DOI] [PubMed] [Google Scholar]
- 3.Aksenov, M. Y., Aksenova, M. V., Butterfield, D. A., Geddes, J. W. & Markesbery, W. R. (2001) Neuroscience 103, 373–383. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, M. L., Rubin, B. R. & Gracy, R. W. (1989) J. Rheumatol. 16, 15–18. [PubMed] [Google Scholar]
- 5.Chapman, M. L., Rubin, B. R. & Gracy, R. W. (1986) J. Rheumatol. 13, 850–852. [PubMed] [Google Scholar]
- 6.Stolzing, A. & Grune, T. (2001) Clin. Exp. Dermatol. 26, 566–572. [DOI] [PubMed] [Google Scholar]
- 7.Yuyama, K., Yamamoto, H., Nishizaki, I., Kato, T., Sora, I. & Yamamoto, T. (2003) J. Neurosci. Res. 73, 351–363. [DOI] [PubMed] [Google Scholar]
- 8.Rojewski, M. T., Baldus, C., Knauf, W., Thiel, E. & Schrezenmeier, H. (2002) Br. J. Haematol. 116, 555–563. [DOI] [PubMed] [Google Scholar]
- 9.Sturlan, S., Baumgartner, M., Roth, E. & Bachleitner-Hofmann, T. (2003) Blood 101, 4990–4997. [DOI] [PubMed] [Google Scholar]
- 10.Asmis, R. & Begley, J. G. (2003) Circ. Res. 92, e20–e29. [DOI] [PubMed] [Google Scholar]
- 11.Zheng, T. S., Hunot, S., Kuida, K., Momoi, T., Srinivasan, A., Nicholson, D. W., Lazebnik, Y. & Flavell, A. R. (2000) Nat. Med. 6, 1241–1247. [DOI] [PubMed] [Google Scholar]
- 12.Sun, Y., Kim, S. H., Zhou, D. C., Ding, W., Paietta, E., Guidez, F., Zelent, A., Ramesh, K. A., Cannizzaro, L., Warrell, R. P. & Gallagher, R. E. (2004) Leukemia 18, 1258–1269. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Calvo, M., Peterson, E. P., Leiting, B., Ruel, R., Nicholson, D. W. & Thornberry, N. A. (1998) J. Biol. Chem. 273, 32608–32613. [DOI] [PubMed] [Google Scholar]
- 14.Levine, R. L., Williams, J. A., Stadtman, E. R. & Schacter, E. (1994) Methods Enzymol. 233, 346–357. [DOI] [PubMed] [Google Scholar]
- 15.Shacter, E., Williams, J. A., Lim, M. & Levine, R. L. (1994) Free Radical Biol. Med. 17, 429–444. [DOI] [PubMed] [Google Scholar]
- 16.Stadtman, E. R. (1990) Free Radical Biol. Med. 9, 315–325. [DOI] [PubMed] [Google Scholar]
- 17.Jing, Y., Dai, J., Chalmers-Redman, R. M. E., Tatton, W. G. & Waxman, S. (1999) Blood 94, 2102–2111. [PubMed] [Google Scholar]
- 18.Dai, J., Weinberg, R. S., Waxman, S. & Jing, Y. (1990) Blood 93, 268–277. [PubMed] [Google Scholar]
- 19.Stadtman, E. R. & Levine, R. L. (2002) Hum. Exp. Toxicol. 21, 83. [DOI] [PubMed] [Google Scholar]
- 20.Clement, M.-V. & Pervaiz, S. (1999) Free Radical Res. 30, 247–252. [DOI] [PubMed] [Google Scholar]
- 21.Burdon, R. H. (1995) Free Radical Biol. Med. 18, 775–794. [DOI] [PubMed] [Google Scholar]
- 22.Sundaresan, M., Yu, Z. X., Ferrans, V. J., Irani, K. & Finkel, T. (1995) Science 270, 296–299. [DOI] [PubMed] [Google Scholar]
- 23.Bae, Y. S., Kong, S. W., Seo, M. S., Baines. I. C., Tekel, E., Chock, P. B. & Rhee, S. G. (1997) J. Biol. Chem. 272, 217–221. [PubMed] [Google Scholar]
- 24.Barrett, W. C., DeGrove, J. P., Keng, Y.-F., Zheng, Z.-Y., Yim, M. B. & Chock, P. B. (1999) J. Biol. Chem. 274, 34543–34546. [DOI] [PubMed] [Google Scholar]
- 25.Starke-Reed, P. E. & Oliver, C. N. (1989) Arch. Biochem. Biophys. 275, 559–567. [DOI] [PubMed] [Google Scholar]
- 26.Brooks, P., Murray, R. Z., Mason, G. G., Hendil, K. B. & Rivett, A. J. (2000) Biochem. J. 352, 611–615. [PMC free article] [PubMed] [Google Scholar]
- 27.Hancock, J. T., Deskan, R. & Niell, S. J. (2001) Biochem. Soc. Trans. 29, 345–350. [DOI] [PubMed] [Google Scholar]
- 28.Chen, Y. C., Lin-Shiau, S. Y. & Lin, J. K. (1998) J. Cell Physiol. 177, 324–333. [DOI] [PubMed] [Google Scholar]
- 29.Akao, Y., Nakagawa, Y. & Akiyama, K. (1999) FEBS Lett. 455, 59–62. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, M., Yang, G., Hong, C., Liu, J., Holle, E., Yu, X., Wagner, T., Vatner, S. F. & Sadoshima, J. (2003) J. Clin. Invest. 112, 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. R., Bar-Noy, S., Kwon, J., Levine, R. L., Stadtman, T. C. & Rhee, S. G. (2000) Proc. Natl. Acad. Sci. USA 97, 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang, W. W., Bains, I. C. & Rhee, S. G. (1998) J. Biol. Chem. 273, 6303–6311. [DOI] [PubMed] [Google Scholar]
- 33.Landino, L. M., Moynihan, K. L., Todd, J. V. & Kennett, K. L. (2004) Biochem. Biophys. Res. Commun. 314, 556–560. [DOI] [PubMed] [Google Scholar]
- 34.Dvorakova, K., Payne, C. M., Tome, M. E., Briehl, M. M., Vasquez, M. A., Waltmire, C. N., Coon, A. & Dorr, R. T. (2002) Mol. Cancer Ther. 1, 185–195. [PubMed] [Google Scholar]
- 35.Berthoux, L., Towers, G. J., Gurer, C., Salomoni, P., Pandolfi, P. P. & Luban, J. (2003) J. Virol. 77, 3162–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cotton, F. A. & Wilkinson, G., eds. (1988) Advanced Inorganic Chemistry (Wiley, New York), p. 401.