Abstract
Nitric oxide ( NO) and its reactive metabolites mediate the oxidation, nitration, and nitrosation of DNA bases, amino acids, and lipids. Here, we report the structural characterization and quantitation of two allylic nitro derivatives of linoleic acid (LNO2), present as both free and esterified species in human red cell membranes and plasma lipids. The LNO2 isomers 10-nitro-9-cis, 12-cis-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid were synthesized and compared with red cell and plasma LNO2 species based on chromatographic elution and mass spectral properties. Collision-induced dissociation fragmentation patterns from synthetic LNO2 isomers were identical to those of the two most prevalent LNO2 positional isomers found in red cells and plasma. By using [13C]LNO2 as an internal standard, red cell free and esterified LNO2 content was 50 ± 17 and 249 ± 104 nM, respectively. The free and esterified LNO2 content of plasma was 79 ± 35 and 550 ± 275 nM, respectively. Nitrated fatty acids, thus, represent the single largest pool of bioactive oxides of nitrogen in the vasculature, with a net LNO2 concentration of 477 ± 128 nM, excluding buffy coat cells. These observations affirm that basal oxidative and nitrating conditions occur in healthy humans to an extent that is sufficient to induce abundant membrane and lipoprotein–fatty acid nitration. Given that LNO2 is capable of mediating cGMP and non-cGMP-dependent signaling reactions, fatty acid nitration products are species representing the convergence of
NO) and its reactive metabolites mediate the oxidation, nitration, and nitrosation of DNA bases, amino acids, and lipids. Here, we report the structural characterization and quantitation of two allylic nitro derivatives of linoleic acid (LNO2), present as both free and esterified species in human red cell membranes and plasma lipids. The LNO2 isomers 10-nitro-9-cis, 12-cis-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid were synthesized and compared with red cell and plasma LNO2 species based on chromatographic elution and mass spectral properties. Collision-induced dissociation fragmentation patterns from synthetic LNO2 isomers were identical to those of the two most prevalent LNO2 positional isomers found in red cells and plasma. By using [13C]LNO2 as an internal standard, red cell free and esterified LNO2 content was 50 ± 17 and 249 ± 104 nM, respectively. The free and esterified LNO2 content of plasma was 79 ± 35 and 550 ± 275 nM, respectively. Nitrated fatty acids, thus, represent the single largest pool of bioactive oxides of nitrogen in the vasculature, with a net LNO2 concentration of 477 ± 128 nM, excluding buffy coat cells. These observations affirm that basal oxidative and nitrating conditions occur in healthy humans to an extent that is sufficient to induce abundant membrane and lipoprotein–fatty acid nitration. Given that LNO2 is capable of mediating cGMP and non-cGMP-dependent signaling reactions, fatty acid nitration products are species representing the convergence of  NO and oxygenated lipid cell-signaling pathways.
NO and oxygenated lipid cell-signaling pathways.
Nitric oxide ( NO) is an endogenous mediator of cell function that acts predominantly by means of the stimulation of soluble guanylate cyclase (1). In addition to regulating vascular relaxation,
NO) is an endogenous mediator of cell function that acts predominantly by means of the stimulation of soluble guanylate cyclase (1). In addition to regulating vascular relaxation,  NO modulates oxidative and free radical reactions, inflammatory cell function, posttranslational protein modification, neurotransmission, and regulation of gene expression (2–5).
NO modulates oxidative and free radical reactions, inflammatory cell function, posttranslational protein modification, neurotransmission, and regulation of gene expression (2–5).  NO exerts a particularly broad influence on oxidative inflammatory reactions by reacting at diffusion-limited rates with superoxide (
NO exerts a particularly broad influence on oxidative inflammatory reactions by reacting at diffusion-limited rates with superoxide ( O2–, k = 1.9 × 1010 M–1·sec–1) to yield peroxynitrite (ONOO–) and its conjugate acid, peroxynitritrous acid (ONOOH), the latter of which undergoes homolytic scission to nitrogen dioxide (
O2–, k = 1.9 × 1010 M–1·sec–1) to yield peroxynitrite (ONOO–) and its conjugate acid, peroxynitritrous acid (ONOOH), the latter of which undergoes homolytic scission to nitrogen dioxide ( NO2) and hydroxyl radical (
NO2) and hydroxyl radical ( OH) (2, 6). Also, biological conditions favor the reaction of ONOO– with CO2, generating nitrosoperoxycarbonate (ONOOCO2–; k = 3 × 104 M–1·sec–1), which yields
OH) (2, 6). Also, biological conditions favor the reaction of ONOO– with CO2, generating nitrosoperoxycarbonate (ONOOCO2–; k = 3 × 104 M–1·sec–1), which yields  NO2 and carbonate (
NO2 and carbonate ( CO3–) radicals by means of homolysis or rearranges to NO3– and CO2 (7). During inflammation, neutrophil myeloperoxidase and heme proteins, such as myoglobin and cytochrome c, form H2O2-dependent Compound I intermediates that are direct oxidants and catalyze the consumption of
CO3–) radicals by means of homolysis or rearranges to NO3– and CO2 (7). During inflammation, neutrophil myeloperoxidase and heme proteins, such as myoglobin and cytochrome c, form H2O2-dependent Compound I intermediates that are direct oxidants and catalyze the consumption of  NO and the oxidation of nitrite (NO2–) to
NO and the oxidation of nitrite (NO2–) to  NO2 (8–11). Although the rate of reaction of
NO2 (8–11). Although the rate of reaction of  NO with O2 is slow (k = 2 × 106 M–2·sec–1), the small molecular radius, uncharged nature, and lipophilicity of
NO with O2 is slow (k = 2 × 106 M–2·sec–1), the small molecular radius, uncharged nature, and lipophilicity of  NO and O2 facilitate their diffusion and concentration in membranes and lipoproteins up to 20-fold (12–14). This “molecular lens” effect that is induced by solvation in hydrophobic cell compartments accelerates the reaction of
NO and O2 facilitate their diffusion and concentration in membranes and lipoproteins up to 20-fold (12–14). This “molecular lens” effect that is induced by solvation in hydrophobic cell compartments accelerates the reaction of  NO with O2 to yield N2O3 and N2O4. As a consequence of this diversity of
NO with O2 to yield N2O3 and N2O4. As a consequence of this diversity of  NO reactions with partially reduced oxygen species, a rich spectrum of products is formed that orchestrates target molecule oxidation, nitrosation, and nitration reactions.
NO reactions with partially reduced oxygen species, a rich spectrum of products is formed that orchestrates target molecule oxidation, nitrosation, and nitration reactions.
Multiple mechanisms account for the nitration of fatty acids by  NO-derived species (15–20). During both basal cell-signaling and tissue-inflammatory conditions,
NO-derived species (15–20). During both basal cell-signaling and tissue-inflammatory conditions,  NO2 that is generated by the aforementioned reactions can react with membrane and lipoprotein lipids. Environmental sources also yield
NO2 that is generated by the aforementioned reactions can react with membrane and lipoprotein lipids. Environmental sources also yield  NO2 as a product of combustion.
NO2 as a product of combustion.  NO2 initiates autooxidation of polyunsaturated fatty acids by means of hydrogen abstraction from the bis-allylic carbon, to form nitrous acid and a resonance-stabilized allylic radical (21). This lipid radical species predominantly reacts with molecular oxygen to form a peroxyl radical. During the unique oxidation–reduction conditions of inflammation or ischemia–reoxygenation, tissue O2 levels are often suppressed and nitrogen oxide levels are elevated, favoring lipid radical reaction with
NO2 initiates autooxidation of polyunsaturated fatty acids by means of hydrogen abstraction from the bis-allylic carbon, to form nitrous acid and a resonance-stabilized allylic radical (21). This lipid radical species predominantly reacts with molecular oxygen to form a peroxyl radical. During the unique oxidation–reduction conditions of inflammation or ischemia–reoxygenation, tissue O2 levels are often suppressed and nitrogen oxide levels are elevated, favoring lipid radical reaction with  NO2 to yield multiple nitration products, including nitrated nitrohydroxy and dinitro fatty acid derivatives (18, 19, 21). Polyunsaturated fatty acids can also be nitrated by acidified nitrite (HNO2), generating a complex mixture of products that are similar to those formed by direct reaction with
NO2 to yield multiple nitration products, including nitrated nitrohydroxy and dinitro fatty acid derivatives (18, 19, 21). Polyunsaturated fatty acids can also be nitrated by acidified nitrite (HNO2), generating a complex mixture of products that are similar to those formed by direct reaction with  NO2, including singly nitrated species that maintain the bis-allylic bond arrangement (18, 19). Acid-catalyzed fatty acid nitration is expected during physiological and pathological conditions in which NO2– is exposed to low pH (e.g., pH < 4.0), such as in the gastric compartment after endosomal or phagolysosomal acidification or in tissues after postischemic reperfusion.
NO2, including singly nitrated species that maintain the bis-allylic bond arrangement (18, 19). Acid-catalyzed fatty acid nitration is expected during physiological and pathological conditions in which NO2– is exposed to low pH (e.g., pH < 4.0), such as in the gastric compartment after endosomal or phagolysosomal acidification or in tissues after postischemic reperfusion.
Nitrated linoleic acid (LNO2)‡ displays robust cell-signaling activities that appear to be antiinflammatory (20, 22–25). LNO2 inhibits platelet function by means of cAMP-dependent mechanisms (26), and it inhibits neutrophil  generation, calcium influx, elastase release, CD11b expression, and degranulation by means of non-cAMP-dependent, non-cGMP-dependent mechanisms (23). LNO2 also induces vessel relaxation, in part by means of cGMP-dependent mechanisms (22, 27). In aggregate, these data infer that nitrated unsaturated fatty acids represent a class of lipid-derived signaling mediators. At present, a lack of clinical quantitation and structural characterization of nitrated fatty acids has limited the establishment of LNO2 derivatives as biologically relevant lipid-signaling mediators. Here, we report that both free and esterified linoleate nitration products are abundant in healthy human red cell and plasma lipids. By using GC-MS and HPLC-MS, 10-nitro-9,12-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid were clinically identified and quantitated.
generation, calcium influx, elastase release, CD11b expression, and degranulation by means of non-cAMP-dependent, non-cGMP-dependent mechanisms (23). LNO2 also induces vessel relaxation, in part by means of cGMP-dependent mechanisms (22, 27). In aggregate, these data infer that nitrated unsaturated fatty acids represent a class of lipid-derived signaling mediators. At present, a lack of clinical quantitation and structural characterization of nitrated fatty acids has limited the establishment of LNO2 derivatives as biologically relevant lipid-signaling mediators. Here, we report that both free and esterified linoleate nitration products are abundant in healthy human red cell and plasma lipids. By using GC-MS and HPLC-MS, 10-nitro-9,12-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid were clinically identified and quantitated.
Experimental Procedures
Materials. Linoleic acid was obtained from Nu Check Prep (Elysian, MN). Phenylselenium bromide, mercury chloride, sodium nitrite, anhydrous tetrahydrofuran, N,N-diisopropylethylamine (99.5%) and acetonitrile were from Sigma–Aldrich. Silica gel HF thin-layer chromatography (TLC) plates (250 μm) were obtained from Analtech. Pentafluorobenzyl (PFB) bromide and methanolic BF3 were obtained from Pierce. The solvents used in syntheses were HPLC grade or better (Fisher Scientific). Solvents for MS analyses were obtained from Burdick and Jackson. We obtained [13C]linoleic acid from Spectra Stable Isotopes (Columbia, MD), and [15N]NaNO2 was obtained from Cambridge Isotope Laboratories (Cambridge, MA). We synthesized [14N]LNO2, [13C]LNO2, and [15N]LNO2 positional isomers, as described in the legends to Figs. 4 and 5, which are published as supporting information on the PNAS web site.
Red Blood Cell and Plasma Lipid Isolation and Extraction. Peripheral blood from healthy human volunteers was collected (with Institutional Review Board approval) by venipuncture in heparinized tubes and centrifuged (1,200 × g for 10 min), and plasma was isolated from red cell pellets from which the buffy coat was removed. Lipid extracts were then prepared from packed red cells and plasma (28) and analyzed by MS. Care was taken to avoid acidification during all steps of plasma fractionation and lipid extraction. Lipid extracts from plasma were fractionated by TLC to separate LNO2 from the bulk of neutral lipids present in plasma, minimizing ionization dampening during LNO2 analysis by MS. To measure the esterified LNO2 content in red cell membranes and plasma lipoproteins, lipid extracts were first hydrolyzed (29), fractionated by TLC, and analyzed by MS.
Analysis of Synthetic LNO2 Methyl and PFB Esters by GC-MS. Methyl esters of synthetic LNO2 isomers were analyzed by GC-MS in both positive- and negative-ion modes. Electron-impact (EI) ionization was used to identify and characterize the fragmentation patterns of the two main positional isomers of LNO2 methyl esters. EI GC-MS was performed by using a Saturn 2000 MS coupled with a 3800 GC (Varian). Samples were ionized by EI at +70 eV and resolved by GC using a CP-7420 capillary column (i.d., 0.25 mm; fused silica, 100 m; Varian). Helium was used as the carrier gas.
Because of the low sensitivity of positive-ion GC-MS to LNO2, negative-ion chemical ionization (NICI) was used to characterize PFB esters of synthetic LNO2 and detect LNO2 species in vivo. PFB esters of synthetic LNO2 were prepared, as well as red cell and plasma lipids (28), with biological lipids first partially purified by TLC. Lipids were then analyzed by NICI GC-MS by using a 5890 GC (Hewlett–Packard) coupled to a single-quadrupole MS (Hewlett–Packard) using a 30-m CP-Sil 8CB MS column (5% phenyl/95% dimethylpolysiloxane; Varian) (30).
Analysis of LNO2 Positional Isomers by ESI Tandem MS (MS/MS). Qualitative analysis of nitrated linoleic acid positional isomers by electrospray ionization (ESI) MS was performed by using a hybrid triple-quadrupole linear ion-trap MS (Applied Biosystems/MDS Sciex, Thornhill, ON, Canada). Positional isomers of LNO2 were resolved by reverse-phase HPLC using a 150 × 2-mm C18 Luna column (particle size, 3 μm; Phenomenex, Belmont, CA). Resolved isomers were detected by MS using a multiple reaction-monitoring (MRM) scan mode by reporting molecules that undergo an m/z 324/277 mass transition. This transition, consistent with the loss of HNO2 ([M-(HNO2)-H]–), is common for all mononitrated isomers of linoleic acid. Concurrent with MRM, enhanced product ion (EPI) analysis was performed to generate characteristic and identifying fragmentation patterns of the eluting species with a precursor mass of m/z = 324.
Detection and Quantitation of LNO2 in Human Red Blood Cells and Plasma. Quantitation of LNO2 was performed as described above, with the following modifications: [13C]LNO2 was added as internal standard to correct for losses during extraction and TLC, and the gradient elution profile was changed so that all LNO2 positional isomers eluted at the same time. The following two MRM transitions were monitored: m/z = 324/277 (LNO2) and m/z = 342/295 ([13C]LNO2). The ratio of analyte to internal standard areas was determined, and LNO2 content was quantitated by using analyst 1.4 (Applied Biosystems).
Results
Synthesis of LNO2. Modification of a nitrosenylation-mediated linoleic acid nitration reaction (Fig. 4A) increased the purity and yield of fatty acid allylic nitration products significantly, facilitating the structural resolution of specific LNO2 positional isomers (22, 31, 32). Preparative TLC permitted the initial resolution of nitrated fatty acids from starting materials and oxidized linoleic acid species (Fig. 4B). These changes in synthetic approaches increased linoleic acid nitration product yield from 4% to >50% (22).
Spectral Analysis of LNO2. Nitrated linoleic acid displays a characteristic absorption profile and maximum, providing determination of an extinction coefficient and a method for measuring concentrations of synthetic LNO2. This species displays a unique absorbance maximum at 329 nm, compared with linoleic acid (Fig. 5A). Plotting absorbance versus concentration profiles for each of the synthetic LNO2 preparations reported here, [14N]LNO2, [15N]LNO2, and [13C]LNO2, generated identical extinction coefficients for all LNO2 derivatives: ε = 10.1 cm–1·M–1 (Fig. 5B).
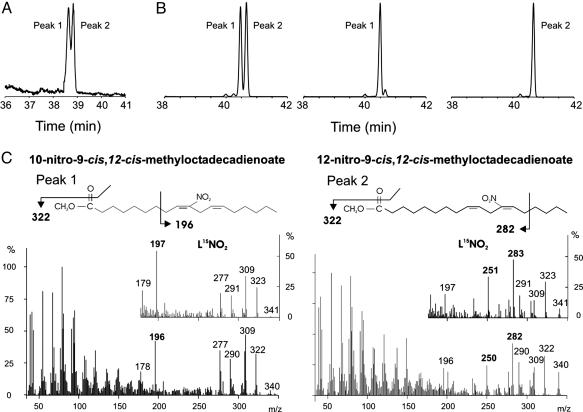
GC-MS Analysis of LNO2. EI GC-MS of LNO2 methyl esters revealed two dominant peaks at 38.75 and 39 min, when resolved by using a 100-m column (Fig. 1A). Product ion analysis of each peak (Fig. 1C) generated fragmentation patterns that were similar to NMR-verified patterns reported (18) for acidic nitration of ethyl linoleate. The first and second peaks correspond to linoleic acid that is nitrated on the C12 and C10, respectively. These two isomers are identified by the unique daughter ions m/z = 250 and m/z = 282 (specific to C12), and m/z = 196 (specific to C10). Analysis of [15N]LNO2 revealed fragmentation patterns with these identifying ions shifted by m/z = +1, further affirming fragments containing the nitro group (Fig. 1C).
Fig. 1.
Characterization of LNO2 by GC MS. (A) LNO2 methyl esters were resolved by using a 100-m fused silica column and detected by total ion-count monitoring after EI ionization. Peak 1 (38.75 min) corresponds to linoleic acid nitrated at the 12-carbon (C12), and Peak 2 (39.00 min) corresponds nitration on the 10-carbon (C10). (B) LNO2 and HPLC-separated positional isomers of LNO2 were derivatized to their PFB esters, resolved on a 30-m CP-Sil 8CB MS, and detected by using NICI GC-MS. Peaks 1 and 2 correspond to C12 and C10 isomers, respectively. These species account for 90–95% of the total peak areas. (C) EI spectra were obtained from the peaks shown in A. Unique fragments (namely, m/z = 196 for C10, and m/z = 250 and m/z = 282 for C12) were detected that enabled structural identification of the isomers.
Although it is useful for structural analysis of synthetic LNO2 isomers, EI GC MS lacks sensitivity for the detection of LNO2 derivatives present in biological samples. Thus, these analyses were performed by NICI GC-MS on PFB derivatives of LNO2 (Fig. 1B). Chromatographic separation of both synthetic LNO2 and TLC-separated red cell lipid extracts resolved two dominant peaks, C12 and C10, and two minor peaks ascribed to either cis-trans or C13 and C9 positional isomers of LNO2. In both cases, >90% of total peak area is accounted for by the C12 and C10 isomers. The C12 and C10 positional isomers of LNO2-PFB derivatives were confirmed to be the same as those identified as LNO2 methyl esters by EI GC-MS (data not shown). Initial detection and quantitation of endogenous levels of LNO2 was performed by using NICI GC-MS; however, extensive sample processing and limited sensitivity motivated an HPLC-MS/MS-based method to characterize and quantitate LNO2 in biological samples.
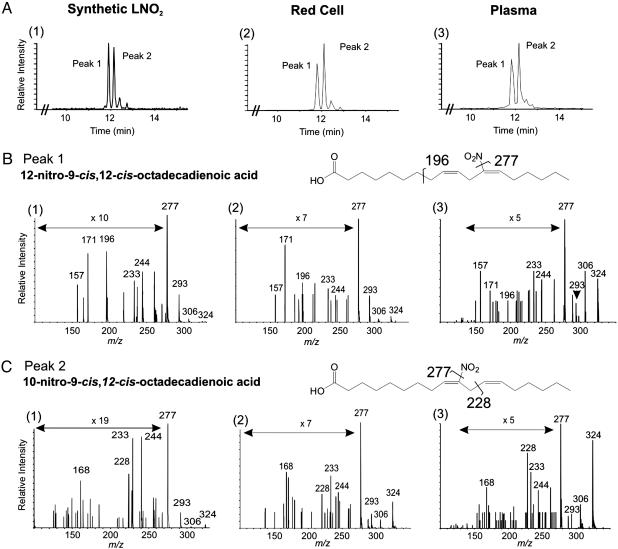
Synthetic and Endogenous LNO2 Isomer Characterization by ESI MS/MS. An HPLC separation strategy that baseline-resolved individual LNO2 isomers permitted the liquid chromatography/MS-based characterization of LNO2 derivatives in biological samples (Fig. 2A). To initially identify individual LNO2 isomers resolved by HPLC separation, the two major species were collected (peaks 1 and 2, Fig. 2A), derivatized to PFB esters, and analyzed by GC-MS. From GC-MS analysis, the first species eluting upon HPLC separation was identified as the C12 isomer, and the second species was identified as the C10 nitro derivative (Fig. 1B). The linear ion-trap mode of the hybrid MS was used to perform EPI analysis on eluting peaks to generate an identifying fragmentation pattern for each LNO2 positional isomer (Fig. 2 B, graph 1, and C, graph 1). MS/MS analysis of C10 LNO2 revealed unique fragments, m/z = 168 and m/z = 228, as well as fragments common to all LNO2 isomers (m/z = 233, 244, 277, 293, and 306; Fig. 2C, graph 1). The m/z = 228 ion was further fragmented in the ion trap by using the MS3 scanning mode, generating a fragment with m/z = 46, indicating the loss of an allylic nitro group and assigning the nitro group to the C9 double bond. MS/MS analysis of C12-LNO2 generated unique fragments (m/z = 196 and m/z =157) and the common fragments listed above (Fig. 2B, graph 1). In aggregate, from the synthetic rationale and GC-MS and ESI MS/MS data, the C10 and the C12 isomers are identified as 10-nitro-9-cis, 12-cis-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid, respectively.
Fig. 2.
Characterization of LNO2 by ESI triple-quadrupole MS. (A) Synthetic LNO2 positional isomers were separated by HPLC and detected by MS/MS in the MRM mode by monitoring the 324/277 collision-induced dissociation transition (graph 1). No other peaks were detected in the chromatogram. The same method was used to resolve LNO2 isomers present in the total lipid extract from 1 ml of packed red blood cells (graph 2) and plasma (graph 3). (B) EPI analysis of Peak 1, revealing fragments unique to the C12 positional isomer of LNO2, m/z = 196 and m/z =157, plus fragments common to all LNO2 isomers (m/z = 233, 244, 277, 293, and 306) (graph 1). EPI analyses of Peak 1 for red cell (graph 2) and plasma (graph 3) lipid extracts gave similar fragmentation patterns. All identifying fragments from graph 1 are present in graphs 2 and 3. (C). In addition to the common fragment ions of LNO2, peak 2 displayed unique fragments m/z = 228 and m/z = 168 (graph 1). These fragments, particularly m/z = 228, indicate nitration of the C10 of LNO2. EPI spectra from Peak 2 of resolved red cell (graph 2) and plasma (graph 3) lipid extracts gave fragmentation patterns similar to the synthetic standard.
ESI MS/MS was used to characterize and quantitate LNO2 species present in human red cells and plasma (Fig. 2). The MRM elution profiles for red cell and plasma lipid extracts were identical to those obtained for synthetic standards (Figs. 2 B, graphs 2 and 3, and C, graphs 2 and 3). All characteristic fragments found in the EPI spectra of the synthetic C10 and C12 LNO2 isomers were present in the resolved biological extracts, affirming that red cells and plasma contain LNO2 positional isomers with chromatographic profiles and fragmentation patterns that are identical to our synthetic standards. It is possible that the configuration of the double bond in endogenous LNO2 may differ from synthetic standards (i.e., the cis-cis configuration may be cis-trans or trans-cis), with a level of structural detail not being provided by EPI analysis.
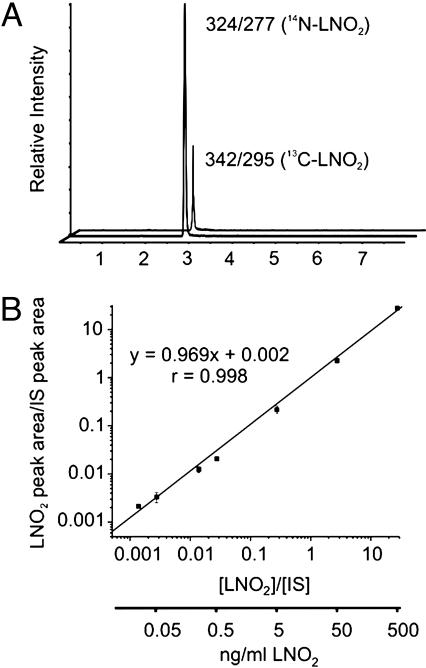
Quantitation of Free and Esterified LNO2 in Red Blood Cells and Plasma. To quantitate net LNO2 species present in red cells and plasma, HPLC gradient conditions were changed so that all positional isomers coeluted (Fig. 3A). Nitrated linoleic acid concentrations were calculated as a function of the ratio of analyte to internal standard peak areas by using an internal standard curve linear over five orders of magnitude (Fig. 3B). Blood samples obtained from 10 healthy human volunteers (five female and five male, ranging 22–45 years of age) revealed free LNO2 in red cells (i.e., LNO2 not esterified to glycerophospholipids or neutral lipids) to be 50 ± 17 pmol/ml packed cells. Total free and esterified LNO2, the amount present in saponified samples, was 249 ± 104 pmol/ml packed cells. Thus, ≈75% of LNO2 in red cells exists in the esterified form. In plasma, free and total (free plus esterified) LNO2 was 79 ± 35 and 630 ± 240 nM, respectively, with free LNO2 representing 85% of total (Table 1) (33–37).
Fig. 3.
Quantitative analysis of red blood cell and plasma LNO2. (A) We added [13C]LNO2 to lipid extractions as an internal standard to quantitate the free and esterified LNO2 content of red cells and plasma obtained from healthy human volunteers. The mean age of the five female and five male subjects was 34 years. The endogenous LNO2 isomers coeluted with the added 13C-labeled LNO2 internal standard, with the internal standard differentiated from endogenous LNO2 by monitoring its unique m/z = 342/295 MRM transition. (B) The internal standard curve for LNO2 reveals linear detector responses over five orders of magnitude. The limit of quantitation (LOQ) for LNO2, as defined by 10 times the SD of the noise, is ≈0.3 fmol (≈100 fg) injected on column.
Table 1. Biologically active nitrogen oxide derivatives in human blood: Comparison with nitrated linoleic acid.
| Species | Compartment | Fraction | Concentration, nM | Source |
|---|---|---|---|---|
| NO2- | Plasma | Total | 205 ± 21 | 35, 36 |
| RSNO | Plasma | Total | 7.2 ± 1.1 | 35, 36 |
| 3-Nitro-tyrosine | Plasma | Total | 0.74 ± 0.30 | 38 |
| LNO2 | Plasma | Free | 79 ± 35 | — |
| Esterified | 550 ± 275 | This article | ||
| Total | 630 ± 240 | |||
| Hb-NO | Blood | Total | <50 | 37 |
| Hb-SNO | Blood | Total | 0-150 | 34 |
| LNO2 | Packed red cells | Free | 50 ± 17 | — |
| Esterified | 199 ± 121 | This article | ||
| Total | 249 ± 104 | |||
| LNO2 | Whole blood* | Total | 477 ± 128* | This article |
Venous blood was obtained from healthy human volunteers and centrifuged (1,200 × g for 10 min), and plasma was isolated from red cell pellets from which the buffy coat was removed. Total lipid extracts were prepared from packed red cells and plasma (28) and analyzed by MS, as described in Experimental Procedures. Total LNO2 (free plus esterified) was determined in lipid extracts after saponification. Free and total LNO2 was quantitated by fitting analyte to internal standard area ratios obtained by MS to an internal standard curve. Concentration values for other bioactive oxides of nitrogen were obtained from the literature. It is noted that nitrite is readily oxidized to nitrite, making precise quantitation of nitrite problematic. Data are expressed as mean ± SD (n = 10; 5 female and 5 male). RSNO S-nitrosothiols; Hb-NO, heme nitrosyl; Hb-SNO, S-nitrosohemoglobin.
Assuming a 40% hematocrit.
Discussion
Cell-signaling studies have revealed that synthetic LNO2 manifests bioactivities that are generally antiinflammatory. Allylic nitro derivatives of linoleic acid inhibit neutrophil degranulation,  O2– formation, CD11b expression, and fMLP-induced Ca2+ influx by means of non-cGMP-dependent, protein kinase-mediated actions (23). In platelets, thrombin-induced aggregation is inhibited upon LNO2-mediated attenuation of cAMP-dependent Ca2+ mobilization and activation of the phosphorylation of vasodilator-stimulated phosphoprotein (VASP) at Ser-157. Current evidence (26) reveals a dual regulation of platelet adenylyl cyclase and phosphodiesterase E activities by LNO2. Additionally, LNO2 has been shown to induce endothelium-independent, cGMP-mediated smooth muscle relaxation (22). These observations reveal that nitrated fatty acids may serve as endogenous cell-signaling molecules, a precept remaining contingent upon the identification of more discrete cell-signaling responses and the clinical detection of these species. The latter issue has been resolved by structurally characterizing linoleic acid nitration products generated by nitrosenylation and those present in human red cell and plasma lipids. By using a 13C-labeled internal standard added during lipid extractions, both free and esterified LNO2 levels were shown to be abundant in the vasculature. Here, we report that the LNO2 positional isomers 10-nitro-9,12-octadecadienoic acid (C10) and 12-nitro-9-cis, 12-cis-octadecadienoic acid (C12) are present as free acids in healthy human blood and as esterified components of red cell membranes and plasma lipoproteins, with a net concentration of ≈500 nM (Table 1). Thus, LNO2 derivatives represent the single largest pool of bioactive oxides of nitrogen in the vascular compartment (33–37). Most of this LNO2 is esterified (≈80%), suggesting that lipases and/or phospholipases will be central for the regulated release of nitrated free fatty acids.
O2– formation, CD11b expression, and fMLP-induced Ca2+ influx by means of non-cGMP-dependent, protein kinase-mediated actions (23). In platelets, thrombin-induced aggregation is inhibited upon LNO2-mediated attenuation of cAMP-dependent Ca2+ mobilization and activation of the phosphorylation of vasodilator-stimulated phosphoprotein (VASP) at Ser-157. Current evidence (26) reveals a dual regulation of platelet adenylyl cyclase and phosphodiesterase E activities by LNO2. Additionally, LNO2 has been shown to induce endothelium-independent, cGMP-mediated smooth muscle relaxation (22). These observations reveal that nitrated fatty acids may serve as endogenous cell-signaling molecules, a precept remaining contingent upon the identification of more discrete cell-signaling responses and the clinical detection of these species. The latter issue has been resolved by structurally characterizing linoleic acid nitration products generated by nitrosenylation and those present in human red cell and plasma lipids. By using a 13C-labeled internal standard added during lipid extractions, both free and esterified LNO2 levels were shown to be abundant in the vasculature. Here, we report that the LNO2 positional isomers 10-nitro-9,12-octadecadienoic acid (C10) and 12-nitro-9-cis, 12-cis-octadecadienoic acid (C12) are present as free acids in healthy human blood and as esterified components of red cell membranes and plasma lipoproteins, with a net concentration of ≈500 nM (Table 1). Thus, LNO2 derivatives represent the single largest pool of bioactive oxides of nitrogen in the vascular compartment (33–37). Most of this LNO2 is esterified (≈80%), suggesting that lipases and/or phospholipases will be central for the regulated release of nitrated free fatty acids.
Recently, nitration products of arachidonic acid (27), linoleic acid (38) and cholesteryl linoleate (39) have been reported in biological samples. Importantly, all of these observations were made after lipid extraction of nitrite-containing specimens under acidic conditions (pH <4), documented to result in HNO2-dependent fatty acid nitration and oxidation (20). This analytical pitfall raises the question of whether the various reported LNO2 species were indeed endogenous, and it impairs an ability to determine concentration and structural characteristics. For this reason, this article includes analysis of potential confounding factors and artifacts in the detection and quantitation of linoleic acid nitration products in clinical specimens and reveals that both acid-catalyzed reactions during lipid processing and the presence of adventitious NO2– induces artifact in LNO2 analysis. Extensive control studies ensured that the LNO2 detected was not a byproduct of lipid extraction, storage, HPLC separation, MS analysis, or the presence of alkyl hydroxy and hydroperoxy derivatives. We added [13C]linoleic acid as a reporter molecule before red cell and plasma lipid purification and analysis during methods development, which permitted the MS detection of any possible 13C-labeled LNO2 products being formed. Also, up to 200 μMNO2– was included in initial lipid extractions to determine whether separations or analysis-induced nitration reactions might be supported by physiological NO2– levels that can exceed 200 nM (34, 35). All critical phases of lipid extraction avoided the use of acidic pH to limit acid-catalyzed, HNO2-dependent lipid nitration reactions. Moreover, the addition and recovery of an LNO2 internal standard to biological lipid extractions showed efficient extraction of lipids of interest at neutral pH. Together, control studies affirmed that in this study, no LNO2 derivatives were generated during the ex vivo processing of clinical samples and subsequent lipid purification and analysis steps.
Nitrated lipids can be synthesized ex vivo by different approaches, with varying degrees of specificity and yield. Acid-catalyzed, HNO2-dependent fatty acid nitration readily nitrates lipids; however, multiple species, including conjugated dienes, nitro, hydroxy, hydroperoxy, nitrohydroxy, and nitrohydroperoxy adducts are formed, with all products displaying nonspecific stereochemistry (18). This reaction pathway may also occur in select biological milieu, such as the low pH environment of the gastric system and upon endosome acidification (40). Linoleic acid nitration by nitrosenylation generates primarily two derivatives of linoleic acid (C10 and C12 LNO2) with no rearrangement of the olefinic groups (41), thus simplifying structural characterization and fortuitously yielding positional isomers that are identical to those of red cell and lipoprotein lipids.
The characterization of synthetic LNO2 species was performed by both GC-MS and HPLC-MS. Analysis of synthetic LNO2 methyl ester products by EI GC-MS indicated that linoleic acid was singly nitrated and not oxidized, with the purified product mixture containing two major nitration products (Fig. 1). Unlike CID fragmentation patterns, which depend on numerous instrument and analysis characteristics, EI fragmentation patterns for a particular molecule remain consistent. Thus, we compared the EI fragmentation patterns of nitrosenylation-induced linoleate nitration products with NMR-verified fragmentation patterns of methyl esters of linoleic acid nitrated by exposure to HNO2 (18). The EI fragmentation patterns of peak 1 and peak 2 upon GC separation were identical to those generated from LNO2 nitrated at the C12 and C10, respectively. Although EI GC-MS provided essential structural information about synthetic LNO2, this technique has limited sensitivity for anionic fatty acids, especially when present in complex biological lipid mixtures. Analysis of PFB esters of LNO2 by NICI GC-MS provided a degree of sensitivity that allowed in vivo detection of LNO2 (data not shown); however, this approach requires extensive sample preparation and has limited quantitative precision. Thus, ESI MS/MS was used to characterize and quantitate synthetic and endogenous LNO2. Unique HPLC retention times and fragmentation patterns for each synthetic LNO2 isomer were obtained by CID, which provided a “molecular fingerprint” that was used to identify LNO2 isomers in biological samples (Fig. 2). These fragmentation patterns were obtained by using an ion-trap mode (EPI) of a hybrid MS, providing enhanced sensitivity to minor fragments and more detailed structural information. The m/z = 228 fragment generated from the second eluting peak, barely detectable without trapping, permitted the assignment of the NO2 functional group to the C9 double bond.
In summary, two positional isomers of LNO2 have been synthesized, resolved, and structurally characterized, with corresponding species identified and quantitated in human blood. Allylic nitro derivatives of fatty acids display unique vascular and inflammatory cell-signaling activities and appear to represent a novel class of lipid-signaling molecules. Whereas  NO-dependent oxidation and nitration reactions will induce nitration of endogenous tissue fatty acids, dietary NO2– or nitrated lipids may also contribute to tissue LNO2 content. In vitro studies presently support the precept that allylic nitro derivatives of fatty acids counter the proinflammatory signaling actions of most eicosanoids, thus these byproducts of inflammatory oxidation and nitration reactions will contribute to the resolution of inflammation. In aggregate, these observations reveal that allylic nitro derivatives of unsaturated fatty acids represent the convergence of
NO-dependent oxidation and nitration reactions will induce nitration of endogenous tissue fatty acids, dietary NO2– or nitrated lipids may also contribute to tissue LNO2 content. In vitro studies presently support the precept that allylic nitro derivatives of fatty acids counter the proinflammatory signaling actions of most eicosanoids, thus these byproducts of inflammatory oxidation and nitration reactions will contribute to the resolution of inflammation. In aggregate, these observations reveal that allylic nitro derivatives of unsaturated fatty acids represent the convergence of  NO and oxidized lipid cell-signaling pathways. Indeed, LNO2 undergoes high-affinity receptor–ligand interactions (F.J.S., Y. Lin, P.R.S.B., Y. E. Chen, and B.A.F., unpublished data) that induce alterations in gene expression and the induction or suppression of multiple cell growth control and inflammatory-related proteins.
NO and oxidized lipid cell-signaling pathways. Indeed, LNO2 undergoes high-affinity receptor–ligand interactions (F.J.S., Y. Lin, P.R.S.B., Y. E. Chen, and B.A.F., unpublished data) that induce alterations in gene expression and the induction or suppression of multiple cell growth control and inflammatory-related proteins.
Acknowledgments
We thank Drs. Sai Chang, Tom Bisenthal, and Nadia Pace (Applied Biosystems/MDS Sciex); Dr. Bruce King (Wake Forest University, Winston-Salem, NC); and Drs. Bill Caufield and Greg Gorman (Southern Research Institute, Birmingham, AL) for their helpful input. This work was supported by National Institutes of Health Grants RO1HL58115 and RO1HL64937. P.R.S.B. was supported by National Institutes of Health Cardiovascular Hypertension Training Grant T32HL07457.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EI, electron impact; EPI, enhanced product ion; ESI, electrospray ionization; MRM, multiple reaction monitor(ing); MS/MS, tandem MS; NICI, negative-ion chemical ionization; PFB, pentafluorobenzyl.
See Commentary on page 11527.
Footnotes
Nitrated linoleic acid is termed LNO2. Nitrohydroxy and dinitro adducts of LNO2 are referred to as L(OH)NO2 and L(NO2)2, respectively. Linoleic acid singly nitrated at either the 10 or 12 carbon, with retention of the cis—cis bis-allylic bond arrangement are referred to as C10-LNO2 and C12-LNO2, respectively. International Union of Pure and Applied Chemistry nomenclature defines these species as 10-nitro-9-cis, 12-cis-octadecadienoic acid and 12-nitro-9-cis, 12-cis-octadecadienoic acid, respectively.
References
- 1.Arnold, W. P., Mittal, C. K., Katsuki, S. & Murad, F. (1977) Proc. Natl. Acad. Sci. USA 74, 3203–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman, J. S., Beckman, T. W., Chen, J., Marshall, P. A. & Freeman, B. A. (1990) Proc. Natl. Acad. Sci. USA 87, 1620–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nathan, C. (1992) FASEB J. 6, 3051–3064. [PubMed] [Google Scholar]
- 4.Jourd'heuil, D., Miranda, K. M., Kim, S. M., Espey, M. G., Vodovotz, Y., Laroux, S., Mai, C. T., Miles, A. M., Grisham, M. B. & Wink, D. A. (1999) Arch. Biochem. Biophys. 365, 92–100. [DOI] [PubMed] [Google Scholar]
- 5.Rubbo, H., Darley-Usmar, V. & Freeman, B. A. (1996) Chem. Res. Toxicol. 9, 809–820. [DOI] [PubMed] [Google Scholar]
- 6.Kissner, R., Nauser, T., Bugnon, P., Lye, P. G. & Koppenol, W. H. (1997) Chem. Res. Toxicol. 10, 1285–1292. [DOI] [PubMed] [Google Scholar]
- 7.Radi, R., Denicola, A. & Freeman, B. A. (1999) Methods Enzymol. 301, 353–367. [DOI] [PubMed] [Google Scholar]
- 8.Baldus, S., Eiserich, J. P., Mani, A., Castro, L., Figueroa, M., Chumley, P., Ma, W., Tousson, A., White, C. R., Bullard, D. C., et al. (2001) J. Clin. Invest. 108, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro, L., Eiserich, J. P., Sweeney, S., Radi, R. & Freeman, B. A. (2004) Arch. Biochem. Biophys. 421, 99–107. [DOI] [PubMed] [Google Scholar]
- 10.Eiserich, J. P., Baldus, S., Brennan, M. L., Ma, W., Zhang, C., Tousson, A., Castro, L., Lusis, A. J., Nauseef, W. M., White, C. R., et al. (2002) Science 296, 2391–2394. [DOI] [PubMed] [Google Scholar]
- 11.Grisham, M. B. (1985) J. Free Radical Biol. Med. 1, 227–232. [DOI] [PubMed] [Google Scholar]
- 12.Liu, X., Miller, M. J., Joshi, M. S., Thomas, D. D. & Lancaster, J. R., Jr. (1998) Proc. Natl. Acad. Sci. USA 95, 2175–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subczynski, W. K., Lomnicka, M. & Hyde, J. S. (1996) Free Radical Res. 24, 343–349. [DOI] [PubMed] [Google Scholar]
- 14.Thomas, D. D., Liu, X., Kantrow, S. P. & Lancaster, J. R., Jr. (2001) Proc. Natl. Acad. Sci. USA 98, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubbo, H., Radi, R., Trujillo, M., Telleri, R., Kalyanaraman, B., Barnes, S., Kirk, M. & Freeman, B. A. (1994) J. Biol. Chem. 265, 26066–26075. [PubMed] [Google Scholar]
- 16.Gallon, A. A. & Pryor, W. A. (1993) Lipids 28, 125–133. [DOI] [PubMed] [Google Scholar]
- 17.Gallon, A. A. & Pryor, W. A. (1994) Lipids 29, 171–176. [DOI] [PubMed] [Google Scholar]
- 18.Napolitano, A., Camera, E., Picardo, M. & d'Ischia, M. (2000) J. Org. Chem. 65, 4853–4860. [DOI] [PubMed] [Google Scholar]
- 19.Napolitano, A., Crescenzi, O., Camera, E., Giudicianni, I., Picardo, M. & d'Ischia, M. (2004) Tetrahedron 58, 5061–5067. [Google Scholar]
- 20.O'Donnell, V. B., Eiserich, J. P., Chumley, P. H., Jablonsky, M. J., Krishna, N. R., Kirk, M., Barnes, S., Darley-Usmar, V. M. & Freeman, B. A. (1999) Chem. Res. Toxicol. 12, 83–92. [DOI] [PubMed] [Google Scholar]
- 21.Pryor, W. A., Lightsey, J. W. & Church, D. F. (1982) J. Am. Chem. Soc. 104, 6685–6692. [Google Scholar]
- 22.Lim, D. G., Sweeney, S., Bloodsworth, A., White, C. R., Chumley, P. H., Krishna, N. R., Schopfer, F., O'Donnell, V. B., Eiserich, J. P. & Freeman, B. A. (2002) Proc. Natl. Acad. Sci. USA 99, 15941–15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coles, B., Bloodsworth, A., Clark, S. R., Lewis, M. J., Cross, A. R., Freeman, B. A. & O'Donnell, V. B. (2002) Circ. Res. 91, 375–381. [DOI] [PubMed] [Google Scholar]
- 24.O'Donnell, V. B. & Freeman, B. A. (2001) Circ. Res. 88, 12–21. [DOI] [PubMed] [Google Scholar]
- 25.O'Donnell, V. B., Chumley, P. H., Hogg, N., Bloodsworth, A., Darley-Usmar, V. M. & Freeman, B. A. (1997) Biochemistry 36, 15216–15223. [DOI] [PubMed] [Google Scholar]
- 26.Coles, B., Bloodsworth, A., Eiserich, J. P., Coffey, M. J., McLoughlin, R. M., Giddings, J. C., Lewis, M. J., Haslam, R. J., Freeman, B. A. & O'Donnell, V. B. (2002) J. Biol. Chem. 277, 5832–5840. [DOI] [PubMed] [Google Scholar]
- 27.Balazy, M., Iesaki, T., Park, J. L., Jiang, H., Kaminski, P. M. & Wolin, M. S. (2001) J. Pharmacol. Exp. Ther. 299, 611–619. [PubMed] [Google Scholar]
- 28.Bligh, E. G. & Dyer, W. L. (1959) Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, T., Bratton, D. L. & Murphy, R. C. (1997) J. Mass Spectrom. 32, 888–896. [DOI] [PubMed] [Google Scholar]
- 30.Wheelen, P., Zirrolli, J. A. & Murphy, R. C. (1994) J. Am. Soc. Mass Spectrom. 6, 40–51. [DOI] [PubMed] [Google Scholar]
- 31.d'Ischia, M., Rega, N. & Barone, V. (1999) Tetrahedron 55, 9297–9308. [Google Scholar]
- 32.Ranu, B. C. & Chakraborty, R. (1991) Tetrahedron Lett. 32, 3579–3582. [Google Scholar]
- 33.Gladwin, M. T., Shelhamer, J. H., Schechter, A. N., Pease-Fye, M. E., Waclawiw, M. A., Panza, J. A., Ognibene, F. P. & Cannon, R. O., III. (2000) Proc. Natl. Acad. Sci. USA 97, 11482–11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rassaf, T., Bryan, N. S., Kelm, M. & Feelisch, M. (2002) Free Radical Biol. Med. 33, 1590–1596. [DOI] [PubMed] [Google Scholar]
- 35.Rassaf, T., Bryan, N. S., Maloney, R. E., Specian, V., Kelm, M., Kalyanaraman, B., Rodriguez, J. & Feelisch, M. (2003) Nat. Med. 9, 481–482. [DOI] [PubMed] [Google Scholar]
- 36.Cosby, K., Partovi, K. S., Crawford, J. H., Patel, R. P., Reiter, C. D., Martyr, S., Yang, B. K., Waclawiw, M. A., Zalos, G., Xu, X., et al. (2003) Nat. Med. 9, 1498–1505. [DOI] [PubMed] [Google Scholar]
- 37.Soderling, A. S., Ryberg, H., Gabrielsson, A., Larstad, M., Toren, K., Niari, S. & Caidahl, K. (2003) J. Mass Spectrom. 38, 1187–1196. [DOI] [PubMed] [Google Scholar]
- 38.Lima, E. S., Di Mascio, P., Rubbo, H. & Abdalla, D. S. (2002) Biochemistry 41, 10717–10722. [DOI] [PubMed] [Google Scholar]
- 39.Lima, E. S., Di Mascio, P. & Abdalla, D. S. (2003) J. Lipid Res. 44, 1660–1666. [DOI] [PubMed] [Google Scholar]
- 40.Knowles, M. E., McWeeny, D. J., Couchman, L. & Thorogood, M. (1974) Nature 247, 288–289. [DOI] [PubMed] [Google Scholar]
- 41.Hayama, T., Tomoda, S., Takeuchi, Y. & Nomura, Y. (1982) Tetrahedron Lett. 23, 4733–4734. [Google Scholar]