Abstract
The de novo design of catalytic proteins provides a stringent test of our understanding of enzyme function, while simultaneously laying the groundwork for the design of novel catalysts. Here we describe the design of an O2-dependent phenol oxidase whose structure, sequence, and activity are designed from first principles. The protein catalyzes the two-electron oxidation of 4-aminophenol (kcat/KM = 1,500 M·1·min·1) to the corresponding quinone monoimine by using a diiron cofactor. The catalytic efficiency is sensitive to changes of the size of a methyl group in the protein, illustrating the specificity of the design.
The de novo design of proteins provides a stringent test of our understanding of their molecular mechanisms of action (1–3). Recently, it has become possible to design proteins with novel three-dimensional structures (4), which has laid the groundwork for the elaboration of function. Catalysis provides a particularly challenging function to achieve, because a successfully designed protein catalyst must bind and precisely orient substrates, transition states, and intermediates adjacent to catalytic groups such as metal ions, general acids, and/or general bases. Two approaches to the design of catalytic proteins include automated sequence design, in which a novel catalytic site is engineered into a natural protein by mutating a subset of its side chains (5–7), and de novo protein design, which requires the simultaneous design of the entire backbone structure and sequence (2). The first method has the advantage of separating the problem of protein design and folding from the more restricted problem of designing an active site. The second approach has the potential advantage of greater applicability in terms of the sizes and shapes of substrates that can be accommodated. Furthermore, de novo protein design critically tests our understanding of how an amino acid sequence dictates both the folding as well as the activity of a protein.
To date, most work on the design of catalytic proteins has focused on hydrolysis of activated 4-nitrophenyl esters, using an active site histidine side chain as a nucleophilic catalyst. Automated sequence design methods have been used to design a variant of thioredoxin that hydrolyses 4-ntirophenyl acetate with a rate enhancement of ≈25-fold (7), when the value of kcat/KM was compared to the second order rate constant for hydrolysis of 4-methyl imidazole. Proteins with similar or greater catalytic efficiencies were observed frequently in a library of four-helix bundle proteins, whose polar exterior residues were randomly selected from Lys, His, Glu, Gln, Asp, and Asn (8). Baltzer and Nilsson (1) have designed a series of helical bundles that employ His residues to promote hydrolysis and intrapeptide acyl transfer reactions. It has also been possible to design helical bundles that catalyze decarboxylation of oxaloacetate (9), in one case by recognizing a key aldamine intermediate in the reaction (10).
Automated sequence design has also been used to design catalytic metalloproteins, by introducing an Fe(II/III)-binding site into various sites within thioredoxin (5). These proteins catalyzed the superoxide dismutase reaction, as well as the formation of diffusable oxygen radical species using hydrogen peroxide and O2 as substrates. An interesting correlation between the steric accessibility, electrochemical midpoint potential, and catalytic potential of the various sites was observed. However, to be biologically and chemically useful, it is important to design an active site that binds organic substrates and utilizes O2 locally within the active site, rather than creating diffusable oxygen radicals, which then react nondiscriminately with organic molecules.
Here, we describe the de novo design of model diiron proteins capable of catalyzing a phenol oxidase reaction. By combining two Fe(II/III) ions within a single site, it is possible to bias the reaction toward two-electron chemistry, thereby avoiding the formation of oxygen radicals. The starting point for the present design is the dueferri (DF) family of de novo-designed diiron proteins (11–13), whose structures have been extensively characterized by NMR and x-ray crystallography (11, 14–16). Here, we focus on DFtet, a four-chain heterotetrameric helical bundle whose structure was originally designed beginning with a mathematical equation describing the positions of the backbone atoms (12). The sequence was designed by using a computational method that not only considered the stabilization of the desired fold, but also the destabilization of likely alternatives. Thus, this protein is a complete product of de novo design, in which the backbone structure, sequence, and catalytic properties are all designed from first principles.
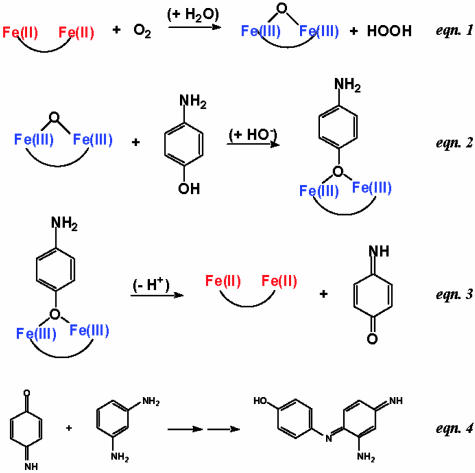
The intended reaction mechanism (Scheme 1) involved the use of O2 to oxidize the diferrous protein to a diferric species. The diferric protein then reacts with the substrate, 4-aminophenol (4AP), producing benzoquinone monoimine. The reduced diferrous form is then oxidized by O2, thereby initiating another catalytic cycle. The released quinone monoimine product is quenched and spectroscopically detected by using a reaction (eqn. 4 in Scheme 1) first studied by Witt (17) and subsequently by Gnaediger (18) and Corbett (19).
Scheme 1.
The ultimate goal is to design an efficient catalyst that does not fall into a deep energy minimum or encounter large energy barriers along any of these steps. Thus, the immediate goal is to find the intersection of sequence space that catalyzes eqn. 1–3 in Scheme 1.
Materials and Methods
Materials. The peptides were synthesized as described (12). All buffers, metal ions, and other chemicals were of the highest quality commercially available. The sequences of the original peptides are
 |
The residues in the A-subunits that were varied are underlined.
General Procedures. CD spectroscopy (12) and size exclusion chromatography (13) were carried out by using methods and equipment described previously.
Size Exclusion Chromatography. Peptides were mixed in the appropriate molar ratios at ≈20 μM (in each peptide) and chromatographed through a Superdex 75 FPLC (Amersham Pharmacia) column equilibrated in 10 mM Mes buffer, pH 6.5, containing 100 mM NaCl at a flow rate of 1.0 ml/min. As a standard for the tetrameric state, we used DFtet, which cleanly forms a tetramer as assessed by analytical ultracentrifugation. The protein is diluted 5- to 10-fold during chromatography, indicating that the protein was tetrameric under concentration range used in the kinetic experiments described below.
Binding and Kinetic Experiments. All experiments were conducted at pH 7.0 (0.15 M Mops/0.15 M NaCl). Kinetic experiments involving Fe(II)-binding/oxidation were performed at 25 μM tetramer concentration under aerobic and anaerobic conditions as described (13). The screening for phenolate binding was initially carried out with phenol, which was added to the diferric species as follows: (i) 2.0 equivalents of ferrous ion were added to the desired tetramer (25 μM tetramer concentration), and the product was incubated at room temperature for 2 h to assure complete oxidation to the diferric species; (ii) phenol was added to a final concentration of 25 mM; (iii) the spectra were measured after an additional 2-h incubation period. Additional experiments were conducted by using 4-cyanophenol (4CP), which was found to bind rapidly to the diferric protein and was resistant to oxidation.
Initial screening experiments measuring the oxidation of 4AP in the presence of 3-phenylene-diamine (MPD) were conducted at 5.0 μM tetramer [diferric formed by preincubation with 2 equivalents Fe(II) for 2 h at room temperature], and the reaction was initiated by addition of MPD (dissolved in dimethylformamide, DMF) to a final concentration of 10 mM, followed by 4AP (in 10% DMF). The final concentration of DMF was kept below 3% in all experiments. The reaction was then monitored by UV–visible spectroscopy, using a diode array spectrophotometer (13), and monitoring the absorption bands of the aminoindoaniline dye that are formed (19).
Experiments at variable substrate concentrations were conducted similarly. Because the extinction coefficient of the product is very sensitive to pH, we measured the product formation at the isosbestic point (528 nm). The extinction coefficient at this wavelength (10,700 M–1·cm–1) was determined under single-turnover conditions in which the substrate was quantitatively converted to product within 15 min. Two equivalents of monoimine are consumed in the formation of a single aminoindoaniline. Kinetics were measured within 1 h to avoid complications associated with conversion of the product to the corresponding phenazines, the accumulation of peroxide, and other decomposition products. Typically, initial rates, νinit, were determined from the first 20 min of the reaction, during which time product formation is linear with respect to time. The Michaelis–Menton constants were obtained from a least-squares fit to the equation (νinit = Vmax/(1 + KM/[4AP]). Competitive inhibition studies were conducted by using variable concentrations of 4CP (preincubated for 5 min), and fixed concentrations of 4AP (0.5 mM) and MPD (10.0 mM). Data were analyzed by using nonlinear least squares and the equation νinit = Vmax/(1 + K′M/[4AP]; (K′M = KM(1 + [4CP]/Ki), which allowed the determination of the dissociation constant for inhibition (Ki) as a function of the concentration of 4CP.
Results
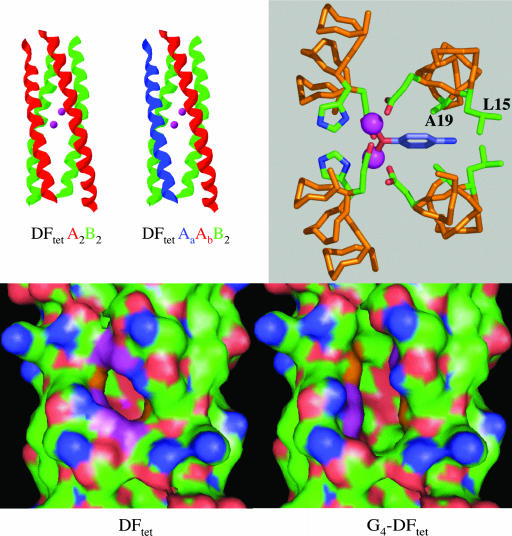
Design of Catalytically Active Variants. The catalytic peptides were designed by varying the sequence of DFtet AaAbB2, which has two identical “B” subunits and two different “A” subunits (Fig. 1 Upper Left). When mixed together in the appropriate stoichiometry, the individual peptides specifically self-assemble into an asymmetric, heterotetrameric helical bundle that binds two metal ions at its active site (13). When identical substitutions are desired in both the A subunits, we instead make the substitutions in the more symmetrical DFtet A2B2 bundle, containing two identical A and B subunits. The advantage of constructing the proteins by noncovalent self-assembly is that large numbers of variants can be formed by mixing various combinations of only a few variants of each noncovalent subunit.
Fig. 1.
Models of DFtet.(Upper Left) Subunit composition of DFtet A2B2 and DFtet AaAbB2.(Upper Right) Model of 4AP bound to DFtet A2B2. The carbons of the phenol are in cyan. (Lower) The solvent-accessible surface associated with DFtet and G4-DFtet.
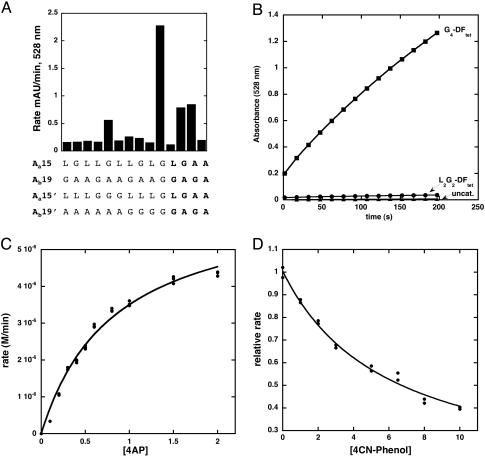
To introduce catalytic activity into these frameworks, we sculpted a pocket capable of binding the substrate 4AP by changing the residues that define the dimensions of the active site pocket. Fig. 1 Upper Right illustrates a model for the active site regions of DFtet A2B2 with 4AP modeled into the active site, and its phenolic oxygen bridging the metal ions (metal–oxygen distance, 2.0 Å). The substrate makes unfavorable contacts with Ala-19 and Leu-15. The steric bulk of these residues was therefore reduced in variants in which Ala-19 is changed to Gly and Leu-15 is changed to either Ala or Gly in both of the A chains.
We also made a number of symmetrical variations based on DFtet-A2B2. A symmetrical variant in which Leu-15 and Ala-19 of both A chains were substituted with Gly is designated G4-DFtet. A second variant, in which Leu-15 was retained and Ala-19 was changed to Gly, is designated DFtet-L2G2 (Fig. 1 Lower). Similarly, DFtet-A2G2 and DFtet-G2A2 have Gly or Ala at the indicated positions.
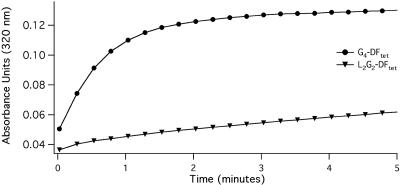
Catalytic Activity. We first examined the ability of the proteins to bind two equivalents of Fe(II) per tetramer, and rapidly oxidize the bound ferrous ions to Fe(III) in the presence of ambient O2, as measured by a strong ligand-to-metal charge transfer band near 320 nm associated with the diferric oxo-bridged species. Some variants produced a relatively stable intermediate with an absorbance near 600 nm (possibly a diferric peroxo species; ref. 13), whereas the variants with the fewest steric restrictions showed increasingly rapid formation of the oxo-species with no detectable intermediates. Fig. 2 compares the rate of formation of the diferric species in G4-DFtet versus L2G2-DFtet. The rate for G4-DFtet (1.5 min–1) is ≈25-fold faster than that observed for the slowest variant (L2G2-DFtet).
Fig. 2.
Oxidation of Fe(II) in the presence of G4-DFtet (circles) and L2G2 DFtet (triangles) (0.15 M Mops, pH 7.0/0.15 M NaCl; 25 μM tetramer; 50 μM iron).
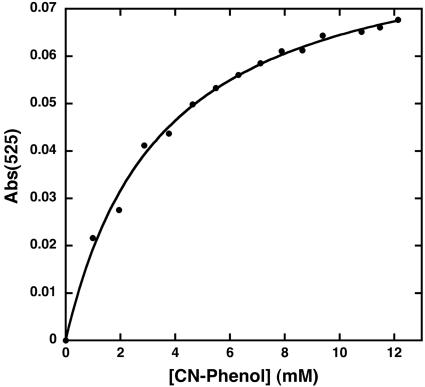
We next measured the ability of the differic forms of the variants to bind to the substrate analogue, phenol, by monitoring the strong ligand to metal charge transfer band associated with a phenolate bound to ferric ion (data not shown). Again, variants with the most open active site gave the greatest binding. Fig. 3 illustrates a binding isotherm for 4CP interacting with diferric G4-DFtet, which was the tetramer with the highest affinity for this phenol. Analysis of the data provides a dissociation constant of 3.5 mM, and an extinction coefficient of 3,500 M–1·cm–1 at 525 nm. Binding was rapid on the second time scale.
Fig. 3.
Binding of 4CP to diferric G4-DFtet (0.15 M Mops, pH 7.0/0.15 M NaCl, 25 μM tetramer).
Finally, we measured the 4AP oxidase activity of the protein in atmospheric O2. Screening was conducted at 5 μM protein concentration. Again, a correlation was observed between catalytic activity (Fig. 4A) and the size of the active site cavity. The most active variant G4-DFtet has the largest active site pocket. This variant showed an ≈1,000-fold rate enhancement, relative to the background reaction when the initial rate of the reaction in the presence and absence of the protein were compared (under conditions described in Fig. 4B). The G4-DFtet catalyzed this reaction for at least 100 turnovers. Changing either of the Gly residues at positions 19 or 15 to Ala gave a protein whose rate was decreased between 2.5- and 5-fold (Fig. 4B). Thus, changes as small as a methyl group had a significant effect on the catalytic activity of the protein.
Fig. 4.
Catalytic activity of G4-DFtet and variants. (A) Oxidation of 4AP catalyzed by variants of DFtet AaAbB2 and DFtet (5 μM tetramer). The last four bars in the histogram are for DFtet-A2B2 (in bold). (B) Catalysis of the oxidation of 4AP by G4-DFtet (squares, 100 μM) and L2G2-DFtet (diamonds, 100 μM) and the background reaction (triangles). (C) Rate of oxidation of 4AP vs. substrate concentration for G4-DFtet (5 μM tetramer). (D) Competitive inhibition of the oxidation of 4AP by 4CP (5 μM tetramer; 0.5 mM). All reactions included 0.15 M Mops (pH 7.0), 0.15 M NaCl, 0.5 mM 4AP, 10 mM MPD, and reactions in A, C, and D included 0.5 mM 4AP.
A measurement of the initial rate of substrate degradation as a function of the concentration of 4AP showed that G4-DFtet displayed saturation kinetics (Fig. 4C), and nonlinear least squares fitting provided Michaelis–Menton parameters, KM = 0.83 ± 0.06 mM; kcat = 1.3 ± 0.1 min–1; kcat/KM = 1,540 M–1·min–1. The protein catalyzed the oxidation of 4AP for >100 turnovers. The finding that kcat was nearly identical to the rate of oxidation of the diferrous to the diferric species (measured above) suggests that, at saturating 4AP concentrations and ambient O2 concentrations, the rate-limiting step is air oxidation of the differous form of the protein (eqn. 1 in Scheme 1). This oxidation may be limited by a conformational change in the protein, because the rate of oxidation of substrate was the same when O2-saturated buffer was used.
Several tests of the specificity and mechanism of the activity of G4-DFtet suggest that it catalyzes the reaction in the expected manner. The initial rate of oxidation of the substrate was unaffected by the addition of superoxide dismutase (25 units/ml), catalase (25 units/ml), or a spin trap (1 mM 1-hydroxy-2,2,5,5-tetramethyl-3-imidazoline-3-oxide), indicating that the reaction did not require the formation of diffusable radicals or peroxide. Also, 4CP is an efficient inhibitor of the reaction (Fig. 4D). The observed rate of oxidation of 4AP depends on the concentration of inhibitor, and the curve is well fit by a competitive inhibition scheme. The computed value of Kinh (3.7 ± 0.3 mM) was within experimental error of the value obtained above by direct titration (3.5 ± 0.3 mM).
The substrate specificity of G4-DFtet is consistent with its proposed mechanism of reaction. The substrate 4-methoxyanaline, in which the hydroxyl group of 4AP is converted to a methoxy, was not a substrate for the protein. Also, 4-aminoaniline, in which the phenolic hydroxyl is replaced by an amino group, was oxidized at a rate only 2-fold greater than the background reaction (5 μM protein, 0.5 mM substrate).
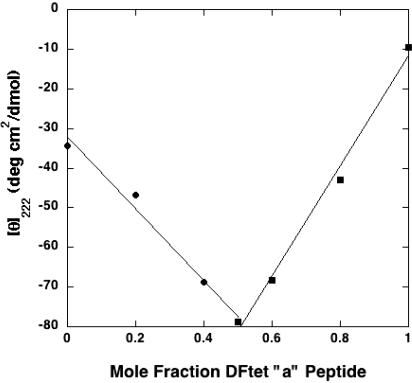
Assembly of G4-DFtet. Previously, we examined the structural and thermodynamic effects of opening the substrate-access cavity of DF1, by changing a pair of residues from Leu to Ala (15), as well as the helix-breaking residue, Gly (16). The mutations had relatively small (<1 Å) effects on the crystal structure of the protein, although they significantly changed its thermodynamic stability (14). We therefore examined the ability of G4-DFtet, which has four helix-destabilizing Gly residues per helical bundle, to fold into a four-helix bundle. At pH 6.5 and in the absence of metal ions, the individual subunits of G4-DFtet gave CD spectra indicative of largely unfolded peptides. However, when mixed in a 1:1 molar ratio, a complex was formed with a helical content identical to that of DFTet-A2B2. Thus, despite the helix-destabilizing Gly mutations, G4-DFtet retained the ability to fold. To confirm that the G4-DFtet-A and DFtet-B peptides associate with a 1:1 stoichiometry, different molar ratios of the peptides (5 μM total peptide concentration) were mixed, and the ellipticity at 222 nm was evaluated. A minimum (signifying maximal helical content) occurred at a molar ratio of precisely 0.5 (Fig. 5), indicating that the stoichiometry of the peptides was 1:1 (20). The G4-DFtet protein also appeared to be tetrameric at low micromolar concentrations, as assessed by size exclusion chromatography as in ref. 13.
Fig. 5.
Titration of G4-DFtet-A into DFtet-B measured by the mean residue ellipticity of the solution at 222 nm. The molar ratio of the two peptides was varied, whereas the total concentration was kept constant at 5.0 μM (100 mM NaCl/25 mM Mes, pH 6.5/1 mM EDTA). The lines shown are based on a linear least square fit of the data from 0 to 0.5 and from 0.5 to 1.0 molar ratio.
Discussion
In conclusion, this work describes an important step forward toward the ultimate goal of designing highly efficient and selective catalysts from scratch. The protein designed here uses a diiron (II/III) site to catalyze oxidation of a phenol with an electrochemical midpoint potentials near that of the cofactor. Other di-Mn and diiron enzymes similarly shuttle between di-Fe(II) and di-Fe(III) or di-Mn(II) and di-Mn(III). These proteins include manganese catalase (21, 22) and rubrerythrin, which is believed to be an NADH-dependent peroxide reductase (23).
Our protein shows many of the key features of biological enzymes in that it has a deeply invaginated active site into which substrates bind, and it displays saturation kinetics. Importantly, the protein is sensitive to changes as small as the size of a single methyl group in the active site residues (1, 7, 8). Although the rate is lower than that observed for many (but not all) highly evolved enzymes, it nevertheless is significantly greater than previous de novo-designed proteins and early catalytic antibodies (8).
Although the three-dimensional structure of the backbone and the sequence of the original DFtet was designed by using computational methods, the subsequent introduction of catalytic activity was accomplished without computational methods or by screening a large number of variants. Advanced methods of computational design (24) and/or screening of larger libraries of variants are quite likely to result in improved catalytic performance.
Abbreviations: DF, dueferri; 4AP, 4-aminophenol; 4CP, 4-cyanophenol; MPD, 3-phenylenediamine.
Note Added in Proof. While this paper was in review, Hellinga and coworkers (25) described and used computational methods to predict mutations that introduce triose phosphate isomerase activity into ribose-binding protein, a receptor that normally lacks enzyme activity.
References
- 1.Baltzer, L. & Nilsson, J. (2001) Curr. Opin. Biotechnol. 12, 355–360. [DOI] [PubMed] [Google Scholar]
- 2.Hill, R. B., Raleigh, D. P., Lombardi, A. & DeGrado, W. F. (2000) Acc. Chem. Res. 33, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffet, D. A. & Hecht, M. H. (2001) Chem. Rev. 101, 3191–3203. [DOI] [PubMed] [Google Scholar]
- 4.Kuhlman, B., Dantas, G., Ireton, G. C., Varani, G., Stoddard, B. L. & Baker, D. (2003) Science 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- 5.Benson, D. E., Wisz, M. S. & Hellinga, H. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D. E., Haddy, A. E. & Hellinga, H. W. (2002) Biochemistry 41, 3262–3269. [DOI] [PubMed] [Google Scholar]
- 7.Bolon, D. N. & Mayo, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei, Y. & Hecht, M. H. (2004) Protein Eng. Des. Sel. 17, 67–75. [DOI] [PubMed] [Google Scholar]
- 9.Johnsson, K., Allemann, R. K., Widmer, H. & Benner, S. A. (1993) Nature 365, 530–532. [DOI] [PubMed] [Google Scholar]
- 10.Allert, M. & Baltzer, L. (2003) Chembiochem 4, 306–318. [DOI] [PubMed] [Google Scholar]
- 11.Lombardi, A., Summa, C. & DeGrado, W. F. (2000) Proc. Natl. Acad. Sci. USA 97, 6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summa, C. M., Rosenblatt, M. M., Hong, J. K., Lear, J. D. & DeGrado, W. F. (2002) J. Mol. Biol. 321, 923–938. [DOI] [PubMed] [Google Scholar]
- 13.Marsh, E. N. & DeGrado, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 5150–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maglio, O., Nastri, F., Pavone, V., Lombardi, A. & DeGrado, W. F. (2003) Proc. Natl. Acad. Sci. USA 100, 3772–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Costanzo, L., Wade, H., Geremia, S., Randaccio, L., Pavone, V., DeGrado, W. F. & Lombardi, A. (2001) J. Am. Chem. Soc. 123, 12749–12757. [DOI] [PubMed] [Google Scholar]
- 16.DeGrado, W. F., Di Costanzo, L., Geremia, S., Lombardi, A., Pavone, V. & Randaccio, L. (2003) Angew. Chem. Int. Ed. Engl. 42, 417–420. [DOI] [PubMed] [Google Scholar]
- 17.Witt, O. N. (1879) Chem. Ber. 12, 933. [Google Scholar]
- 18.Ullmann, F. & Gnaedinger, J. (1912) Chem. Ber. 45, 3437–3445. [Google Scholar]
- 19.Corbett, J. F. (1969) J. Chem. Soc. B 8, 823–826. [Google Scholar]
- 20.Davies, D. R. & Rich, A. (1958) J. Am. Chem. Soc. 80, 1003–1004. [Google Scholar]
- 21.Law, N., Caudle, M. T. & Pecoraro, V. L. (1999) Adv. Inorg. Chem. 46, 305–439. [Google Scholar]
- 22.Dismukes, G. C. (1996) Chem. Rev. 96, 2909–2926. [DOI] [PubMed] [Google Scholar]
- 23.Lumppio, H. L., Shenvi, N. V., Summers, A. O., Voordouw, G. & Kurtz, D. M., Jr. (2001) J. Bacteriol. 183, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Looger, L. L., Dwyer, M. A., Smith, J. J. & Hellinga, H. W. (2003) Nature 423, 185–190. [DOI] [PubMed] [Google Scholar]
- 25.Dwyer, M. A., Looger, L. L. & Hellinga, H. W. (2004) Science 304, 1967–1971. [DOI] [PubMed] [Google Scholar]