SUMMARY
DNA methylation directed by 24-nucleotide (nt) small interfering RNAs (siRNAs) plays critical roles in gene regulation and transposon silencing in Arabidopsis. Twenty-four-nt siRNAs are known to be processed from double-stranded RNAs by Dicer-like 3 (DCL3) and loaded into the effector Argonaute 4 (AGO4). Here we report a distinct class of siRNAs independent of DCLs (sidRNAs). sidRNAs are present as ladders of ~20 to 60 nt in length, often having same 5’ ends but differing in 3’ ends by 1-nt steps. We further show that sidRNAs are associated with AGO4 and capable of directing DNA methylation. Finally we show that sidRNA production depends on distributive 3’-5’ exonucleases. Our findings suggest an alternative route for siRNA biogenesis: precursor transcripts are bound by AGO4 and subsequently subjected to 3’-5’ exonucleolytic trimming for maturation. We propose that sidRNAs generated through this route are the initial triggers of de novo DNA methylation.
INTRODUCTION
Cytosine DNA methylation is an epigenetic modification that regulates gene expression, represses transposable elements (TEs), and maintains genome integrity. In plants, DNA methylation is mainly found in TEs and occurs in three sequence contexts: CG, CHG and CHH (where H is either A, T, or C) (Law and Jacobsen, 2010). Arabidopsis thaliana has evolved specific DNA methyltransferases to maintain methylation in each context. Methylation at CG and CHG sites is maintained primarily by Methyltransferase 1 (MET1, an ortholog of DNMT1 in mammals) and Chromomethylase 3 (CMT3), respectively; whereas the maintenance of CHH methylation is catalyzed by Domain Rearranged Methyltransferase 2 (DRM2, an ortholog of DNMT3 in mammals) and CMT2 (Stroud et al., 2013; Zemach et al., 2013).
The establishment of DNA methylation in all sequence contexts is mediated by a specialized RNA interference (RNAi) pathway referred to as RNA-directed DNA methylation (RdDM) (Law and Jacobsen, 2010; Matzke et al., 2009). RdDM entails the transcription of target loci by the plant-specific RNA polymerase IV (Pol IV) (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005). The resulting transcripts (P4RNAs) serve as templates for RNA-dependent RNA polymerase 2 (RDR2) to make double-stranded RNAs (dsRNAs) (Haag et al., 2012; Xie et al., 2004), which are processed by Dicer-Like 3 (DCL3) into 24-nt small interfering RNAs (siRNAs) (Blevins et al., 2015; Qi et al., 2005; Xie et al., 2004; Zhai et al., 2015). These Pol IV-dependent siRNAs (P4siRNAs) are loaded into Argonaute 4 (AGO4) (Li et al., 2006; Qi et al., 2006; Ye et al., 2012; Zilberman et al., 2003). Likely through base-pairing between siRNAs and nascent scaffold transcripts generated by another plant specific RNA polymerase, Pol V (Wierzbicki et al., 2008), AGO4/siRNA complexes are recruited to target loci to direct DRM2-catalyzed DNA methylation (Wierzbicki et al., 2009; Zhong et al., 2014).
In addition to DCL3, Arabidopsis contains three other DCLs for generating distinct small RNA (sRNA) species. DCL1 acts in the maturation of 21-nt microRNAs (miRNAs) or siRNAs from hairpin-structured precursors (Kurihara and Watanabe, 2004; Rogers and Chen, 2013), DCL2 functions primarily in the production of 22-nt viral siRNAs (vsiRNAs) (Blevins et al., 2006; Bouche et al., 2006; Xie et al., 2004), whereas DCL4 mainly acts in the biogenesis of 21-nt trans-acting siRNAs (ta-siRNAs) (Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005). DCL2, DCL3 and DCL4 also function partially redundantly in P4siRNA production as well as the establishment and maintenance of DNA methylation (Henderson et al., 2006).
In this study, we identified a class of siRNAs that are independent of DCLs (named “sidRNA” for siRNAs independent of DCLs). sidRNAs are mainly originated from transposons and intergenic sequences as well as transgenes. sidRNAs interact with and recruit AGO4 to target loci to direct DNA methylation. Intriguingly, sidRNAs require 3’-5’ exonucleases for their production. Our study reveals an unexpected siRNA biogenesis pathway, which may act in the initiation of RdDM.
RESULTS
Genome-Wide Identification of Small RNAs that Are Independent of DCL Proteins in Arabidopsis Seedlings
The biogenesis of all sRNA species discovered so far in plants requires DCL proteins. In an attempt to examine whether there are DCL-independent sRNAs in Arabidopsis, we gel-purified RNAs between 18 and 30 nt from three-week-old seedlings of wild-type (Col-0), the triple mutant dcl2-1 dcl3-1 dcl4-2 (referred to as dcl2/3/4 hereafter) carrying null alleles of DCL2, DCL3 and DCL4 (Henderson et al., 2006), and the quadruple mutant dcl1-9 dcl2-1 dcl3-1 dcl4-2 (referred to as dcl1/2/3/4 hereafter) carrying an additional hypomorphic allele of DCL1 (Jacobsen et al., 1999) (Figure S1A), and prepared cDNA libraries for deep sequencing on an Illumina high-throughput sequencing platform (Table S1).
Analyses of sequenced reads revealed that miRNAs were reduced by ~98% in abundance in dcl1/2/3/4 relative to Col-0 or dcl2/3/4, and ta-siRNAs were reduced by ~99% in abundance in both dcl2/3/4 and dcl1/2/3/4 relative to Col-0 (Figure S1B), consistent with the demonstrated roles of DCL1 and DCL4 in processing miRNAs and ta-siRNAs, respectively (Kurihara and Watanabe, 2004; Xie et al., 2005). Intriguingly, considerable amounts of other siRNAs including P4siRNAs were still detected in dcl2/3/4 and dcl1/2/3/4 (Figure S1B). This was surprising because the biogenesis of P4siRNAs is known to be dependent mainly on DCL3 and partially on DCL2 and DCL4 (Henderson et al., 2006). In order to examine the origins of their biogenesis, after the removal of miRNAs and ta-siRNAs, siRNAs obtained from dcl1/2/3/4 were mapped to the Arabidopsis nuclear genome, and 14,360 siRNA-producing loci were identified using the following criteria: 1) length ≥ 100 nt; 2) number of unique sRNAs ≥ 5, each separated from nearest neighbors by a maximum of 50 nt; 3) expression level ≥ 15 RPKM (reads per kilobase per million) (Figure 1A; Table S2). These loci were mainly located in annotated TEs and repeats (74.4%) as well as intergenic regions (14.5%) (Figure 1A). Distinct from the siRNAs in Col-0 that are predominantly 24 nt in length, siRNAs in dcl1/2/3/4 and dcl2/3/4 exhibited a relatively even size distribution ranging from 18 to 30 nt, with a mild peak at 21 nt (Figure 1B). This suggests that the detected siRNAs were unlikely produced by DCLs, as DCLs are known to generate sRNAs of discrete sizes (Qi et al., 2005; Xie et al., 2004). Moreover, in contrast to the dramatic reduction of miRNA accumulation in dcl1/2/3/4 relative to dcl2/3/4, siRNAs generated from these loci accumulated at comparable levels in dcl2/3/4 and dcl1/2/3/4 (Figure 1C), suggesting that they were not products of the residual DCL1 activity in dcl1/2/3/4. We therefore named them sidRNAs for siRNAs independent of DCLs.
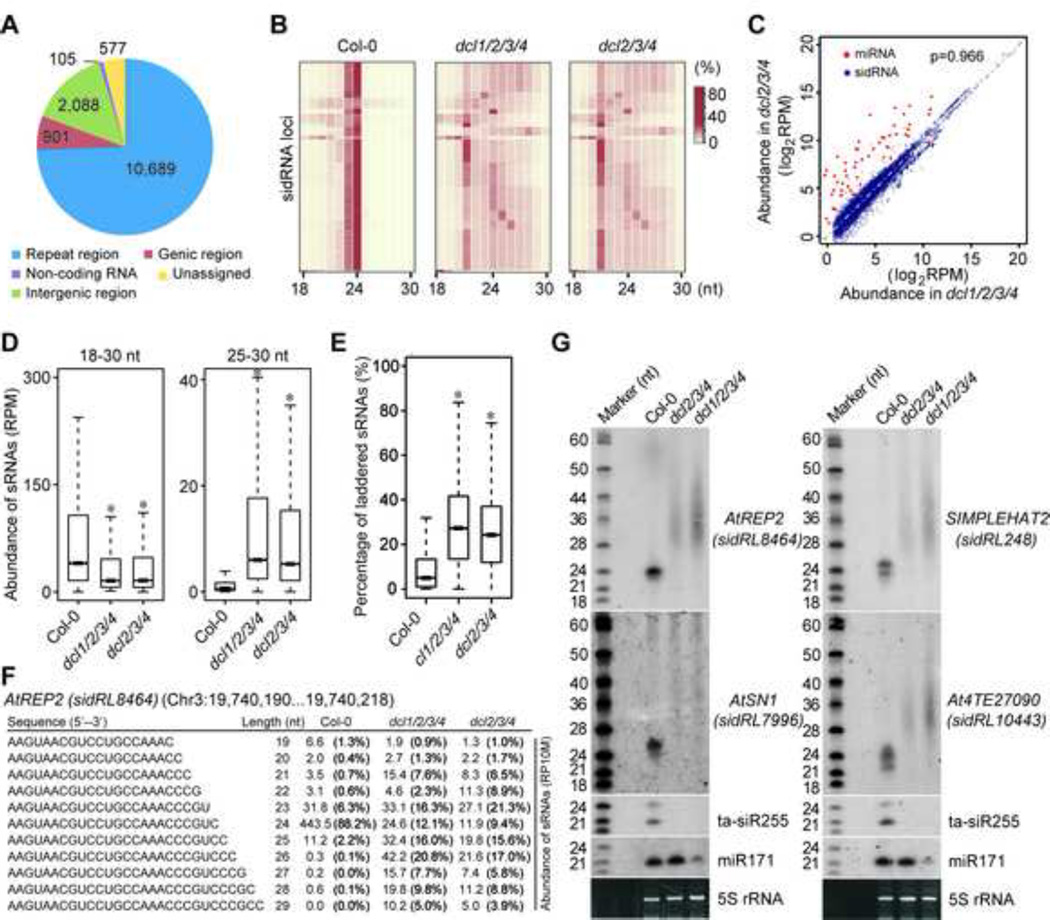
Figure 1. Identification of sidRNAs in Arabidopsis.
(A) Pie chart summarizing the numbers of sidRNA loci in the indicated categories.
(B) Heatmap showing the size distribution of sequenced sRNAs from sidRNA-producing loci in Col-0, dcl2/3/4 and dcl1/2/3/4. Color intensity represents the fraction of sRNAs of different sizes.
(C) Scatter plot showing the abundance (log2RPM) of each miRNA (red) and sidRNA locus (blue) in dcl1/2/3/4 versus that in dcl2/3/4. A reference line of slope=1 is shown. The Spearman’s correlation coefficient (ρ) for sidRNAs in dcl1/2/3/4 and dcl2/3/4 mutants is indicated.
(D) Box plots of levels of sRNAs of the indicated sizes produced from sidRNA loci in Col-0, dcl2/3/4 and dcl1/2/3/4. Asterisks indicate a significant difference between Col-0 and the mutants (P < 10−15, Mann–Whitney U test).
(E) Box plots of the percentages of laddered sRNAs produced from sidRNA loci in Col-0, dcl2/3/4 and dcl1/2/3/4. Asterisks indicate a significant difference between Col-0 and the mutants (P < 10−15, Mann–Whitney U test).
(F) A representative group of laddered sRNAs produced from AtREP2. Numbers represent the abundance (RPM) of sRNAs with different lengths ranging from 19 to 29 nt. Values in the parentheses indicate the percentages of sRNAs with different lengths.
(G) Detection of sRNA production at representative sidRNA loci in the indicated plants by Northern blot. 5S rRNAs stained with ethidium bromide were used as loading controls. A set of 32P-labeled RNA oligos were electrophoresed in parallel and used as size markers. The blots were stripped and re-probed.
See Figures S1, Table S1 and Table S2 for additional information about sidRNA identification in seedling, Figure S2, Figure S3 and Table S4 for information about detection of sidRNAs in other systems.
Further analyses indicated that total siRNAs produced from the sidRNA-generating loci in Col-0 were about five times more than those in dcl2/3/4 and dcl1/2/3/4 (Figure 1D). Interestingly, siRNAs of 25 nt or larger that very likely represent the production of sidRNAs, were also detected in Col-0, albeit at much lower levels than those in dcl2/3/4 and dcl1/2/3/4 (Figure 1D). Most intriguingly, considerable amounts of sidRNAs produced from each locus appeared as ladders in size, often having the same 5’ ends but differing in 3’ ends by singlent steps (Figure 1E and F). Northern blot analysis confirmed that sidRNAs generated from several representative loci (AtREP2, SIMPLEHAT2, AtSN1, and At4TE27090) over-accumulated in dcl2/3/4 and dcl1/2/3/4 and displayed a wide size distribution between 20 and 60 nt, with most being in the range of 30–40 nt (Figure 1G). RNA species of 30–60 nt were also detected in dcl2/3/4 and dcl1/2/3/4 by deep sequencing analysis (Figure S1C).
Effects of Pol IV/RDR2 Mutations on sidRNA Production
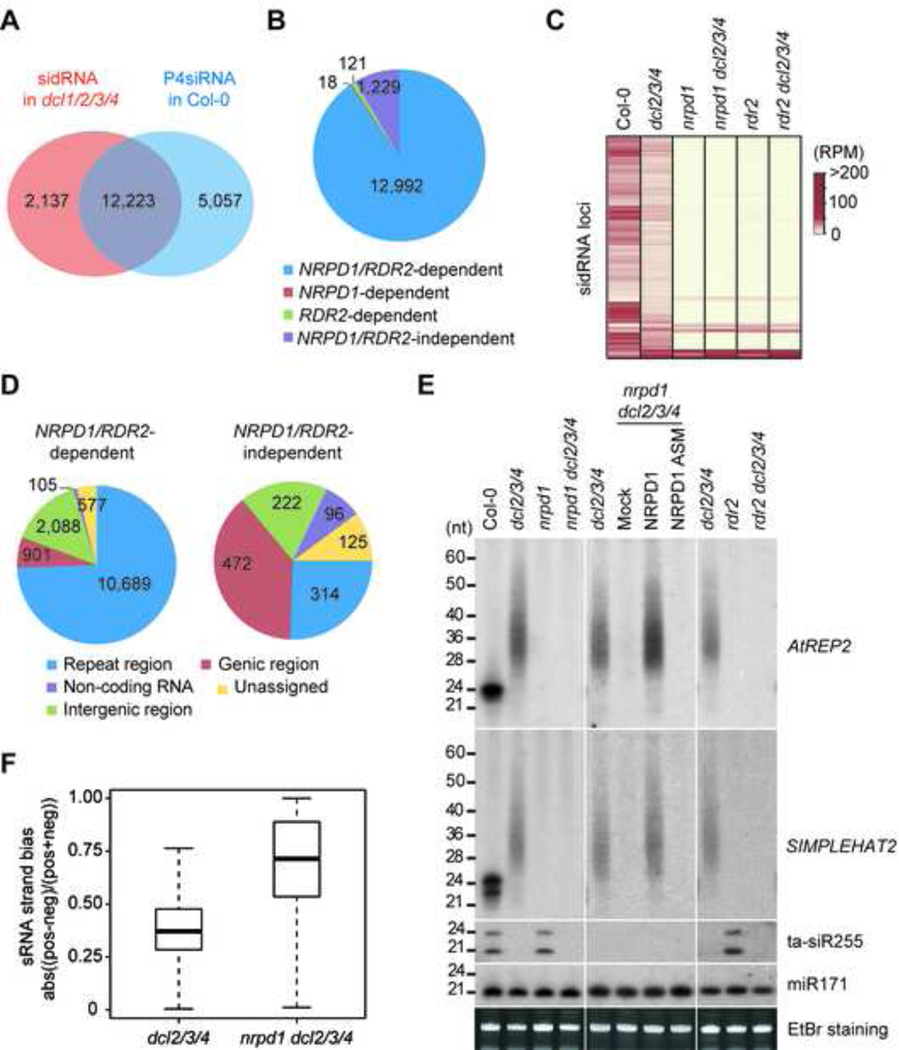
A comparison of profiles of sidRNAs in dcl1/2/3/4 and P4siRNAs in Col-0 revealed that over 85% of the sidRNA loci overlapped with the loci that produced P4siRNAs in Col-0 (Figure 2A). The generation of P4siRNAs relies on coupled actions of Pol IV and RDR2 (Haag et al., 2012). To examine whether sidRNA production is also dependent on Pol IV and RDR2, we introduced null mutation of NRPD1 (encoding the largest subunit of Pol IV) or RDR2 into the dcl2/3/4 mutant. Considering that dcl2/3/4 produces nearly equal amounts of sidRNAs as dcl1/2/3/4 and it is more convenient to combine mutations in dcl2/3/4 (Figure 1C, D and G), we used dcl2/3/4, instead of dcl1/2/3/4, in the following studies. The deep sequencing results showed that sidRNA production at 1,229 loci was independent of NRPD1 and RDR2, whereas that at 12,992 loci was dependent on both NRPD1 and RDR2 (Figure 2B and Table S3) and dysfunction of either NRPD1 or RDR2 resulted in severe reduction of sidRNA production at these loci (Figure 2C and Table S3). Intriguingly, vast majority of the NRPD1/RDR2-dependent sidRNA loci were located in repeat regions and only 7% of them resided in genic regions, whereas 38% of NRPD1/RDR2-independent sidRNA loci were located in genic regions (Figure 2D). Northern blot results confirmed that mutation in Pol IV or RDR2 had a dramatic effect on the generation of sidRNAs at representative loci AtREP2 and SIMPLEHAT2 (Figure 2E). Expression of wild-type NRPD1, but not an active site mutant (ASM) of NRPD1 (Haag et al., 2009), was able to restore the sidRNA levels in dcl2/3/4 (Figure 2E), further indicating that functional NRPD1 is essential for sidRNA generation at these loci. We noticed that, however, low levels of sidRNAs could still be detected at many of the NRPD1/RDR2-dependent sidRNA loci in nrpd1 dcl2/3/4 or rdr2 dcl2/3/4 and they appear to have a strong strand bias (Figure 2F).
Figure 2. Effects of Pol IV/RDR2 Mutations on sidRNA Production.
(A) Venn diagram showing the overlap between sidRNA loci in dcl1/2/3/4 and P4siRNA loci in Col-0.
(B) Pie chart showing the numbers of sidRNA loci dependent on NRPD1 and/or RDR2.
(C) Heat map of the abundance of sRNAs generated from sidRNA loci in the indicated plants.
(D) Pie chart summarizing the numbers of NRPD1/RDR2-dependent and -independent sidRNA loci in the indicated categories.
(E) Detection of sRNA production at representative sidRNA loci in the indicated plants by Northern blot. 5S rRNAs stained with ethidium bromide were used as loading controls.
(F) Box plots showing the strand bias of sidRNAs in dcl2/3/4 and nrpd1 dcl2/3/4 mutants. Strand bias value was calculated as the absolute (abs) percentage differences between sRNAs reads produced from the positive (pos) and negative (neg) strands of sidRNA loci.
See also Table S3.
Detection of sidRNAs in Other Systems
The existence of sidRNAs in Arabidopsis seedlings prompted us to determine whether sidRNAs are also prevalent in other systems. First, Arabidopsis endosperm is a specialized tissue that supplies nutrients for the developing embryo. There exists a large population of P4siRNAs in Arabidopsis endosperm (Mosher et al., 2009). These P4siRNAs are predominantly, if not exclusively, derived from the maternal allele and the expression of a subset of these siRNAs is restricted to flowers and young siliques (Mosher, 2010; Mosher et al., 2009). Through deep sequencing of endosperm and embryo tissues of Col-0, dcl2/3/4 and nrpd1 dcl2/3/4 (Table S1), we found the existence of Pol IV-dependent and endosperm-specific sidRNAs at 140 loci (Table S3), represented by a population of laddered 25–30-nt siRNAs (Figure S2A–C).
Second, the production of siRNAs can be induced by actively transcribed transgenes (Beclin et al., 2002; Mourrain et al., 2000). To explore whether sidRNA production can be detected in a transgene system, we transformed Arabidopsis with a 35S:GUS construct and monitored the production of sidRNAs derived from the transgenic region over six generations by deep sequencing (Table S1). sidRNAs expressed in the same ladder-like pattern were detected in each generation of transgenic lines (Figure S3A–C), suggesting that, like endogenous loci, transgenic loci can also produce sidRNAs. Intriguingly, Pol IV was dispensable for the generation of sidRNAs at the transgenic loci (Figure S3A and B), hinting that other RNA polymerase, presumably Pol II, is responsible for transcribing these sidRNAs.
Third, active transposons often trigger the production of sRNAs that direct the establishment of DNA methylation and inactivation of their transposition (Ito, 2012; Kim and Zilberman, 2014). To determine whether sidRNAs can be derived from active transposable elements, we took advantage of an epigenetic recombinant inbred line (epi15), in which a retrotransposon called EVD became active and invasive from the eighth generation (F8) of inbreeding due to the loss of DNA methylation at its 5’ long-terminal repeat (LTR) region (Mari-Ordonez et al., 2013). DCL-dependent siRNAs that are 21–22 nt or 24 nt long were found to be produced to initiate DNA methylation and suppress EVD mobilization from F8 to F14 (Mari-Ordonez et al., 2013). Analyses of the published sRNA datasets (Mari-Ordonez et al., 2013) revealed that high levels of laddered sidRNAs were also detected in the generations F11 and F14 (Figure S3D–F). The generation of sidRNAs derived from 3’ gag region appeared to precede the production of sidRNAs transcribed from 5’ LTR (Figure S3E), coinciding with the 3’-5’ spreading of DNA methylation.
sidRNAs Are Associated with AGO4
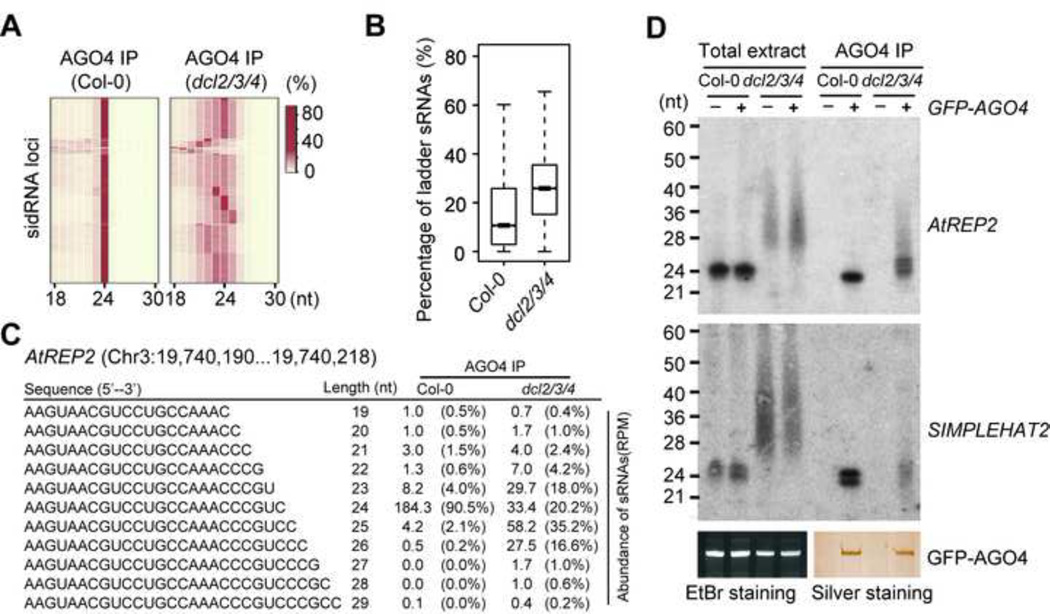
In light of the prevalence of sidRNA expression, we asked whether sidRNAs could be functional via associating with AGO proteins. Since the loci generating sidRNA and 24-nt siRNA largely overlap, and the majority of sidRNAs depend on Pol IV and RDR2 for their biogenesis, we hypothesized that a large proportion of sidRNA may be channeled to binding with AGO4. To test this, we immunoprecipitated AGO4 from transgenic lines expressing N-terminally GFP-tagged AGO4 (GFP-AGO4) in Col-0 or dcl2/3/4 background and purified sRNAs associated with GFP-AGO4. Deep sequencing of purified sRNAs revealed that AGO4 predominantly bound 24-nt siRNAs in Col-0 but bound sidRNAs that were mainly 21–26 nt in length in dcl2/3/4 (Figure 3A). A considerable fraction of AGO4-associated sidRNAs generated from each locus also appeared as ladders (Figure 3B and C), albeit to a lesser extent compared to the bulk of sidRNAs (Figure 1E and F) . Confirming the deep sequencing results, Northern blot analysis revealed that AGO4 bound sidRNAs at AtREP2 and SIMPLEHAT2 loci, although the sizes of AGO4-associated sidRNAs were smaller, compared to those detected in total extract in dcl2/3/4 (Figure 3D). We speculate that the longer forms of RNA species are precursors of mature sidRNAs associated with AGO4. It is noteworthy that a small fraction of presumed sidRNA precursors was also detected in AGO4 immunoprecipitates (Figure 3D), suggesting that these transcripts are loaded onto AGO4 before being processed into mature sidRNAs. In summary, our findings suggest that AGO4 is an effector protein of sidRNAs.
Figure 3. sidRNAs Are Associated with AGO4.
(A) Heatmap showing the size distribution of AGO4-associated sRNAs from sidRNA-producing loci in Col-0, dcl2/3/4 and dcl1/2/3/4. Color intensity represents the fraction of sRNAs of different sizes.
(B) Box plots of the percentages of laddered AGO4-associated sRNAs produced from sidRNA loci in Col-0 and dcl2/3/4.
(C) A representative group of laddered AGO4-associated sRNAs produced from AtREP2. Numbers represent the abundance (RPM) of sidRNAs with different lengths ranging from 19 to 29 nt. Values in the brackets indicate the percentages.
(D) Detection of sRNAs at representative sidRNA loci in total extracts and AGO4 immunoprecipitates by Northern blot. 5S rRNAs stained with ethidium bromide were used as loading controls for total extracts. A silver-stained gel shows that comparable amounts of AGO4 immunoprecipitates were used for sRNA extraction.
sidRNAs Are Capable of Directing DNA Methylation
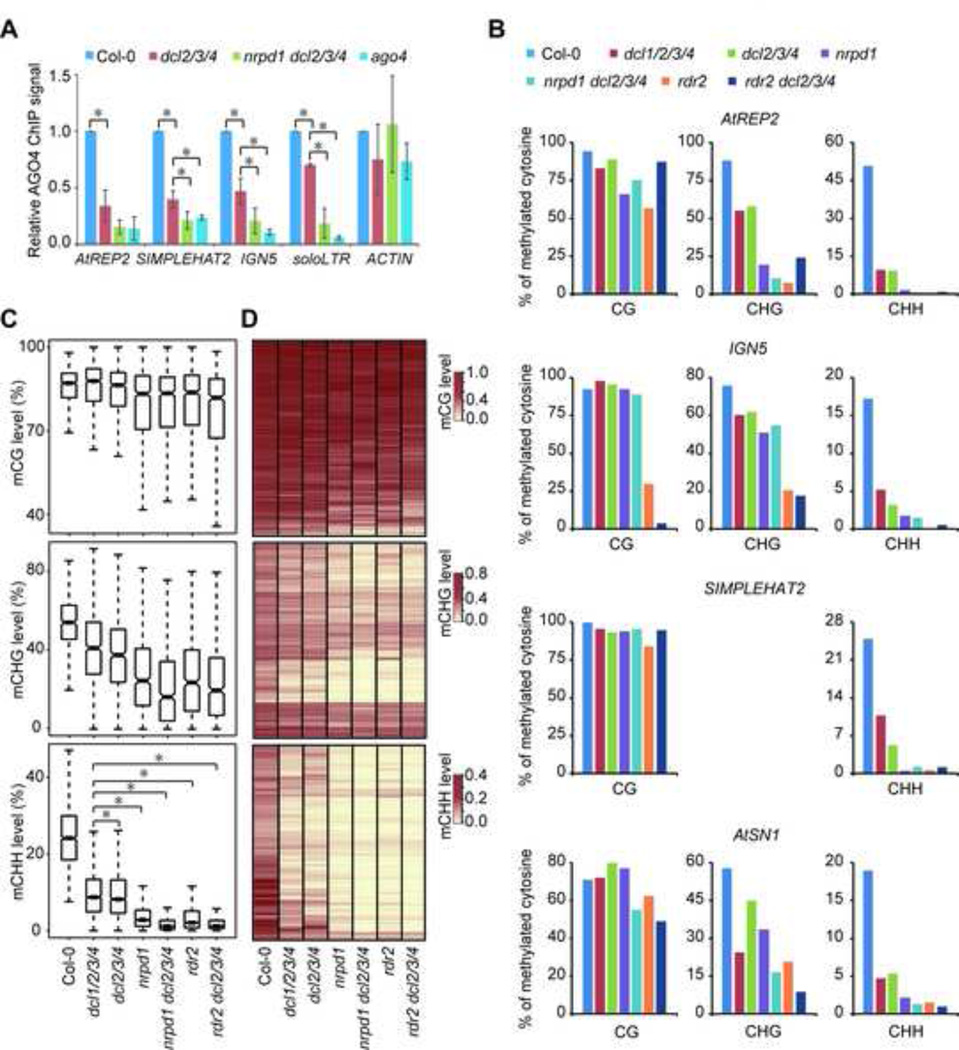
The association of sidRNAs with AGO4 prompted us to examine whether sidRNAs are capable of guiding AGO4 to target loci. To this end, we performed ChIP-qPCR to assess the occupancy of AGO4 at several representative loci in Col-0 as well as in mutants including dcl2/3/4, nrpd1 dcl2/3/4 and ago4. As expected, AGO4 was enriched at these loci in Col-0 (Figure 4A). However, locus-specific AGO4 occupancy in dcl2/3/4 was moderately reduced, but not eliminated (Figure 4A), suggesting that factors other than 24-nt siRNAs, presumably sidRNAs, are involved. Indeed, AGO4 occupancy was further decreased by mutation of NRPD1 that is important for sidRNA biogenesis at these loci and by mutation of AGO4 that binds sidRNAs (Figure 4A). Thus, our data suggest that sidRNAs are capable of targeting AGO4 to chromatin.
Figure 4. sidRNAs Are Capable of Directing DNA Methylation.
(A) Detection of AGO4 occupancy at the indicated loci in Col-0 and the indicated mutants by ChIP analysis. AGO4 ChIP signals in the mutants are presented in relation to those in Col-0 (arbitrarily set to 1.0). Error bars indicate standard deviations of three biological replicates. Asterisks indicate a significant difference between the indicated groups (P < 0.05, t test ).
(B) Analysis of DNA methylation at the indicated loci in Col-0 and the indicated mutants by bisulfite sequencing. Presented is overall percent methylation of cytosine sites in different sequence contexts. More than 20 clones were sequenced for each sample
(C) Box plots of CG, CHG and CHH methylation levels at sidRNA loci in Col-0 and the indicated mutants. Asterisks indicate a significant difference between the indicated groups (P < 10−15, Mann–Whitney U test).
(D) Heat map of CG, CHG and CHH methylation levels at sidRNA loci in Col-0 and the indicated mutants.
We next examined whether sidRNAs play a role in directing DNA methylation. By performing locus-specific bisulfite sequencing, we found that nrpd1 dcl2/3/4 and rdr2 dcl2/3/4, mutants deficient in both 24-nt siRNA and sidRNA biogenesis, exhibited greater defects in DNA methylation than dcl1/2/3/4 and dcl2/3/4 (Figure 4B), suggesting that sidRNAs play a role in directing DNA methylation. Although the extent of reduction in DNA methylation caused by loss of sidRNAs appeared to vary in a locus-specific manner, for all loci examined, DNA methylation in CHG and CHH contexts was more severely affected than that in CG context (Figure 4B).
To investigate the impact of sidRNA deficiency on DNA methylation genome-wide, we carried out whole genome bisulfite sequencing and found that the overall CHG and CHH DNA methylation levels in nrpd1 dcl2/3/4 and rdr2 dcl2/3/4 are significantly lower than those in dcl1/2/3/4 and dcl2/3/4 (Figure 4C and D). The level of CG methylation was slightly reduced by NRPD1 or RDR2 mutation (Figure 4C and D). Here our findings are in agreement with previous methylome surveys showing that dcl2/3/4 is a mutant causing reduced CHH methylation whereas nrpd1 and rdr2 are mutants leading to eliminated CHH methylation (Stroud et al., 2013). Taken together our data suggest that sidRNAs primarily contribute to CHG and CHH methylation.
Probing the Mechanism Underlying sidRNA Biogenesis
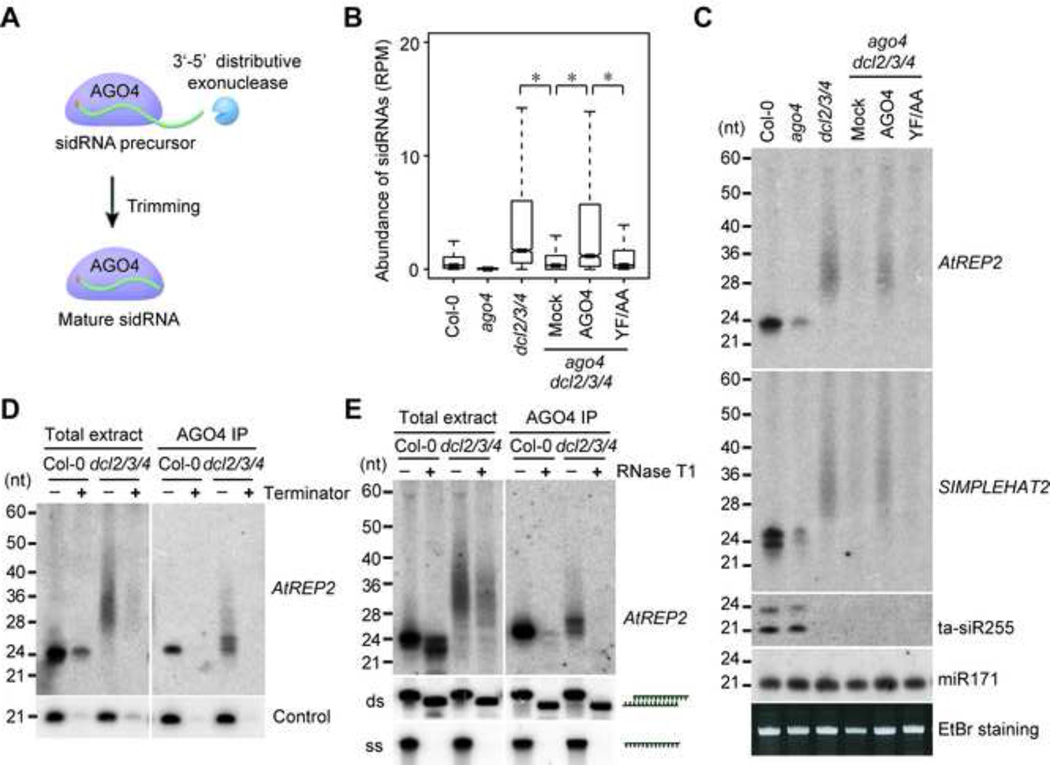
The ladder-like expression pattern of sidRNAs is reminiscent of the products of distributive 3’-5’ exonucleases. Distributive exonuclease are known to cleave, release, and engage another substrate, creating laddered products (Deutscher and Li, 2001). We thus hypothesize that the biogenesis of sidRNAs might involve binding of the 5’-end of sidRNA precursor transcripts to AGO4 and subsequent trimming from the 3’-end by a 3’-5’ distributive exonuclease (Figure 5A).
Figure 5. Probing The Mechanism Underlying sidRNA Biogenesis.
(A) A model for sidRNA maturation. A sidRNA precursor transcript is bound by AGO4 and subsequently trimmed from the 3’-end by a 3’-5’ distributive exonuclease to yield a mature sidRNA.
(B) Box plots of the abundance of sidRNAs in the indicated plants. Asterisks indicate a significant difference between the indicated groups (P < 10−15, Mann–Whitney U test).
(C) Detection of sRNA production at representative sidRNA loci in the indicated plants by Northern blot. 5S rRNAs stained with ethidium bromide were used as loading controls.
(D) Sensitivity of sidRNAs to Terminator exonuclease. sRNAs in total extracts and AGO4 immunoprecipitates prepared from Col-0 and dcl2/3/4 were treated with Terminator exonuclease and AtREP2 sRNAs were probed by Northern blot. A 21-nt spike-in RNA with 5’ monophosphate was added in the reactions and used as a positive control.
(E) Sensitivity of sidRNAs to RNase T1. sRNAs in total extracts and AGO4 immunoprecipitates prepared from Col-0 and dcl2/3/4 were treated with RNase T1 and AtREP2 sRNAs were probed by Northern blot. Spike-in RNAs including a 50-bp double-stranded RNA with 2-nt 3’ over-hang (ds) and a 50-nt single-stranded (ss) RNA were added in the reactions and used as controls for RNase T1 digestion.
In accordance with this hypothesis, we found that the sidRNA level in dcl2/3/4 was dramatically reduced by mutation of ago4, suggesting the involvement of AGO4 in sidRNA production (Figure 5B). Moreover, transgenic expression of wild-type AGO4, but not a mutant AGO4 (AGO4YFAA) that is defective in sRNA binding (Ye et al., 2012), was able to restore the generation of sidRNAs in ago4 dcl2/3/4 (Figure 5B), suggesting that the sRNA binding activity of AGO4 is important for sidRNA generation. Locus-specific examination of sidRNA production by northern blot also revealed that sidRNA generation required AGO4 and its sRNA binding activity (Figure 5C). It is known that some components acting at the effector stage of RdDM pathway are also required for siRNA production, likely through a feedback regulation. We cannot exclude the possibility that the role of AGO4 in sidRNA production could also be indirect,
To gain insights into the mechanism of sidRNA biogenesis, we investigated the features of sidRNAs. We first examined whether sidRNAs have 5’ triphosphate or monophosphate ends. Total RNAs or AGO4-bound RNAs were prepared from Col-0 and dcl2/3/4 and treated with a 5’ to 3’ exonuclease Terminator that specifically degrades RNAs with 5’ monophosphate ends. sRNAs generated from AtREP2 locus were detected by northern blot. As expected, Terminator treatment dramatically reduced the abundance of 24-nt siRNAs in Col-0 that bear a 5’ monophosphate (Figure 5D). Interestingly, sidRNA levels in dcl2/3/4 were also greatly reduced by Terminator treatment (Figure 5D), suggesting that sidRNAs primarily have 5’ monophosphate ends.
We next examined whether sidRNAs are double-stranded or single-stranded by treating sidRNAs with RNase T1, an endoribonuclease that degrades single-stranded RNAs. Control double-stranded RNA (dsRNA) migrated faster in gel because of the degradation of 2-nt 3’ overhangs whereas control single-stranded RNAs (ssRNAs) became disappeared after RNase T1 treatment (Figure 5E). Consistent with our previous finding that 24-nt siRNAs in total extract are mainly double-stranded and become single-stranded when associated with AGO4 (Ye et al., 2012), 24-nt siRNAs in total extract but not those associated with AGO4 were resistant to RNase T1 digestion (Figure 5E). Interestingly, we found that a fraction of the sidRNAs in total extract were single-stranded, whereas AGO4-associated sidRNAs were exclusively single-stranded (Figure 5E), suggesting that the process of sidRNA maturation involves the conversion of dsRNAs to ssRNAs and subsequent loading of ssRNAs onto AGO4. Taken together, we uncovered several important features of sidRNAs, which support our trimming model.
Distributive 3’-5’ Exonucleases Are Required for sidRNA Accumulation and DNA Methylation
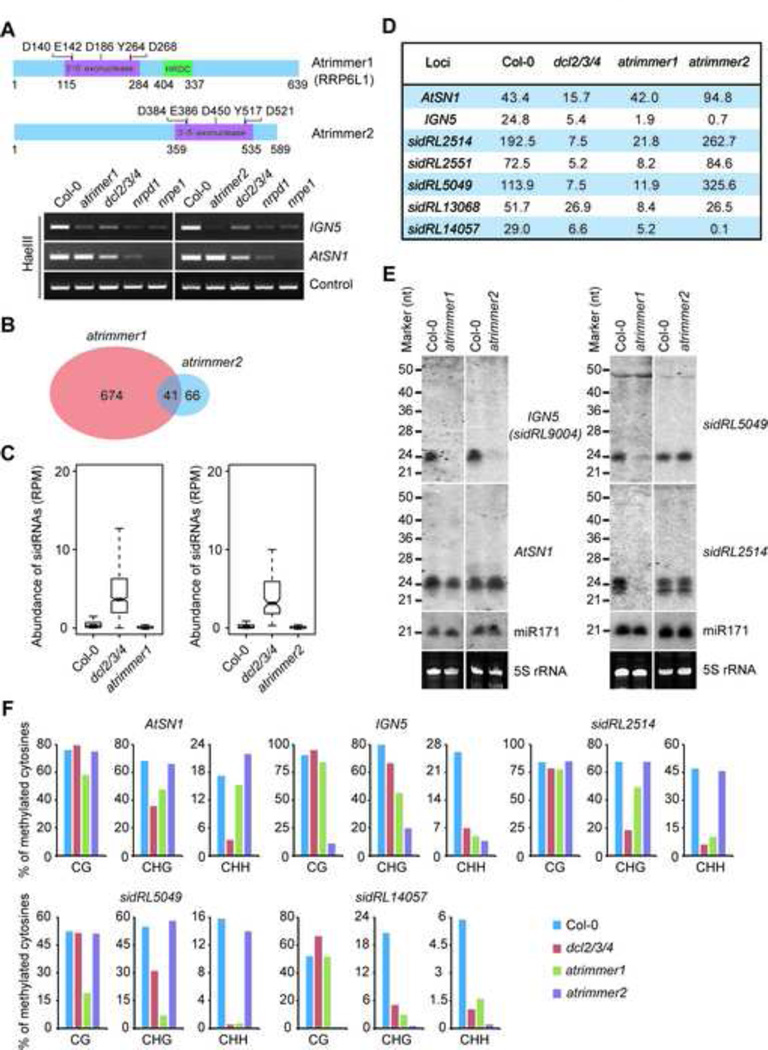
As proposed above, the production of sidRNAs needs 3’-5’ distributive exonucleases for trimming, we thus sought to identify the exonucleases that are required for sidRNA biogenesis. Since sidRNAs play a role in CHG and CHH DNA methylation, we attempted to search for candidate exonucleases involved in sidRNA production by performing methylation-specific Chop-PCR, a rapid way to examine CHH methylation levels at representative loci (Zhang et al., 2014b). Among thirteen mutants carrying null mutations of 3’-5’ distributive exonucleases, two mutants, with mutations mapped to At1g54440 and At1g56310, respectively, had reduced CHH methylation levels at IGN5 (Figures 6A and S4). At1g54440 encodes AtRRP6L1, a nuclear subunit that associates with the exosome core complex. AtRRP6L1 has been reported to function in the processing of long non-coding RNAs (lncRNAs) involved in epigenetic silencing (Shin and Chekanova, 2014). It also participates in the retention of Pol V transcribed scaffold lncRNAs through a pathway independent of exosome-mediated RNA degradation (Zhang et al., 2014a). At1g56310 encodes an uncharacterized exonuclease. Considering their possible involvement in trimming sidRNAs, we named AtRRP6L1 and the uncharacterized ribonuclease Atrimmer1 and Atrimmer2, respectively. Both Atrimmer1 and Atrimmer2 contain a conserved 3’-5’ exonuclease domain (Figure 6A).
Figure 6. Distributive 3’-5’ Exonucleases Are Required for siRNA Accumulation and DNA Methylation.
(A) Identification of distributive 3’-5’ exonucleases required for DNA methylation. Diagrams show the structure of Atrimmer1/RRP6L1 and Atrimmer2 (Upper panel). The positions of residues forming the conserved DEDD-Y active site are indicated. The bottom panel shows DNA methylation levels at the AtSN1 and IGN5 loci in the indicated plants as measured by Chop-PCR. The methylation-sensitive restriction enzyme HaeIII was used to specifically cleave unmethylated DNA in CHH context. A fragment of Actin that lacks HaeIII sites was amplified as a control. See Figure S4 for Chop-PCR results from all tested 3’-5’ exonuclease mutants.
(B) Venn diagram showing the numbers of sidRNA loci affected by atrimmer1/rrp6l1, atrimmer2 or both.
(C) Box plots of the levels of sidRNAs generated from Atrimmer1- (Left panel) and Atrimmer2- (Right panel) affected sidRNA loci in the indicated plants.
(D) Relative levels (shown in RPMs) of representative Atrimmer1/2-controlled sidRNA loci in the indicated plants measured by deep sequencing.
(E) Detection of sRNA production at representative sidRNA loci in the indicated plants by Northern blot. 5S rRNAs stained with ethidium bromide were used as loading controls.
(F) Analysis of DNA methylation at the indicated loci in Col-0 and the indicated mutants by bisulfite sequencing. Presented is overall percent methylation of cytosine sites in different sequence contexts. More than 20 clones were sequenced for each sample.
To better investigate the functions of Atrimmer1 and Atrimmer2 in sidRNA biogenesis, we carefully examined the change of DNA methylation levels at sidRNA-generating loci in several mutants. The CHH methylation levels at IGN5 and AtSN1 in nrpd1 and nrpe1 (NRPE1 encodes the largest subunit of Pol V) are lower than those in dcl2/3/4 (Figure 6A), suggesting that IGN5 and AtSN1 are sidRNA-generating loci. Surprisingly, the extents of reduction in IGN5 CHH DNA methylation levels in atrimmer1 and atrimmer2 were greater than that in dcl2/3/4 and comparable to those in nrpd1 and nrpe1 (Figure 6A), indicating that Atrimmer1 and Atrimmer2 dysfunction likely abolished DCL-dependent DNA methylation as well as sidRNA-mediated DNA methylation. One explanation for our results could be that sidRNA biogenesis is required for DCL-dependent biogenesis of 24-nt siRNAs.
To further explore whether Atrimmer1 and Atrimmer2 are required for sidRNA biogenesis genome-wide, we performed sRNA deep sequencing analysis with atrimmer1 and atrimmer2 mutants to identify Atrimmer1 and/or Atrimmer2 dependent sidRNA loci. We found that Atrimmer1 and Atrimmer2 regulated the production of sidRNAs at 715 and 107 loci, respectively, with 41 loci being regulated by both (Figure 6B). In atrimmer1 and atrimmer2, the levels of sidRNAs at the regulated loci were dramatically reduced (Figure 6C). To determine whether Atrimmers regulate sidRNA production and eventually regulate DNA methylation, we chose several loci with sidRNA production impaired by Atrimmer dysfunction (Figure 6D and E) and performed locus-specific bisulfite sequencing. Importantly, we found that dysfunction of Atrimmer1 or Atrimmer2 caused further reduction of DNA methylation levels at some loci compared to dysfunction of DCLs. CHG and CHH DNA hypomethylation was always detected at loci where sidRNAs were produced at low levels in atrimmer1 and atrimmer2 (Figure 6F). Our data suggest that Atrimmer1 and Atrimmer2 promote locus-specific DNA methylation through regulating sidRNA production. Taken together, we identified Atrimmer1 and Atrimmer2 as potential trimmers of sidRNAs at a subset of loci.
DISCUSSION
Our study demonstrates the existence of a distinct class of sRNAs (sidRNAs) that are generated via an alternative route independent of DCLs in Arabidopsis. The biogenesis of sidRNAs likely entails 5’-end binding of sidRNA precursor transcripts to AGO4 followed by trimming by 3’-5’ exonucleases (Atrimmers). Intriguingly, DNA methylation mediated by sidRNAs appears to be required for the canonical RdDM. We propose that sidRNAs are the initial triggers of RdDM.
sidRNA Biogenesis
In this study, we identified 14,360 loci that produce sidRNAs in Arabidopsis seedlings (Figure 1A). The majority of the identified sidRNA loci overlap with P4siRNA loci (Figure 2A), and dysfunction of NRPD1 or RDR2 largely reduced sidRNA production (Figure 2C and E), suggesting that Pol IV/RDR2 transcripts are precursors of sidRNAs at these loci. This is also supported by the fact that DNA regions producing Pol IV/RDR2-dependent transcripts in dcl2/3/4 strongly overlap with P4siRNA loci (Li et al., 2015).
RDR2 dysfunction, which was thought to cause accumulation of single-stranded nascent Pol IV transcripts, surprisingly resulted in dramatic loss of sidRNA signals (Figure 2C and E). A previous study also showed that RDR2 has the same effect on the production of Pol IV-dependent transcripts, as does Pol IV (Li et al., 2015). One possible explanation is that RDR2 may play a role in stabilizing Pol IV transcripts and, once RDR2 binds to Pol IV transcripts, the stabilization facilitates the conversion of Pol IV transcripts into dsRNAs. However, in light of the weak Pol IV transcriptional activity but robust RDR2 activity (Haag et al., 2012), we favor another possibility that RDR2 converts single-stranded Pol IV transcripts into dsRNAs, which are then unwound and separated into ssRNAs. These ssRNAs then serve as RDR2 substrates and undergo exponential amplification by RDR2. In this case, a large amount of single-stranded sidRNA precursors are generated and RDR2 dysfunction leads to loss of most, if not all, RNA signal. This process resembles the amplification of single-stranded priRNA precursors by a RDRC in fission yeast (Halic and Moazed, 2010) and replication of ssRNA viruses from limited amounts of viral genomic RNAs by viral RdRPs (Ahlquist, 2002).
Nascent transcripts of Pol IV/RDR2 are expected to have a 5’ triphosphate. However, recent studies showed that Pol IV/RDR2 transcripts bear 5’ monophosphate (Li et al., 2015; Zhai et al., 2015). We found that sidRNAs and their precursors also bear 5’ monophosphate (Figure 5D), suggesting that Pol IV/RDR2 transcripts are likely processed by a yet-to-be identified mechanism before they are loaded onto AGO4. The absence of double-stranded sidRNA precursors in AGO4 immunocomplex (Figure 5E) suggests that sidRNA precursors are loaded onto AGO4 in single-stranded forms or in double-stranded forms but one of the strands is quickly dissociated from the complex. The anchoring of single-stranded sidRNA precursors then facilitates the trimming process as discussed below.
The majority of AGO4-associated sidRNAs are shorter than sidRNAs in total cellular extract (Figure 3), suggesting that sidRNAs are further processed upon anchoring to AGO4. The laddered pattern of AGO4-associated sidRNAs is in favor of an evolutionarily conserved trimming mechanism involving the actions of 3’-5’ distributive exonucleases. Exonucleases have been found to participate in RNAi, transposon silencing and heterochromatin formation in multiple species. For instance, in fission yeast, RNA degradation products associate with AGO1 to recruit Triman for their 3’ end processing, resulting in priRNAs that initiate heterochromatin formation (Halic and Moazed, 2010; Marasovic et al., 2013). In Neurospora, the exonuclease QIP mediates 3’-5’ trimming and maturation of miRNA-like sRNAs (Xue et al., 2012). The exonuclease MUT-7 has been reported to interact with protein complex that generate sRNAs and thus be required for RNAi and transposon silencing in worm (Ketting et al., 1999; Tops et al., 2005). In silkworm, primary piRNA precursors are trimmed to the mature piRNA size by the activity of an unknown 3’-5’ exonuclease (Kawaoka et al., 2011). In this study, we showed that two 3’-5’ exonucleases, Atrimmer1 and Atrimmer2, are required for sidRNA generation in Arabidopsis (Figure 6). It appeared that Atrimmer1 and Atrimmer2 merely regulate sidRNA generation at ~5% of the sidRNA loci that we have identified. Since our screening is based on methylation status of two loci, exonucleases responsible for sidRNA generation at other loci were very likely neglected. Also, functional redundancies probably hindered the isolation of more Atrimmer mutants. It is very likely that there are other exonucleases that control sidRNA generation at different loci. It will be interesting to identify these exonucleases in the future.
Loss of Atrimmers unexpectedly resulted in diminished sidRNA production instead of accumulation of longer sidRNA precursors (Figure 6E), suggesting that, in addition to performing their trimming functions, Atrimmers may also stabilize sidRNA precursors. Without Atrimmers, sidRNA precursors would dissociate from AGO4 and undergo degradation. Although Atrimmer1/RRP6L1 is a nuclear auxiliary subunit of core complex, the trimming function of Atrimmer1 seems to be independent of exosome function because depletion of two exosome subunits (RRP4 and RRP41) caused neither defective sRNA accumulation nor defects in DNA methylation (Shin et al., 2013; Zhang et al., 2014a).
In addition to Pol IV/RDR2-dependent sidRNA, sidRNA production at 1,229 loci appears to be independent of NRPD1 and RDR2 (Figure 2B and C). We also found that low levels of sidRNAs were still produced from many of the Pol IV/RDR2-dependent loci in the absence of Pol IV or RDR2 and they are mainly derived from one of the DNA strands (Figure 2F). In light of the fact that Pol II-dependent RNAs with a strong strand bias have been previously detected at loci that generate P4siRNAs (Li et al., 2015), we propose that Pol II contributes to the production of sidRNA precursors. Supporting this notion, we found that sidRNAs were produced from a transgenic locus under the control of a 35S promoter that is transcribed by Pol II (Figure S3A–C).
A proposal for sidRNAs as Initial Triggers of DNA Methylation
The canonical RdDM pathway involves 24-nt siRNAs that are produced by coordinated activities of Pol IV, RDR2 and DCL3. These 24-nt siRNAs are bound by AGO4 and used as a guide through base pairing with Pol V-generated scaffold transcripts, to recruit DRM2 that catalyzes de novo DNA methylation (Law and Jacobsen, 2010; Matzke et al., 2009). Targeting of Pol IV and Pol V is thus critical for the specificity of RdDM. It has been shown that the recruitment of Pol IV is assisted by two Pol-IV-interacting proteins, Classy 1 (CLSY1) (Law et al., 2011) and Sawadee Homeodomain Homolog 1(SHH1)/DNA-Binding Transcription Factor 1 (DTF1) (Law et al., 2013; Zhang et al., 2013). SHH1/DTF1 is capable of binding methylated H3K9 and such binding activity is required for its role in targeting Pol IV to RdDM targets (Law et al., 2013; Zhang et al., 2013), suggesting the involvement of H3K9 methylation at RdDM targets in efficient Pol IV occupancy. Recently, it has been shown that Pol V is recruited to pre-methylated DNA through the methyl-DNA binding proteins SUVH2 and SUVH9 (Johnson et al., 2014; Liu et al., 2014). This is likely achieved through the interaction between SUVH2/SUVH9 and the Pol V-interacting DMS3/DRD1/RDM1 (DDR) complex (Johnson et al., 2014; Law et al., 2010; Liu et al., 2014; Zhong et al., 2012). Thus, the recruitment of Pol IV and Pol V appears to require pre-existing epigenetic modifications (H3K9 and DNA methylation, respectively) at RdDM targets. However, this raises an important question as to how RdDM is initiated in the first place when a locus (for example, a newly integrated transgene or TE) originally does not have any epigenetic marks.
Methylation at subsets of TEs has been shown to be dependent on Pol II (Stroud et al., 2013) and RDR6 (Nuthikattu et al., 2013; Pontier et al., 2012). It has been proposed that some 21–22-nt siRNAs processed by DCL2 and DCL4 from Pol II/RDR6-generated dsRNAs initiate low levels of de novo DNA methylation in an AGO2- and/or AGO6-dependent manner, and such initial methylation activates the canonical RdDM pathway (Kim and Zilberman, 2014; Matzke and Mosher, 2014; McCue et al., 2015; Panda and Slotkin, 2013). Another study utilizing a unique system that reconstructed de novo silencing of an active retrotransposon suggests that dsRNAs produced by Pol II/RDR6, when present at high levels, could be processed by DCL3 into 24-nt siRNAs to induce de novo methylation (Mari-Ordonez et al., 2013).
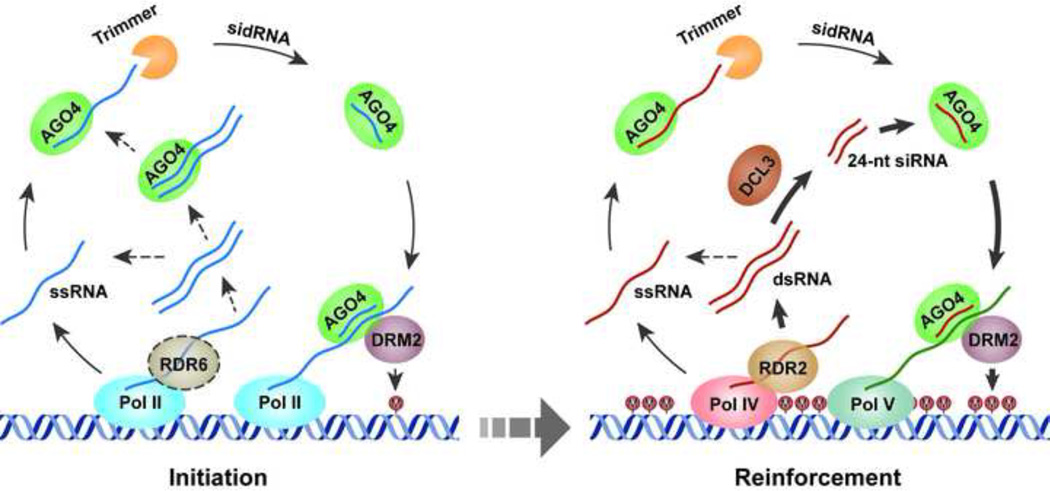
Our finding of sidRNAs leads us to propose an alternative model for the initiation of RdDM (Figure 7). In this model, Pol II transcripts (possibly amplified by RDR6) associate with AGO4 and undergo 3’-5’ trimming by Atrimmers. The resulting sidRNAs then mediate the initiation of DNA methylation at locus without any epigenetic modification by recruiting DRM2. Low levels of DNA methylation established by Pol II-dependent sidRNAs may mark the locus for subsequent recruitment of Pol IV and RDR2, which produces either double-stranded or single-stranded RNA precursors. Double-stranded precursors are processed by DCL3 to generate 24-nt siRNAs that are subsequently loaded onto AGO4, whereas single-stranded precursors directly bind to AGO4 and undergo 3’-5’ trimming. Despite being generated through different routes, both 24-nt siRNAs and sidRNAs then recruit DRM2 by base pairing with scaffold transcripts generated by Pol V, to reinforce DNA methylation. The maintenance of CG is then controlled by MET1. By contrast, the maintenance of CHG and CHH methylation might still rely on coordinated actions of 24-nt siRNA and sidRNAs. The role of sidRNAs in maintaining DNA methylation is supported by the results that sidRNAs mediate enrichment of AGO4 on chromatin and CHG/CHH DNA methylation (Figure 4).
Figure 7. A Model for Biogenesis of sidRNAs and Initiation of DNA Methylation.
In this model, an active locus (for example, a newly integrated transgene or transposon) is transcribed by Pol II. Pol II transcripts (possibly amplified by RDR6). associate with AGO4 and undergo 3’-5’ trimming by Atrimmers. The resulting sidRNAs then mediate the initiation of DNA methylation by recruiting DRM2. Low levels of DNA methylation established by sidRNAs may mark the locus for subsequent recruitment of Pol IV and RDR2. The vast majority of the Pol IV/RDR2 transcripts are processed by DCL3 to generate 24-nt siRNAs that are subsequently loaded onto AGO4, while a small fraction of them are directly recruited by AGO4 and undergo 3’-5 trimming to produce sidRNAs. 24-nt siRNAs and sidRNAs then recruit DRM2 by base pairing with scaffold transcripts generated by Pol V, to reinforce DNA methylation.
In our model, DCL3-dependent biogenesis of canonical 24-nt siRNAs and Atrimmer-dependent biogenesis of sidRNAs are not mutually exclusive. Instead, they act in parallel in wild type plants to produce sRNAs from TEs at which RdDM has been established. At loci with established RdDM, the vast majority of Pol IV/RDR2 transcripts are processed by DCL3 into canonical 24-nt siRNAs and a small fraction of them are processed into sidRNAs by the trimming mechanism. In the dcl2/3/4 mutant, where DCL3 is absent, a larger fraction of the Pol IV/RDR2 transcripts are processed by the trimming mechanism, as evident from the over-accumulation of laddered sRNAs in these mutants (Figure 1E–G). At active loci, Pol II/RDR6 transcripts are processed by the trimming mechanism to produce sidRNAs, which trigger RdDM. In summary, we propose that sidRNAs produced by the trimming mechanism have a minor contribution to the maintenance of DNA methylation at established RdDM loci, but play key roles in the initiation of RdDM.
EXPERIMENTAL PROCEDURES
Plant Materials and Oligonucleotides
All Arabidopsis lines used in this study are in Columbia (Col-0) background. Detailed information about mutants, transgenic lines, and oligonucleotides used in this study can be found in the Extended Experimental Procedures.
Deep Sequencing and Data Analysis
sRNAs of 18–30 nt or 30–60 nt were gel-purified and subjected to library construction as described (Mi et al., 2008). Bisulfite sequencing libraries were constructed using the NEXTflex Bisulfite-Seq kit (BIOO SCIENTIFIC) according to the manufacturer’s instructions. Libraries were sequenced on the Illumina HiSeq 2000 platform by Bionova (Beijing) Biotech Company. Detailed protocols for library construction and methods for sequencing data analyses can be found in the Extended Experimental Procedures.
sRNA Northern Blot
sRNAs were purified from total extracts or AGO4 immunoprecipitates and subjected to Northern blot analysis as described (Qi et al., 2006).
Immunoprecipitation
Immunoprecipitation of GFP-AGO4 complexes from 3-week-old seedlings was performed as described previously (Ye et al., 2012). The quality of purification was examined by SDS-PAGE followed by silver staining.
AGO4 ChIP
ChIP assay was performed with anti-AGO4 antibody (αAGO4) as described (Rowley et al., 2011).
Bisulfite Sequencing and Chop-PCR
Genomic DNA was extracted from 3-week-old seedlings using the DNeasy Plant Mini kit (QIAGEN). 500 ng of DNA was used for bisulfite sequencing analysis as previously described (Wu et al., 2010). 500 ng of DNA was used for Chop-PCR as described (Zhang et al., 2014b) after digestion with HaeIII.
Analyses of Features of sidRNAs
Sensitivities of sidRNAs to RNase T1 and Terminator exonuclease were examined to determine whether sidRNAs are single stranded and contain 5’ monophosphate ends. Detailed protocols can be found in the Extended Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (Grant Nos. 31225015, 31330042 and 31421001) to Y. Q. We’d like to acknowledge the assistance of Computing Platform of National Protein Science Facility (Beijing).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Sequencing data have been deposited in Gene Expression Omnibus under accession number GSE74398.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, four Supplemental Figures, and seven Supplemental Tables and can be found with this article online at XXXXXX.
AUTHOR CONTRIBUTIONS
R.Y., Z.C., B.L., M.J.R., and N.X. conducted the experiments, R.Y., Z.C., B.L., M.J.R., Y.L., J.C., X.H., A.T.W., and Y.Q analyzed the data, R.Y., Z.C., X.H., A.T.W., and Y.Q. designed the experiments. R.Y., Y.L., and Y.Q. wrote the paper.
REFERENCES
- Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1270–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Current biology : CB. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife. 2015:4. doi: 10.7554/eLife.09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Jr, Hohn T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic acids research. 2006;34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. The EMBO journal. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Progress in nucleic acid research and molecular biology. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Current biology : CB. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PloS one. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Molecular cell. 2012;48:811–818. doi: 10.1016/j.molcel.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-Independent Primal RNAs Trigger RNAi and Heterochromatin Formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Zhang X, Lu C, Johnson L, Meyers BC, Green PJ, Jacobsen SE. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nature Genetics. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Ito H. Small RNAs and transposon silencing in plants. Development, growth & differentiation. 2012;54:100–107. doi: 10.1111/j.1440-169X.2011.01309.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- Johnson LM, Du J, Hale CJ, Bischof S, Feng S, Chodavarapu RK, Zhong X, Marson G, Pellegrini M, Segal DJ, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y. 3’ end formation of PIWI-interacting RNAs in vitro. Molecular cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends in plant science. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu JK, Wohlschlegel JA, Jacobsen SE. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Current biology : CB. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature reviews Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS genetics. 2011;7:e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, Chen X. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome research. 2015;25:235–245. doi: 10.1101/gr.182238.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZW, Shao CR, Zhang CJ, Zhou JX, Zhang SW, Li L, Chen S, Huang HW, Cai T, He XJ. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS genetics. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Molecular cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Mari-Ordonez A, Marchais A, Etcheverry M, Martin A, Colot V, Voinnet O. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Current opinion in cell biology. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature reviews Genetics. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- McCue AD, Panda K, Nuthikattu S, Choudury SG, Thomas EN, Slotkin RK. ARGONAUTE 6 bridges transposable element mRNA-derived siRNAs to the establishment of DNA methylation. The EMBO journal. 2015;34:20–35. doi: 10.15252/embj.201489499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA. Maternal control of Pol IV-dependent siRNAs in Arabidopsis endosperm. The New phytologist. 2010;186:358–364. doi: 10.1111/j.1469-8137.2009.03144.x. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, DeFraia C, Thomas EN, Slotkin RK. The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21–22 nucleotide small interfering RNAs. Plant physiology. 2013;162:116–131. doi: 10.1104/pp.113.216481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Panda K, Slotkin RK. Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant signaling & behavior. 2013;8:e25206. doi: 10.4161/psb.25206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Picart C, Roudier F, Garcia D, Lahmy S, Azevedo J, Alart E, Laudie M, Karlowski WM, Cooke R, et al. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Molecular cell. 2012;48:121–132. doi: 10.1016/j.molcel.2012.07.027. [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Molecular cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang X-J, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. The Plant cell. 2013;25:2383–2399. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS genetics. 2011;7:e1002120. doi: 10.1371/journal.pgen.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Chekanova JA. Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS genetics. 2014;10:e1004612. doi: 10.1371/journal.pgen.1004612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Wang HL, Lee J, Dinwiddie BL, Belostotsky DA, Chekanova JA. The role of the Arabidopsis Exosome in siRNA-independent silencing of heterochromatic loci. PLoS genetics. 2013;9:e1003411. doi: 10.1371/journal.pgen.1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tops BBJ, Tabara H, Sijen T, Simmer F, Mello CC, Plasterk RHA, Ketting RF. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic acids research. 2005;33:347–355. doi: 10.1093/nar/gki183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Molecular cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS biology. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Yuan H, Guo J, Liu Y. Reconstitution of an Argonaute-dependent small RNA biogenesis pathway reveals a handover mechanism involving the RNA exosome and the exonuclease QIP. Molecular cell. 2012;46:299–310. doi: 10.1016/j.molcel.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Molecular cell. 2012;46:859–870. doi: 10.1016/j.molcel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & development. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell. 2015;163:445–455. doi: 10.1016/j.cell.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ma ZY, Zeng L, Tanaka K, Zhang CJ, Ma J, Bai G, Wang P, Zhang SW, Liu ZW, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tang K, Qian W, Duan CG, Wang B, Zhang H, Wang P, Zhu X, Lang Z, Yang Y, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Molecular cell. 2014a;54:418–430. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tang K, Wang B, Duan CG, Lang Z, Zhu JK. Protocol: a beginner’s guide to the analysis of RNA-directed DNA methylation in plants. Plant methods. 2014b;10:18. doi: 10.1186/1746-4811-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157:1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nature Structural & Molecular Biology. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.