Abstract
The HIV-1 matrix protein p17, excised proteolytically from the N terminus of the Gag polyprotein, forms a protective shell attached to the inner surface of the plasma membrane of the virus. During the late stages of the HIV-1 replication cycle, the N-terminally myristoylated p17 domain targets the Gag polyprotein to the host-cell membrane for particle assembly. In the early stages of HIV-1 replication, however, some p17 molecules dissociate from the viral membrane to direct the preintegration complex to the host-cell nucleus. These two opposing targeting functions of p17 require that the protein be capable of reversible membrane interaction. It is postulated that a significant structural change in p17 triggered by proteolytic cleavage of the Gag polyprotein sequesters the N-terminal myristoyl group, resulting in a weaker membrane binding by the matrix protein than the Gag precursor. To test this “myristoyl switch” hypothesis, we obtained highly purified synthetic HIV-1 p17 of 131 amino acid residues and its N-myristoylated form in large quantity. Both forms of p17 were characterized by circular dichroism spectroscopy, protein chemical denaturation, and analytical centrifugal sedimentation. Our results indicate that although N-myristoylation causes no spectroscopically discernible conformational change in p17, it stabilizes the protein by 1 kcal/mol and promotes protein trimerization in solution. These findings support the premise that the myristoyl switch in p17 is triggered not by a structural change associated with proteolysis, but rather by the destabilization of oligomeric structures of membrane-bound p17 in the absence of downstream Gag subdomains.
The HIV-1 matrix protein p17 is the N-terminal domain of a larger precursor polyprotein encoded by the HIV-1 gag gene (1). After ribosomal translation of viral genes, the p17 domain directs HIV-1 Gag polyproteins to the plasma membrane of the host cell, where assembly and budding of virions occur (2–4). Excised proteolytically during and immediately after budding of virus particles, p17 forms a protective shell associated directly with the inner leaflet of the viral membrane (5, 6). In the early stages of the HIV-1 life cycle, p17 also can dissociate from the viral membrane and direct the core-derived preintegration complex to the host-cell nucleus (7–9), although the precise role of p17 in regulating HIV-1 nuclear import and enabling the virus to replicate in nondividing cells remains controversial (10–14). More recently, isolated p17 has been shown to enhance HIV-1 infection and replication in activated T lymphocytes by up-regulating the secretion of proinflammatory cytokines through an extracellular receptor-mediated signaling pathway (15).
Viral maturation and nuclear targeting apparently require that p17 bind the cell membrane reversibly. p17 is posttranslationally myristoylated at the N terminus (2, 4). The interaction of p17 with the plasma membrane is mediated by this 14-carbon fatty acid moiety and a cluster of positively charged amino acid residues located in the N-terminal region of the protein (16–19). Ample evidence shows that p17, upon cleavage from Gag by the HIV-1 protease, has a markedly reduced binding affinity for the plasma membrane as compared with the unprocessed polyprotein (18, 20, 21). Further, deletions in the C-terminal α-helical region of the p17 protein dramatically enhance its membrane binding (20). These findings have led to the hypothesis that the proteolytic event is accompanied by a significant structural change in p17 that results in sequestration of the N-terminal myristoyl group by the C-terminal α-helix, thus weakening the membrane-binding capacity of p17 (20). Subsequent studies seem to support this hypothesis (18, 22), even though the precise region of the protein involved in the myristate sequestration remains controversial (18, 20).
The myristoyl group can adopt two different conformations in a protein: exposed and hidden (or sequestered). The transition between alternating orientations of the myristoyl moiety relative to the protein, known as the “myristoyl switch,” can be triggered by certain biochemical and biophysical events including proteolysis (23). In the case of some proteins, myristate sequestration causes significant protein structural changes (23, 24). For these reasons, the postulated myristoyl switch model for p17 is thought to be a possible regulatory mechanism by which the matrix protein dictates its subcellular localization to exert different biological functions in HIV-1 pathogenesis. However, definitive structural evidence to support this hypothesis is lacking for p17. The question of how proteolytic processing of the Gag polyprotein during viral maturation induces a significant structural change in p17, and thus sequestration of the myristoyl moiety, remains unanswered. Although both NMR and crystal structures of recombinant nonmyristoylated p17 have been determined (25–29), these studies shed little light on the structural basis of the myristoyl switch hypothesis. Comparative studies of both myristoylated and nonmyristoylated p17 are needed to better understand the molecular mechanism by which it functions in the early and late stages of the HIV-1 life cycle.
Here we report total chemical synthesis of the 131-residue p17 and its myristoylated form, i.e., myristoylated HIV-1 matrix protein p17 [myr(+)-p17] and nonmyristoylated HIV-1 matrix protein p17 [myr(–)-p17], by using a three-segment native chemical ligation approach (30). Hundreds of milligrams of highly pure and correctly folded synthetic proteins were readily prepared. Both forms of HIV-1 p17 were characterized by circular dichroism (CD) spectroscopy, protein chemical denaturation, and analytical centrifugal sedimentation. Our results indicate that the addition of the N-terminal myristoyl group resulted in no major conformational change in p17. Instead, myristoylation enhanced the stability and promoted trimerization of the matrix protein. We propose that the myristoyl switch in p17 is triggered by destabilization of oligomeric structures of membrane-bound p17 as a result of the proteolytical processing of the HIV-1 Gag polyprotein during viral maturation.
Materials and Methods
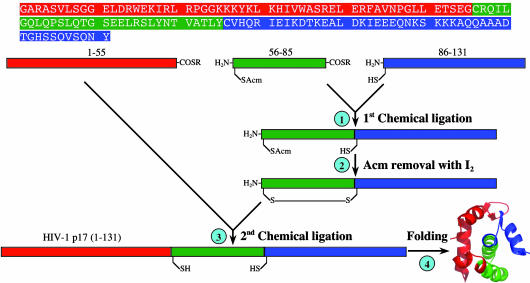
Peptide Synthesis, Chemical Ligation, and Protein Folding. The amino acid sequence of and the synthetic strategy for the 131-residue HIV-1 matrix protein p17 are outlined in Fig. 1. Four peptides (three peptide thioesters and one peptide acid) were independently synthesized on appropriate phenylacetamidomethyl resins by using a custom-designed, machine-assisted chemistry tailored from the 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate activation/N,N-diisopropylethylamine in situ neutralization protocol developed by Kent and coworkers (31, 32) for Boc solid-phase peptide synthesis. The four peptides are: H-(1–55)-S-acetyl-leucinyl-OH or (1–55), myr-(1–55)-S-acetyl-leucinyl-OH or myr-(1–55), H-Cys(Acm)-(57–85)-S-acetyl-leucinyl-OH (ACM, acetamidomethyl) or (56–85), and H-Cys-(87–131)-OH or (86–131). All crude peptides, after cleavage and deprotection in HF, were purified to homogeneity by preparative C18 reversed-phase (RP) HPLC, and their molecular masses were verified by electrospray ionization MS.
Fig. 1.
Strategy for the synthesis of the HIV-1 matrix protein p17 by means of sequential native chemical ligation of three peptide fragments at the ligation sites Gly-55–Cys-56 and Tyr-85–Cys-86. The N-terminal Cys residue of the second peptide thioester was temporarily protected by Acm to prevent intramolecular head-to-tail cyclization. A 0.25-mmol-scale synthesis resulted in ≈275 mg of folded protein, i.e., an overall yield of 7.5%.
Native chemical ligation of H-Cys(Acm)-(57–85)-S-acetylleucinyl-OH and H-Cys-(87–131)-OH went to completion within 8 h at a total concentration of 20 mg/ml in 0.2 M phosphate buffer containing 6 M guanidine hydrochloride (GuHCl) and 2% thiophenol (vol/vol) (pH 7.5). Removal of the Acm group from the purified ligation product, H-Cys(Acm)-(57–131)-OH, was achieved quantitatively by dissolving the Cys-protected peptide at 0.5 mg/ml in 0.1 M citric acid/0.2 M HCl, to which 5 mM iodine (in MeOH) was added dropwise to a final concentration of 0.05 mM or until a stable yellow color was observed. The reaction proceeded for 30 min before being quenched by ascorbic acid. The major product, an intramolecularly disulfide-bridged H-Cys-(57–131)-OH, was purified to homogeneity and used directly in the subsequent ligation reaction. Ligation of H-(1–55)-S-acetyl-leucinyl-OH or myr-(1–55)-S-acetyl-leucinyl-OH and H-Cys-(57–131)-OH was carried out essentially as described above, resulting in two full-length peptides of 131-aa residues.
Protein folding was carried out by dissolving a purified and lyophilized p17 polypeptide at 1 mg/ml in 0.2 M phosphate buffer containing 6 M GuHCl and 10 mg/ml DTT (pH 7.5), followed by extensive dialysis against acidified water. Folded proteins were quantified by UV absorbance measurements at 280 nm by using a molar extinction coefficient (16,960) calculated according to an algorithm published in ref. 33. All analytical RP-HPLC runs were performed on a Waters Alliance system (Milford, MA) or an Agilent 1100 series (Palo Alto, CA) equipped with a Waters Symmetry 300 C18 column (5 μm, 4.6 × 150 mm) at a flow rate of 1 ml/min and a temperature of 40°C. Solvent A was water containing 0.1% trifluoroacetic acid, and solvent B was acetonitrile containing 0.1% trifluoroacetic acid.
Analytical Ultracentrifugation. Sedimentation equilibrium experiments were run at 35°C on a Beckman XL-A Optima system by using a four-hole An-60 rotor and double-sector center-pieces of 12-mm path length. Myr(–)-p17 and myr(+)-p17 were prepared at a concentration of 100 μM each in 50 mM sodium phosphate buffer containing 100 mM NaCl (pH 5.5). Buffer density and partial specific volume (v-bar) were calculated by using the program sednterp (www.jphilo.mailway.com/download.htm). The molar extinction coefficient was calculated from UV absorbance scans of the samples used for analytical ultracentrifugation. The rotor speed range for myr(+)-p17 and myr(–)-p17 data collection was 18,000–21,000 and 18,000–50,000 rpm, respectively. Scans used absorption optics at settings of 0.002-cm step size, three averages per point, and 295 nm. Data analysis was performed with at least four rotor speeds per sample by using the programs ultrascan (Version 6.0, www.ultrascan.uths.csa.edu) and winnonlin (Version 1.06, Pharsight, Mountain View, CA).
CD Spectroscopy and Chemical Denaturation of p17. CD spectra of myr(–)-p17 and myr(+)-p17 at 0.5 mg/ml in 10 mM phosphate buffer (pH 6.8) were obtained at room temperature on a J-810 spectropolorimeter (Jasco, Easton, MD) by using a 1-mm cuvette. Protein denaturation by GuHCl was carried out at room temperature and monitored at 222 nm by CD spectroscopy. Specifically, an initial 2.4 ml of protein solution prepared at 0.1 mg/ml in 20 mM phosphate buffer (pH 6.8) was aliquotted into a 10-mm cuvette. An increasing amount of aliquot was withdrawn, followed immediately by addition of an equal volume of denatured protein of the same concentration, prepared in 20 mM phosphate buffer containing 7.6 M GuHCl (pH 6.8). This procedure generated a stepwise increase (0.25 M) in the concentration of GuHCl in the cuvette from 0 to 4 M after 16 withdrawal/addition cycles. The solution in the cuvette was thoroughly mixed before signals at 222 nm were recorded at different GuHCl concentrations. The experimental data were subjected to a six-parameter nonlinear regression analysis by using an equation published in ref. 34 that was derived from a two-state protein denaturation model, yielding the free energy change of unfolding, ΔG° (kcal/mol), at zero GuHCl concentration.
Results
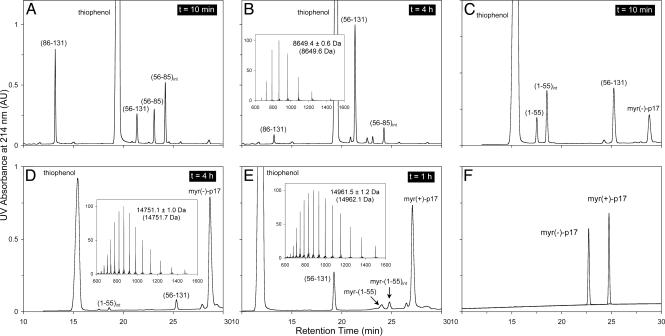
The HIV-1 Matrix Protein p17 Can Be Synthesized Readily by Means of Native Chemical Ligation. The protein p17 contains two conveniently located Cys residues in the sequence, Cys-56 and Cys-86, making it an ideal protein molecule to be synthesized by using sequential, two-step native chemical ligation. As illustrated in Fig. 2 A and B, the first ligation reaction by using the C-terminal peptide (86–131) and the middle fragment (56–85) was almost complete in 4 h, yielding a readily separable ligation product (56–131) with a molecular mass of 8,649.4 ± 0.6 Da. The experimentally determined mass value agrees with the expected value of 8,649.6 Da calculated on the basis of the average isotopic compositions of the ligation product. Notably, a stable intermediate H-(56–85)-S-phenyl accumulated as the result of a nucleophilic substitution reaction between H-(56–85)-S-acetylleucinyl-OH and thiophenol, as identified by electrospray ionization MS. This finding suggests that thiophenol-catalyzed native chemical ligation is achieved through formation of a more reactive peptide thioester species with a presumably better leaving group–S-phenyl.
Fig. 2.
Analysis of the synthesis and native chemical ligation of myr(–)-p17 and myr(+)-p17. (A and B) Native chemical ligation of peptide fragments (56–85) and (86–131) monitored by analytical C18 RP-HPLC (5–65% solvent B over 30 min) at different time intervals. (C and D) Ligation of peptide fragments (1–55) and (56–131) monitored by HPLC (20–45% solvent B over 30 min) at 10 min and 4 h. (E) Ligation of peptide fragments myr-(1–55) and (56–131) monitored by HPLC (25–50% solvent B over 30 min) at 1 h. (F) Folded and purified HIV-1 p17 and its myristoylated form analyzed by C18 RP-HPLC (5–65% solvent B over 30 min). The molecular masses determined by electrospray ionization MS for the three ligation products, H-Cys(Acm)-(57–131)-OH, myr(–)-p17, and myr(+)-p17, were in agreement with the expected values (in parentheses) calculated based on the average isotopic compositions of the peptides. Finally, the three transient ligation intermediates, (56–85)int, (1–55)int, and myr-(1–55)int, were the products derived from a nucleophilic substitution reaction of corresponding peptide thioesters with the catalyst thiophenol.
For the first ligation reaction, the sulfhydryl group of the N-terminal Cys-56 in the peptide thioester H-(56–85)-S-acetylleucinyl-OH was protected by Acm to prevent intramolecular ligation, i.e., head-to-tail backbone cyclization. Because intramolecular ligation proceeds much faster than intermolecular reactions due to a much higher effective concentration of reactants, peptide thioesters with unprotected N-terminal Cys have been constructed for efficient synthesis of circular peptides and proteins (35). Cys-56(Acm) of the first ligation product, H-Cys(Acm)-(57–131)-OH, was deprotected before the second ligation reaction. Methods for Cys(Acm) deprotection in peptides are well established in the literature. We chose I2 treatment of the peptide in acidic aqueous solution to simplify subsequent workup steps. Removal of Acm proceeded quantitatively, yielding the major product: an intramolecularly disulfide-bridged H-(56–131)-OH as suggested by electrospray ionization MS analysis (data not shown). Although the reaction also generated two minor products, i.e., a reduced monomeric H-(56–131)-OH and its intermolecularly disulfide-bridged dimer, all three products could be used for the second ligation reaction where reducing thiophenol would render them chemically identical.
The second ligation between H-(1–55)-S-acetyl-leucinyl-OH and H-(56–131)-OH proceeded as efficiently as the first one (Fig. 2 C and D). A thiophenol-derived peptide thioester intermediate H-(1–55)-S-phenyl, similar to the one observed previously, also accumulated stably during the ligation reaction. The full-length ligation product, H-(1–131)-OH, gave rise to a molecular mass of 14,751.1 ± 1.0 Da, in agreement with the expected value of 14,751.7 Da calculated on the basis of the average isotopic compositions of myr(–)-p17. In a parallel experiment, myristoylated (1–55)-S-acetyl-leucinyl-OH was ligated with H-(56–131)-OH, and similar results were obtained (Fig. 2E). The observed molecular mass of myr-(1–131)-OH, i.e., 14,961.5 ± 1.2 Da, is within experimental error of the calculated value of 14,962.1 Da based on the average isotopic compositions of myr(+)-p17.
On a typical 0.25-mmol synthetic scale, 275 mg of HPLC-purified and folded protein was obtained, representing an overall yield of synthesis of 7.5%. Shown in Fig. 2F are the two final products of p17 analyzed by RP-HPLC under the same chromatographic conditions. Myristoylation of the HIV-1 matrix protein caused a shift of retention time of >2 min on the C18 RP column, presumably because of the presence of the 14-carbon hydrophobic fatty acid at the N terminus.
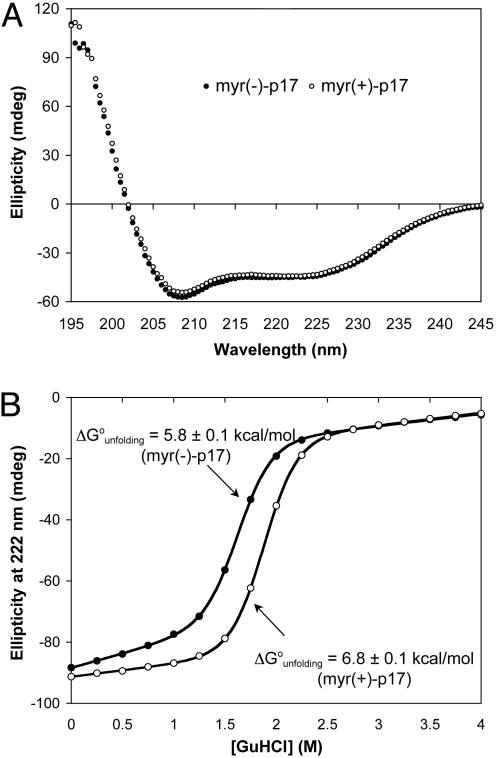
Myristoylation of p17 Results in Little Structural Change and Stabilizes the Matrix Protein by 1 kcal/mol. Both forms of synthetic HIV-1 p17 adopt indistinguishable, predominantly α-helical structures in solution, as indicated by CD spectroscopic analysis (Fig. 3A). This finding, although in line with known structural features of recombinant HIV-1 p17 characterized by NMR spectroscopy and x-ray crystallography (28, 29), also supports the conclusion that synthetic p17 is correctly folded. Further evidence for the correct folding of synthetic p17 comes from studies of GuHCl-induced denaturation (Fig. 3B), where cooperative unfolding with an apparent two-state transition is indicative of a compact, single-domain globular structure of the protein. Significantly, myr(+)-p17 is more stable than myr(–)-p17 by 1 kcal/mol, suggesting that the myristoyl moiety forms energetically favorable hydrophobic contacts with surrounding residues.
Fig. 3.
CD spectra of myr(–)-p17 and myr(+)-p17 (A) and their two-state, GuHCl-induced denaturation monitored at 222 nm by CD spectroscopy (B). Both proteins adopted a similar, if not identical, predominantly α-helical structure, as evidenced by the dual peaks at 208 and 222 nm, characteristic for α-helices. Myr(+)-p17 was 1 kcal/mol more stable than its nonmyristoylated form in PBS.
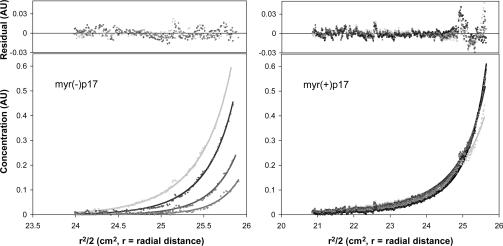
Myristoylation of p17 Promotes Protein Trimerization. Analytical ultracentrifugation was used as a tool for evaluating high-order assembly of the synthetic proteins in solution (Fig. 4). Data analysis by using the program winnonlin indicated that although myr(+)-p17 at 100 μM and 35°C was present in a monomer–trimer equilibrium with an association constant of 1.2 ± 0.4 × 108 M–2, myr(–)-p17 existed as a monomer with no significant amount of trimeric species present under the same conditions. The experimental data did not fit equilibria between monomer and dimer or other higher-order oligomers. These results suggest that myristoylation of p17 promotes protein trimerization in solution. It is worth noting that for myr(+)-p17 at 100 μM, about half of the protein formed trimer. In contrast, >99% of myr(+)-p17 used in the protein denaturation experiment was in the monomeric form because the protein concentration was much lower (0.1 mg/ml).
Fig. 4.
Sedimentation equilibrium for myr(–)-p17 and myr(+)-p17 at 100 μM and 35°C. The data for myr(–)-p17 were collected at eight different rotor speeds ranging from 18,000 to 50,000 rpm; only the high-speed sedimentation equilibrium data are shown (38,000, 42,000, 46,000, and 50,000 rpm). The data for myr(+)-p17 were collected at five different rotor speeds: 18,000, 19,000, 20,000, 21,000, and 22,000 rpm. Myr(–)-p17 existed as a monomer under the experimental conditions, whereas myr(+)-p17 was in a monomer–trimer equilibrium with a Keq value of 1.2 × 108 M–2.
Discussion
N-terminal myristoylation is an important posttranslational processing event that targets proteins to the inner surface of the plasma membrane by inserting the 14-carbon hydrophobic anchor into the lipid bilayer (23). Because the energetic strength of such interaction alone is insufficient to hold lipid-linked proteins at the membrane surface (36, 37), additional stabilizing interactions between the protein and membrane are needed. Despite the multipoint anchoring, lipid proteins can still dissociate from the plasma membrane, an event coupled to functions important in HIV-1 pathogenesis in the case of p17. Understanding the molecular determinants governing the reversible membrane-binding process will require comprehensive structural and functional studies of lipid-linked proteins such as p17, which cannot be readily produced in large quantities in heterologous expression systems lacking intracellular machineries for proper posttranslational modifications.
Several techniques for site-specific unnatural modifications of proteins are currently in practice, including total chemical synthesis (32, 38, 39), in vitro translation (40–42), auxotrophic bacterial expression (43–46), enzymatic semisynthesis (47–49), native chemical ligation (30, 50), expressed protein ligation (51), and, more recently, expression in Escherichia coli by using novel orthogonal tRNA/synthetase pairs (52, 53). Among these methods, total chemical synthesis coupled with native chemical ligation has proven to be a robust and versatile technique for the production of small- to medium-sized proteins (54).
Native chemical ligation, developed by Kent and coworkers (30), allows two fully unprotected synthetic peptides to react chemoselectively in aqueous solution to assemble a longer polypeptide chain linked by a new peptide bond. The method requires the presence of a C-terminal α-thioester moiety on the first peptide and an N-terminal cysteine residue on the second one. When multistep ligation strategies are used (55), large proteins can be assembled in solution. The 131-residue HIV-1 p17 contains two Cys residues conveniently situated in the sequence. Our experimental results clearly demonstrate that the matrix protein and its unnaturally modified forms can be readily synthesized by means of two-step, sequential ligation of three peptide segments. This methodology has afforded us a unique opportunity to investigate how the hypothesized myristoyl switch model for p17 works at the molecular level in the lipid-mediated membrane-binding process.
The myristoyl switch model stipulates that the p17 domain, as part of the HIV-1 Gag polyprotein, adopts a structure that exposes the N-terminal myristoyl group for interaction with the plasma membrane. However, upon cleavage by the HIV-1 protease during viral maturation, the p17 domain undergoes a significant conformational change, resulting in burial or sequestration of the fatty acid moiety and, thus, a weakened membrane-binding capacity. Several lines of evidence derived from cryoelectron microscopic characterizations of virus particles and functional studies seem to support this hypothesis (18, 20–22, 56), yet a definitive structural proof remains elusive. Recently, the solution structure of a 283-residue N-terminal fragment of the immature HIV-1 Gag polyprotein comprising both the p17 domain and the downstream capsid protein has been determined (57). The p17 domain in the immature Gag polyprotein assumed an essentially identical structure to that observed for the cleaved form determined by x-ray crystallography and NMR spectroscopy (28, 29), suggesting that proteolysis did not induce structural changes in p17. The proteins used for structural determination were nonmyristoylated, leaving room for uncertainty as to whether proteolysis leads to structural changes in myristoylated proteins. Nevertheless, if the myristoyl switch were the result of a significant structural rearrangement after cleavage and involving a distal C-terminal α-helix, as originally postulated for p17, myristoylated p17 would adopt a different conformation than unmodified p17. Our findings seem to suggest otherwise.
Both forms of p17 gave rise to virtually identical CD spectra, suggesting that N-myristoylation has little impact on the overall structure of the protein. Notably, myristoylated p17 is more stable than the nonmyristoylated protein by 1 kcal/mol. Solvent-exposed hydrophobic side chains in a protein are, in general, destabilizing because they can be buried in the unfolded state, thus favoring protein denaturation (58). A solvent-exposed myristoyl group is expected to further destabilize the protein because of an additional entropy penalty associated with the long flexible hydrocarbon chain (58). Our finding indicates the existence of an energetically favorable interaction of a hydrophobic nature between the 14-carbon fatty acid moiety and proximal residues in the protein. The N terminus of p17 shown in various structural analyses is highly mobile (27–29), suggesting that it would not interfere with such an interaction. Whereas our data identify a “myristoylhidden” interaction mode for p17 in the absence of any spectroscopically discernible structural change in the protein, residues involved in the myristoyl sequestration remain to be structurally identified.
The central question remains: What triggers the burial of the myristoyl group in p17 in the reversible membrane binding process? A partial answer may lie in the destabilization of oligomeric structures of p17 at the membrane surface. Nonmyristoylated p17, at up to millimolar concentrations in solution, adopts a monomeric structure composed of five α-helices connected by short loops, as determined by NMR spectroscopy (25, 28). In contrast, x-ray diffraction shows that nonmyristoylated p17 crystallizes as a trimer, similar in other respects to the NMR structure (29). Trimers and/or higher-order oligomers of p17 are believed to be the quaternary structure relevant to membrane binding. In fact, mutations detrimental to p17 trimerization have been shown to prevent viral particle assembly (59, 60). Our results obtained from the sedimentation experiments indicate that at concentrations >100 μM, myristoylated p17 predominantly exists as a trimer, whereas the nonmyristoylated protein stays as a monomer under identical conditions. We speculate that p17 trimerization in solution is mediated by the clustering of exposed myristoyl groups. It is plausible that the myristoyl switch in p17 at the membrane surface, although not caused by any structural change associated with proteolysis, may be triggered energetically. Because regions of the HIV-1 Gag polyprotein outside the p17 domain contribute to membrane binding as well as to protein oligomerization (18, 21, 61–64), the departure of downstream Gag subdomains upon proteolytic cleavage predictably weakens oligomeric structures of p17 at the membrane surface. Such destabilization may lead to the dissociation of “clustered” myristoyl moieties or p17 molecules from each other and, ultimately, the detachment of some p17 molecules from the membrane. This reversible membrane-binding event is energetically favored to some extent because the myristoyl moiety in dissociated monomeric p17 molecules can adopt a thermodynamically more stable, sequestered conformation. A structural proof is needed to support this possibility by demonstrating the existence of myristoyl-sequestered monomeric and myristoyl-exposed trimeric structures of the p17 protein. Crystallographic studies of both forms of synthetic p17 are needed. Finally, it is worth noting that phosphorylation at the C-terminal Tyr of p17 has been suggested to be an important regulatory event in facilitating import of the viral preintegration complex into the cell nucleus (9, 65, 66). Tyr phosphorylation induces the binding of p17 to the core domain of integrase (65). It remains to be seen whether Tyr phosphorylation also promotes dissociation of trimeric p17 in solution and destablization of multimeric p17 at the membrane surface.
Shortly before we submitted this manuscript, Tang, Summers, and coworkers (67) published the NMR structure of myristoylated HIV-1 p17, and it seems to be in agreement with the conclusions we have reached based on our own studies. The structure reveals that the myristoyl group is partially sequestered in a hydrophobic cavity that requires only minor conformational adjustments for insertion. Their work further suggests that the exposure of the myristoyl group is coupled with protein trimerization, which is enhanced by the presence of other Gag subdomains. They concluded that the HIV-1 myristoyl switch is not induced by conformational changes but through entropic modulation of a preexisting equilibrium between myristoyl-sequestered monomer and myristoyl-exposed trimer. Notably, although the NMR study did not obtain myristoyl-exposed trimer because of protein aggregation at high concentrations, indirect spectroscopic evidence supported its existence in solution.
Acknowledgments
We thank Drs. Mika Popovic and Fabio Romerio of the Institute of Human Virology (Baltimore) for useful discussions. This work was supported by National Institutes of Health Grants AI056264 and AI058939 (to W.L.) and Maryland Cancer Research Grant CH649CRF and supported in part by the Intramural AIDS Targeted Antiviral Program of the Office of the Director, the National Institutes of Health (J.L.).
Abbreviations: Acm, acetamidomethyl; CD, circular dichroism; GuHCl, guanidine hydrochloride; myr(+)-p17, myristoylated HIV-1 matrix protein p17; myr(–)-p17, nonmyristoylated HIV-1 matrix protein p17; RP, reversed-phase.
References
- 1.Freed, E. O. (1998) Virology 251, 1–15. [DOI] [PubMed] [Google Scholar]
- 2.Bryant, M. & Ratner, L. (1990) Proc. Natl. Acad. Sci. USA 87, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gheysen, D., Jacobs, E., de Foresta, F., Thiriart, C., Francotte, M., Thines, D. & De Wilde, M. (1989) Cell 59, 103–112. [DOI] [PubMed] [Google Scholar]
- 4.Gottlinger, H. G., Sodroski, J. G. & Haseltine, W. A. (1989) Proc. Natl. Acad. Sci. USA 86, 5781–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nermut, M. V., Hockley, D. J., Jowett, J. B., Jones, I. M., Garreau, M. & Thomas, D. (1994) Virology 198, 288–296. [DOI] [PubMed] [Google Scholar]
- 6.Gelderblom, H. R. (1991) AIDS 5, 617–637. [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., Haggerty, S., Dempsey, M. P., Sharova, N., Adzhubel, A., Spitz, L., Lewis, P., Goldfarb, D., Emerman, M. & Stevenson, M. (1993) Nature 365, 666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Schwedler, U., Kornbluth, R. S. & Trono, D. (1994) Proc. Natl. Acad. Sci. USA 91, 6992–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallay, P., Swingler, S., Aiken, C. & Trono, D. (1995) Cell 80, 379–388. [DOI] [PubMed] [Google Scholar]
- 10.Reil, H., Bukovsky, A. A., Gelderblom, H. R. & Gottlinger, H. G. (1998) EMBO J. 17, 2699–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haffar, O. K., Popov, S., Dubrovsky, L., Agostini, I., Tang, H., Pushkarsky, T., Nadler, S. G. & Bukrinsky, M. (2000) J. Mol. Biol. 299, 359–368. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O., Englund, G. & Martin, M. A. (1995) J. Virol. 69, 3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier, R. A., Meyer, B. E., Simon, J. H., Fischer, U. & Malim, M. H. (1997) EMBO J. 16, 4531–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont, S., Sharova, N., DeHoratius, C., Virbasius, C. M., Zhu, X., Bukrinskaya, A. G., Stevenson, M. & Green, M. R. (1999) Nature 402, 681–685. [DOI] [PubMed] [Google Scholar]
- 15.De Francesco, M. A., Baronio, M., Fiorentini, S., Signorini, C., Bonfanti, C., Poiesi, C., Popovic, M., Grassi, M., Garrafa, E., Bozzo, L., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono, A., Orenstein, J. M. & Freed, E. O. (2000) J. Virol. 74, 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan, X., Yu, X., Lee, T. H. & Essex, M. (1993) J. Virol. 67, 6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spearman, P., Horton, R., Ratner, L. & Kuli-Zade, I. (1997) J. Virol. 71, 6582–6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, W., Parent, L. J., Wills, J. W. & Resh, M. D. (1994) J. Virol. 68, 2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou, W. & Resh, M. D. (1996) J. Virol. 70, 8540–8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scarlata, S., Ehrlich, L. S. & Carter, C. A. (1998) J. Mol. Biol. 277, 161–169. [DOI] [PubMed] [Google Scholar]
- 22.Hermida-Matsumoto, L. & Resh, M. D. (1999) J. Virol. 73, 1902–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Resh, M. D. (1999) Biochim. Biophys. Acta 1451, 1–16. [DOI] [PubMed] [Google Scholar]
- 24.Valentine, K. G., Mesleh, M. F., Opella, S. J., Ikura, M. & Ames, J. B. (2003) Biochemistry 42, 6333–6340. [DOI] [PubMed] [Google Scholar]
- 25.Matthews, S., Barlow, P., Boyd, J., Barton, G., Russell, R., Mills, H., Cunningham, M., Meyers, N., Burns, N., Clark, N., et al. (1994) Nature 370, 666–668. [DOI] [PubMed] [Google Scholar]
- 26.Matthews, S., Barlow, P., Clark, N., Kingsman, S., Kingsman, A. & Campbell, I. (1995) Biochem. Soc. Trans. 23, 725–729. [DOI] [PubMed] [Google Scholar]
- 27.Massiah, M. A., Worthylake, D., Christensen, A. M., Sundquist, W. I., Hill, C. P. & Summers, M. F. (1996) Protein Sci. 5, 2391–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massiah, M. A., Starich, M. R., Paschall, C., Summers, M. F., Christensen, A. M. & Sundquist, W. I. (1994) J. Mol. Biol. 244, 198–223. [DOI] [PubMed] [Google Scholar]
- 29.Hill, C. P., Worthylake, D., Bancroft, D. P., Christensen, A. M. & Sundquist, W. I. (1996) Proc. Natl. Acad. Sci. USA 93, 3099–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawson, P. E., Muir, T. W., Clark-Lewis, I. & Kent, S. B. (1994) Science 266, 776–779. [DOI] [PubMed] [Google Scholar]
- 31.Schnolzer, M., Alewood, P., Jones, A., Alewood, D. & Kent, S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193. [DOI] [PubMed] [Google Scholar]
- 32.Kent, S. B. (1988) Annu. Rev. Biochem. 57, 957–989. [DOI] [PubMed] [Google Scholar]
- 33.Pace, C. N., Vajdos, F., Fee, L., Grimsley, G. & Gray, T. (1995) Protein Sci. 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, W. Y., Starovasnik, M. A., Dwyer, J. J., Kossiakoff, A. A., Kent, S. B. & Lu, W. (2000) Biochemistry 39, 3575–3584. [DOI] [PubMed] [Google Scholar]
- 35.Botos, I., Wu, Z., Lu, W. & Wlodawer, A. (2001) FEBS Lett. 509, 90–94. [DOI] [PubMed] [Google Scholar]
- 36.Buser, C. A., Sigal, C. T., Resh, M. D. & McLaughlin, S. (1994) Biochemistry 33, 13093–13101. [DOI] [PubMed] [Google Scholar]
- 37.Peitzsch, R. M. & McLaughlin, S. (1993) Biochemistry 32, 10436–10443. [DOI] [PubMed] [Google Scholar]
- 38.Wlodawer, A., Miller, M., Jaskolski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. (1989) Science 245, 616–621. [DOI] [PubMed] [Google Scholar]
- 39.Milton, R. C., Milton, S. C. & Kent, S. B. (1992) Science 256, 1445–1448. [DOI] [PubMed] [Google Scholar]
- 40.Noren, C. J., Anthony-Cahill, S. J., Griffith, M. C. & Schultz, P. G. (1989) Science 244, 182–188. [DOI] [PubMed] [Google Scholar]
- 41.Mendel, D., Ellman, J. A., Chang, Z., Veenstra, D. L., Kollman, P. A. & Schultz, P. G. (1992) Science 256, 1798–1802. [DOI] [PubMed] [Google Scholar]
- 42.Bain, J. D., Switzer, C., Chamberlin, A. R. & Benner, S. A. (1992) Nature 356, 537–539. [DOI] [PubMed] [Google Scholar]
- 43.Beiboer, S. H., van den Berg, B., Dekker, N., Cox, R. C. & Verheij, H. M. (1996) Protein Eng. 9, 345–352. [DOI] [PubMed] [Google Scholar]
- 44.Budisa, N., Minks, C., Alefelder, S., Wenger, W., Dong, F., Moroder, L. & Huber, R. (1999) FASEB J. 13, 41–51. [DOI] [PubMed] [Google Scholar]
- 45.Hamano-Takaku, F., Iwama, T., Saito-Yano, S., Takaku, K., Monden, Y., Kitabatake, M., Soll, D. & Nishimura, S. (2000) J. Biol. Chem. 275, 40324–40328. [DOI] [PubMed] [Google Scholar]
- 46.van Hest, J. C. M., Kiick, K. L. & Tirrell, D. A. (2000) J. Am. Chem. Soc. 122, 1282–1288. [Google Scholar]
- 47.Bigler, T. L., Lu, W., Park, S. J., Tashiro, M., Wieczorek, M., Wynn, R. & Laskowski, M. (1993) Protein Sci. 2, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson, D. Y., Burnier, J., Quan, C., Stanley, M., Tom, J. & Wells, J. A. (1994) Science 266, 243–247. [DOI] [PubMed] [Google Scholar]
- 49.Wallace, C. J. (1995) Curr. Opin. Biotechnol. 6, 403–410. [DOI] [PubMed] [Google Scholar]
- 50.Lu, W., Qasim, M. A. & Kent, S. B. (1996) J. Am. Chem. Soc. 118, 8518–8523. [Google Scholar]
- 51.Muir, T. W., Sondhi, D. & Cole, P. A. (1998) Proc. Natl. Acad. Sci. USA 95, 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L., Brock, A., Herberich, B. & Schultz, P. G. (2001) Science 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L., Brock, A. & Schultz, P. G. (2002) J. Am. Chem. Soc. 124, 1836–1837. [DOI] [PubMed] [Google Scholar]
- 54.Dawson, P. E. & Kent, S. B. (2000) Annu. Rev. Biochem. 69, 923–960. [DOI] [PubMed] [Google Scholar]
- 55.Bang, D., Chopra, N. & Kent, S. B. (2004) J. Am. Chem. Soc. 126, 1377–1383. [DOI] [PubMed] [Google Scholar]
- 56.Fuller, S. D., Wilk, T., Gowen, B. E., Krausslich, H. G. & Vogt, V. M. (1997) Curr. Biol. 7, 729–738. [DOI] [PubMed] [Google Scholar]
- 57.Tang, C., Ndassa, Y. & Summers, M. F. (2002) Nat. Struct. Biol. 9, 537–543. [DOI] [PubMed] [Google Scholar]
- 58.Eisenberg, D. S. & Richards, F. M., eds. (1995) Adv. Protein Chem. 46.
- 59.Morikawa, Y., Zhang, W. H., Hockley, D. J., Nermut, M. V. & Jones, I. M. (1998) J. Virol. 72, 7659–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cannon, P. M., Matthews, S., Clark, N., Byles, E. D., Iourin, O., Hockley, D. J., Kingsman, S. M. & Kingsman, A. J. (1997) J. Virol. 71, 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandefur, S., Varthakavi, V. & Spearman, P. (1998) J. Virol. 72, 2723–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Platt, E. J. & Haffar, O. K. (1994) Proc. Natl. Acad. Sci. USA 91, 4594–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Craven, R. C., Leure-duPree, A. E., Weldon, R. A., Jr., & Wills, J. W. (1995) J. Virol. 69, 4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bennett, R. P., Nelle, T. D. & Wills, J. W. (1993) J. Virol. 67, 6487–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallay, P., Swingler, S., Song, J., Bushman, F. & Trono, D. (1995) Cell 83, 569–576. [DOI] [PubMed] [Google Scholar]
- 66.Camaur, D., Gallay, P., Swingler, S. & Trono, D. (1997) J. Virol. 71, 6834–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang, C., Loeliger, E., Luncsford, P., Kinde, I., Beckett, D. & Summers, M. F. (2004) Proc. Natl. Acad. Sci. USA 101, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]