Abstract
Viruses in the rhinovirus C species (RV-C) can cause severe respiratory illnesses in children including pneumonia and asthma exacerbations. A transduced cell line (HeLa-E8) stably expressing the CDHR3-Y529 receptor variant, supports propagation of RV-C after infection. C15 clinical or recombinant isolates replicate in HeLa-E8, however progeny yields are lower than those of related strains of RV-A and RV-B. Serial passaging of C15 in HeLa-E8 resulted in stronger cytopathic effects and increased (≥ 10-fold) virus binding to cells and progeny yields. The adaptation was acquired by two mutations which increased binding (VP1 T125K) and replication (3A E41K), respectively. A similar 3A mutation engineered into C2 and C41 cDNAs also improved viral replication (2–8 fold) in HeLa but the heparan sulfate mediated cell-binding enhancement by the VP1 change was C15-specific. The findings now enable large-scale cost-effective C15 production by infection and the testing of RV-C infectivity by plaque assay.
Keywords: Rhinovirus C, Pathogenesis, Adaptation, Receptor specificity, Heparan sulfate
1. Introduction
Rhinoviruses (RV) are classified into three species (A, B and C) of the Picornaviridae family. They subdivide further into more than 160 genotypes that are responsible for the majority of upper respiratory tract infections (common colds), and also many lower respiratory tract illnesses (Hayden, 2004; Gern, 2010). The first isolates in the RV-C species were described in 2006 (Lamson et al., 2006; Arden et al., 2006). Consequently, 55 genotypes have been reported, now believed to be synonymous with serotypes, according to sequence diversity thresholds observed in the capsid-coding proteins, VP1 and VP4 (Simmonds et al., 2010; McIntyre et al., 2013). The RV-A and RV-C tend to cause more severe illnesses in children compared to RV-B; however, RV-C infections are more closely linked with childhood asthma exacerbations (Calvo et al., 2010; Bizzintino et al., 2011; Lee et al., 2012; Cox et al., 2013; Drysdale et al., 2014; Fawkner-Corbett et al., 2015).
Prototype RV-A and RV-B laboratory strains representing major and minor receptor groups, are commonly used in vitro to study virus biology and host cell responses. These include A1 (subtypes a and b), A2, A16, A39 and B14 (Stanway et al., 1984; Skern et al., 1985; Hughes et al., 1988; Kim et al., 1989; Lee et al., 1995), most of which are available as fully-sequenced, infectious cDNA reagents. To achieve full viability in culture, these strains were typically passaged multiple times in continuous HeLa cell line after their initial clinical isolations but before recombinant cloning. The resulting adapted forms of these viruses are preferred for experiments because they replicate to high titers and induce strong cytopathic effect (CPE) in cell culture (Conant & Hamparian, 1968). In contrast, most initial, unpassaged clinical RV-A and RV-B isolates generally replicate less efficiently and induce milder CPE in HeLa cells, even though their infectivity to natural host cells (differentiated airway epithelium) remains quite robust (Nakagome et al., 2014).
Human cadherin-related family member 3 (CDHR3) is a cell surface protein that mediates RV-C binding to cells, and consequently allows virus entry and replication when it is induced to express in cell culture. A transduced HeLa cell line derivative (HeLa-E8) which stably expresses the CDHR3-Y529 variant supports infection and propagation of multiple RV-C isolates in such cultures (Bochkov et al., 2015). C15, a recombinant derivative of a clinical RV-C isolate (Bochkov et al., 2011) replicates to detectable titers in HeLa-E8, yet the yields of progeny virus never reach the levels achieved with parallel infections by RV-A or RV-B HeLa-adapted strains. Moreover, C15 infections (clinical or recombinant isolates) of HeLa-E8 cells rarely produce visible CPE, a useful phenotypic marker of effective, productive viral synthesis.
We now report the isolation of a C15a strain which was adapted to growth in cultured HeLa-E8 cells by serial passaging. The progeny, as a heterotypic population, or as cloned recombinant derivatives, give enhanced virus yields after infection and induce visible CPE in these cells, validating their use as effective new reagents for RV-C investigations. Genome sequencing of passaged C15a populations identified two mutations which recapitulated the complete adaptive phenotype, and, moreover, could partially confer adaptation to other recombinant RV-C types.
2. Results
2.1. C15 adaptive passage
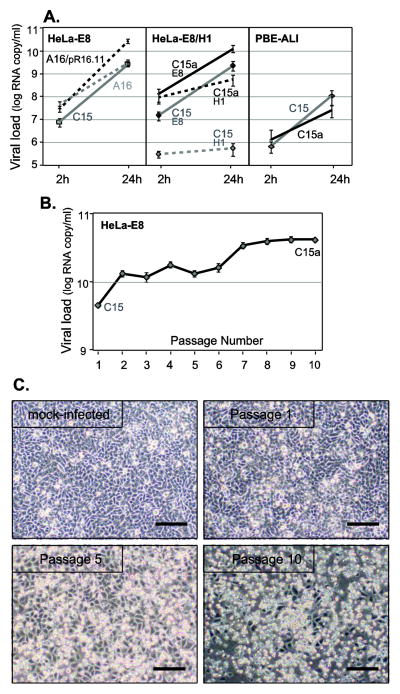
Preparations of wild-type C15 or A16 viruses will infect HeLa-E8 cells, however, the progeny yields after 24 h are typically about 10-fold lower than for A16/pR16.11 (Lee & Wang, 2003), an A16 recombinant virus previously adapted to HeLa cell growth (Fig. 1A). To improve these yields for an RV-C, HeLa-E8 cells were infected with a high dose of recombinant C15 (about 2,000 viral (v) RNA copies per cell). The virus inoculum was replaced with fresh growth medium 2 h post-infection (p.i.) after which the cells were incubated for 72 h (34°C), and then harvested. The clarified cell lysates after three freeze / thaw cycles were used for the next round of infection in a blind-passage serial process repeated a total of 10 times (P1 to P10). At each step, the virus yield was tested by quantitative (q) RT-PCR and by P10, this value had increased significantly relative to the P1 starting sample (Fig. 1B). After only five of these passages however, visual cell monitoring clearly showed evidence of stronger cytopathic effects (with detached and rounded cells). In the P10 infected cells, almost complete cell lysis was evident at 72 h p.i. (Fig. 1C).
Figure 1.
C15 adaptation. (A) Monolayers of HeLa-E8 and HeLa-H1, or differentiated cultures of PBE-ALI cells from three donors, grown in 12-well plates were infected with A16, A16/pR16.11, C15 or C15a virus at 4 × 108 vRNA copies per well. After 2 h (input virus to estimate binding) or 24 h (replication) incubation at 34°C, the cells were analyzed for viral RNA signals by RT-qPCR (means ± s.d., n ≥ 3). (B) C15 virus was passaged serially in HeLa-E8 cells as described in Results and each passage was evaluated 72 h p.i. for virus load (viral RNA copies per ml) by RT-qPCR (means ± s.d., n = 3). Polyclonal C15a is defined as the Passage 10 material. (C) Passage 1, Passage 5 and Passage 10 and mock-infected cells were visualized (72 h p.i.) and photographed by light microscopy. Scale bar is 100 μm.
Serial passage adaptation ultimately resulted in virus samples (C15a) with increased (≥ 10-fold) binding titers (cell-associated input virus) in HeLa-E8 cells (2 h samples), and progeny yields (24 h samples) that were about 10-fold higher than the initial C15 material (Fig. 1A). Surprisingly, when the C15a population was tested for binding to non-transduced HeLa-H1 cells (parental line to HeLa-E8), the adaptation had affected this condition too, and the virus reacted nearly equivalently with both cell lines. However, progeny yields in HeLa-E8 were consistently more than 1.5 log higher when compared to the HeLa-H1 cells. Visual inspection showed C15a infections caused lysis of HeLa-E8 cells, but the infected HeLa-H1 cells did not lyse (data not shown). Adaptation to HeLa-E8 was cell-type specific, because the C15a virus samples tended to have a lowered ability to replicate in fully-differentiated cultures of human primary bronchial epithelial cells grown at an air-liquid interface (PBE-ALI, Fig. 1A). While C15a maintained a C15-like cell-binding potential in PBE-ALI, the relative progeny yield in these cultures was now 5–10-fold lower.
2.2. C15a genome sequencing
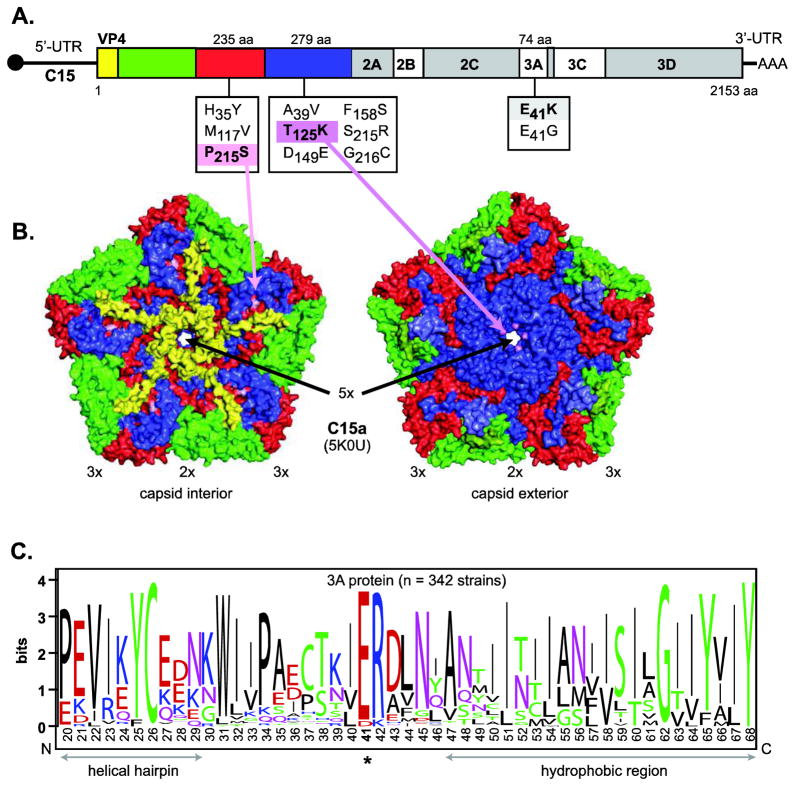
Total RNA was extracted from a sample of the P10 lysate, amplified into 14 overlapping viral amplicons by RT-PCR, as predicted by the parental C15 sequence, and sequenced. Although no mutations were found in the 5′ and 3′ UTRs, fragment analysis showed most shared a subset of three high frequency polyprotein missense mutations (Fig. 2A). A few additional low frequency missense mutations were also observed within the P10 population, all of which were contained exclusively within the VP3 and VP1 capsid genes, or in the 3A gene. No other nonsynonymous substitutions were detected elsewhere in the genome. However, two silent mutations were found in VP1 and one in the 3C gene (data not shown). The high frequency mutations altered VP3 amino acid Pro215 to Ser (P215S), VP1 amino acid Thr125 to Lys (T125K), and 3A protein Asp41 to Lys (E41K).
Figure 2.
C15a mutations. (A) Schematic of the C15 genome shows all missense mutations identified in structural (VP1 and VP3) and non-structural (3A) regions of the C15a population. The dominant mutations (found in >5 out of 10 clones) are highlighted. (B) Depiction of a C15a capsid pentamer structure (PDB 5KOU) localizes VP3 P215 and VP1 T125 to the interior and exterior surfaces, respectively. The locations of the 5-fold, 3-fold, and 2-fold symmetry axes are indicated by 2×, 3×, and 5×, respectively. (C) WebLogo (Crooks et al., 2004) depiction for an amino acid sequence alignment of RV sequences (n = 342) shows E41 as the dominant residue in this highly conserved position of the 3A protein. Color coding in this panel is by residue type, the overall height of the stack indicates the sequence conservation, while the height of symbols within the stack indicates the relative frequency of each amino acid at that position.
Within the repertory of sequenced RV-C genomes (n = 67), VP3 P215 is the dominant amino acid in 70% of the isolates (S215 represents 29%). The recent cryoEM structure of C15a (Liu et al., 2016) locates this position on the inner surface of the C15a capsid shell adjacent to the packaged viral RNA (Fig. 2B, interior). The VP1 T125 sequence represents 48% of the known RV-C. K125 is also found here natively in 12% of the sequenced isolates, as are Gln, Ala, Arg, Ser, His and Pro, in diminishing frequency (sum of 40%). This amino acid is on the outside of the capsid structure, where individual protomers cluster their contributed residues within a tight “well” or depression-like feature surrounding each 5-fold axis of virion symmetry (Fig. 2B, exterior). 3A E41 (86%) shares its polyprotein position exclusively with D41 (Glu, 14%), and these are the only amino acids found at this site in all sequenced rhinoviruses, including the RV-A (n = 208) and RV-B (n = 74). From the partially determined structure of poliovirus 3A (Strauss et al., 2003), the D/E41 locale contributes to a strongly conserved sequence motif marking the junction of helical hairpin and a hydrophobic domain (Fig. 2C). We speculate that this region may promote attachment of 3A protein to internal cellular membranes during viral RNA synthesis.
2.3. C15a reconstruction
Logically, capsid outer surface changes (i.e. VP1 T125K), or replication complex changes (i.e. 3A E41K) were the best prospective contributors to the C15a adaptation phenomenon, so these mutations were engineered singly and in tandem back into the parental C15 cDNA. The resultant viruses (C15 K125, C15 K41, C15 K125/K41), isolated after RNA transfection of WisL cells, were tested for cell binding and replication phenotypes in both HeLa-E8 and HeLa-H1 cells (Fig. 3A). All 3 preparations which harbored the K125 mutation (C15a, C15 K125 and C15 K125/K41) showed about a 10-fold increase in binding to HeLa-E8 cells relative to C15, and moreover, these same viruses now collectively interacted with HeLa-H1 cells at levels more than 100-fold more effectively than C15. The 3A K41 mutation did not influence the binding levels for either cell line, but when this mutation was present singly (C15 K41) or in combination with the capsid mutation (C15 K125/K41) the progeny yields from those infected cells were consistently more than 5–10-fold higher than from C15 alone. The enhanced replication induced by 3A K41 was observed even in HeLa-H1 cells, where this mutation alone induced about 7-fold increase in vRNA levels at 24 h p.i., despite the poor initial binding to the surface of these cells (Fig. 3A).
Figure 3.
C15a recombinants. (A) Recombinant C15 and recombinant C15 harboring one or both dominant adaptive mutations (VP1 T125K, 3A T41K) and polyclonal C15a were tested for relative binding affinities (cell-associated input virus at 2 h) and replication potential (24 h) in HeLa-E8 and HeLa-H1 cells. Infections per well were initiated with equivalent 4 × 108 vRNA copies per well. Viral load was measured by RT-qPCR (means ± s.d., n = 5). (*) indicates p < 0.05 significance (t-test) vs. C15. (B) Infected cells were visualized and photographed by light microscopy at 48 h p.i. Scale bar is 100 μm. (C) Cellular cytotoxicity (LDH release) caused by recombinant and polyclonal C15a viruses in HeLa-E8 cells over the time-course of infection (OD490, means ± s.d., n = 3). (*) indicates p < 0.05 significance (t-test) vs. C15.
Microscopic inspection of infected cells showed C15 K125 induced mild but visible CPE after 48 h p.i. (Fig. 3B). Parallel infection with C15 K125/K41 showed even stronger cell lysis which could not be distinguished from that of C15a. To quantitatively assess cellular cytotoxicity, LDH concentrations were also measured in growth medium 24, 48 and 72 h after infection. In agreement with the development of differential cytopathic effects, C15 preparations with the K125 mutation (C15a, C15 K125 and C15 K125/K41) released more LDH than did C15 or C15-K41 over the entire time course of infection; however, C15 K41 was more cytotoxic than C15 at 48 and 72 h p.i. (Fig. 3C). The results clearly indicate that C15 adaptation in HeLa-E8 cells was acquired by two key mutations which contributed to increased cell-binding (T125K in VP1) and replication (E41K in 3A), respectively. The combination, when engineered as cDNA recapitulated the polyclonal C15a growth phenotypes for cell binding, RNA replication and CPE. The third dominant amino acid change observed in the C15a population (VP3 P215S), was also tested singly and in tandem with VP1 T125K for possible effects on virus entry or cell binding. No growth phenotype of any kind could be associated with this mutation, which presumably only arose as a fortuitous change acquired during serial passaging (data not shown).
2.4. VP1 K125 mediates heparan sulfate binding
To understand the underlying mechanisms of increased C15a binding to HeLa cells, we examined glycan contribution to the novel receptor specificity. Although the cryoEM structure of C15a predicts a sialylated glycan interaction donated by the CDHR3 receptor (Liu et al., 2016), a preliminary screen for possible di- o trisaccharide binding partners or for related inhibitors added to the virus/cell binding reactions failed to demonstrate binding to glycans (data not shown). In parallel experiments, however, the recombinant and adapted C15a mutant panels were also tested for heparan sulfate (HS) and heparin (as a less expensive substitute for HS) inhibitory effects. A16 and A1a viruses, which use respectively ICAM-1 and LDLR receptors, were not affected by heparin in their interactions with HeLa-E8 cells (Fig. 4A). The same was true for the unadapted C15 virus which recognizes cells through CDHR3-mediated reactions (Bochkov et al., 2015). But the adaptive mutations within both C15a and C15 K125, now made both viruses susceptible to heparin, reducing their binding titers 44 and 17-fold, respectively, or back down to the values without the adaptive mutation. Heparin and HS (1–2 mg/ml) both had this effect (Fig. 4B), and the observed degree of inhibition was dependent upon the dose of administered glycan (Fig. 4C).
Figure 4.
Heparin and heparan sulfate effects. (A) Virus samples (108 vRNA copies) were incubated with heparin (2 mg/ml) or medium alone (no heparin) for 30 min at 34°C, before inoculation of HeLa-E8 cells. After an attachment period (30 min at room temp, 30 min at 34°C) the cells were washed (3× PBS), harvested and tested by RT-qPCR for the attached virus RNA signals (means ± s.d., n ≥ 3). (*) indicates p < 0.05 significance (t-test). (B) C15a and C15 K125 (108 vRNA copies) were incubated with heparin or heparan sulfate (1 mg/ml) as in A, before HeLa-E8 infections and virus binding assays. (C) The C15a experiment in A was repeated using different doses of heparin in the preincubation. (D) HeLa-E8 cells (12 well-plates) were treated (37°C, 2 h) with the indicated dose (U) of heparinase I before inoculation with C15a virus (108 vRNA copies) and tested for virus binding as in A. (E) C15a and recombinant C15 K125/K41 viruses (108 vRNA copies) were incubated with heparin (1 mg/ml, 30 min, 34°C) before inoculation of HeLa-E8 or HeLa-H1 cells. After an attachment period (30 min at room temp, 30 min at 34°C), the cells were washed with PBS (3×). Cells were harvested and assayed for the input virus (1 h p.i.) and progeny virus (24 h p.i.) RNA signals by RT-qPCR (means ± s.d., n = 3). In panels B, C, and D, (*) indicates p < 0.05 significance (t-test) vs. medium control sample.
In agreement with other described properties of sulfated proteoglycans, enzymatic pretreatment of HeLa-E8 cells with heparinase I reduced C15a binding more than 7-fold, because presumably, the adapted virus now had fewer cellular-displayed HS binding sites available to it (Fig. 4D). Interestingly, when the adapted viruses (polyclonal population or recombinant) were tested comparatively, pretreatment with heparin abolished both binding and replication in the HeLa-H1 cells, but nonetheless, both viruses still replicated (to a degree) in HeLa-E8 cells despite the heparin treatment (Fig. 4E). This indicates that adapted virus could bind and enter cells through either of two non-competing, independent routes, native CDHR3 binding (HeLa-E8 cells), or HS binding (HeLa-E8 and H1) conferred through the VP1 T125K mutation.
2.5. Infectivity assay for RV-C
RV-C infections with native virus have been difficult to quantitate other than by qRT-PCR assays (Bochkov et al., 2011; McLeish et al., 2012; Schibler et al., 2012; Brebion et al., 2015). Given the enhanced growth of C15a and strong CPE in HeLa-E8 cells, alternative techniques could be evaluated for direct plaque assays. Indeed, this virus formed coherent, well-isolated small plaques on this cell line after 96 h of incubation (Fig. S1A). As a condition of this assay, the overlay needed to contain agarose, not agar, to avoid endogenous heparin contamination and subsequent virus-binding inhibition. Infectivity tests with cloned recombinant C15 preparations demonstrated that a combination of both VP1 T125K and 3A E41K mutations is required for plaque formation (Fig. S1B) since no isolated plaques were observed after infection with viruses possessing single mutations (data not shown). After careful evaluation of absorbance values (OD), PFU, and vRNA measurements from several independent virus preparations, the ratio of total viral particles (or viral RNA copies) to plaque-forming infectious particles in C15a suspensions purified by sucrose-cushion centrifugation, was estimated at about 200:1 (all data not shown). This value is quite similar to that reported for a laboratory strain of A16 type (Lee & Wang, 2003).
2.6. Recombinant C2 and C41 viruses
The 3A amino acid responsible for increased viral replication titers is universally conserved as E/D41 among native RV isolates. The VP1 T125 location conferring HS binding in addition to CDHR3 binding, is in a region more highly variable in sequence. Both of these mutations or their structural analogs (VP1 Q122K in C2 virus, and VP1 T124K in C41 virus) were engineered into full-length C2 or C41 cDNAs. The resultant recombinant viruses allowed testing of individual and combined mutational effects in genome contexts other than C15. All these viruses were able to interact with HeLa-E8 cells through the receptor-mediated native CDHR3 mechanism (input virus signals of more than 107 vRNA copies in these assays), but none of the new preparations acquired enhanced HeLa-H1 binding properties or enhanced HeLa-E8 binding properties (Fig. 5A,B). In neither case, did the recombinant VP1 changes now confer adaptive HS binding, indicating that the C15 VP1 K125 mutation was clearly genotype-specific. On the other hand, the 3A mutation effects were more ubiquitous. This change improved viral replication levels by 2–7 fold in both C2 and C41 contexts, regardless of whether it was tested singly or in tandem with the corresponding VP1 mutation. Moreover, even in HeLa-H1 cells, where the virus binding levels are always extremely low regardless of virus genotype, there was a weak but reproducibly discernable a 3–8-fold increase in viral RNA levels by 24 h p.i. This mutation, regardless of HeLa cell type or virus genotype, always increased the RV-C replication fecundity, independent of any other tested parameters.
Figure 5.
C2 and C41 recombinants. Recombinant (A) C2 and (B) C41 viruses engineered to express the indicated VP1 mutations (K122 or K124 respectively) and/or 3A K41 mutation, were inoculated into HeLa-E8 or HeLa-H1 cells (12-well plates, 4 × 108 vRNA copies per well). Virus titer was measured by RT-qPCR (means ± s.d., n ≥ 3) as in Fig 1A. (*) indicates p < 0.05 significance (t-test) vs. C2 or C41.
2.7. Adaptation of A16 virus
HeLa cell adapted variant of A16 virus (pR16.11) has been described (Lee et al., 1995; Lee & Wang, 2003), however, the use of HS-displaying surface proteins in addition to their native receptor (ICAM-1) for cell attachment has not been reported. To test mechanisms of adaptation of RV-A vs. RV-C, another recombinant A16 virus (pR16.939), derived from a clinical isolate (Nakagome et al., 2014), was serially passaged in HeLa-H1 cells. As with C15, strong visible CPE was noticed after only five serial passages. Almost complete cell lysis was evident at P10 (Fig. 6A). The virus progeny yield had also increased significantly relative to the P1 starting sample (Fig. 6B,C).
Figure 6.
A16 adaptation. A recombinant A16 virus (pR16.939), derived from a clinical isolate, was passaged serially in HeLa-H1 similar to Fig 1C. (A) At the indicated passages (72 h p.i.), cells were visualized by light microscopy. Scale bar, 100 μm. (B) A16 virus at each passage was evaluated 72 h p.i. for virus load (viral RNA copies per ml) by RT-qPCR (means ± s.d., n = 3). Polyclonal A16a is defined as the Passage 10 material. (C) Engineered recombinant viruses with the indicated mutation(s) were tested relative to A16 and A16a (adapted population) samples at 2 h (input virus to estimate binding) and 24 h (replication) in HeLa-H1 cells (12-well plates, 4 × 108 viral RNA copies per well). M1: Q261R; M2: Q261R, K281R; M3: Q261R, K268R, K281R; M4: D264Y. Viral load was determined by RT-qPCR (means ± s.d., n = 3). (*) indicates p < 0.05 significance (t-test) vs. A16. (D) Two regional PCR-derived cDNA amplicons (PCR1 and PCR2) from the A16 passage 10 population were sequenced to identify adaptation-specific mutations. Those identified in the 2C gene are indicated; high frequency mutations (found in > 5 out of 10 clones) are boldface.
Population sequencing was focused on the VP3–VP1 and 2C–3C genome regions (Fig. 6C), looking specifically for the analogous changes in structural and non-structural genes as in the C15a virus. But these changes were never observed. Instead, A16a harbored a handful of 2C gene coding changes (Fig. 6D) including Q261R, D264Y, K268R, and K281R. These were detected in cloned cDNA amplicons as individual mutations or in various linked combinations (M1–M4). Relative to the starting virus, the adapted A16a population typically gave about one log higher virus progeny yields (Fig. 6C), but without a concurrent, measurable increase in cell binding affinity. Infectivity tests with reconstructed recombinant A16 viruses carrying these mutation combinations showed that each of them was capable of enhanced viral replication matching the levels of the population A16a (Fig. 6C). Observation of the Q261R mutation is not unique to this adaptation. It was previously reported when an A16 isolate (A16/pR16.11) was also adapted for growth in HeLa-H1 cells (Lee et al., 1995; Lee & Wang, 2003) so it may represent a key adaptive position unique to this particular type of virus. The other observed mutations in the new A16a population, cluster nearby to 2C Q261, perhaps fortuitously, or contributing to the same presently unknown replication enhancement mechanism.
3. Discussion
Virus strains adapted to tissue culture are valuable reagents for the synthesis of high-titer preparations, infectivity assays, and molecular pathogenesis experiments. For maximum effectiveness however, it is important to understand how the adaptation products differ from native isolates. Serial passage of C15 in transduced HeLa-E8 cells, expressing surface-dominant variant (Y529) of CDHR3, identified two independent, yet synergistic adaptive mutations, T125K in VP1 and E41K in 3A. Their effectiveness in increased virus yields and CPE is documented by the new feasibility for RV-C plaque assays and the preparation of sufficient virus stock for the first high resolution RV-C capsid structure determination (Liu et al., 2016)
Some enteroviruses, including select RV-A, can use sialic acid glycans or other polysaccharides as functional receptors (Zautner et al., 2003; Khan et al., 2007; Israelsson et al., 2010; Tan et al., 2013; Liu et al., 2015; Liu et al., 2016). Moreover, cell adaptation through serial passaging in vitro sometimes changes receptor specificity to include HS proteoglycans (Sa-Carvalho et al., 1997; Klimstra et al., 1998; Smit et al., 2002; Vlasak et al., 2005). When this happens, the combination of native receptor binding and heparin binding synergistically can boost the total virus interactions with cells, and consequently may lead to increased titers in tissue culture.
VP1 T125 is a capsid surface residue. It clusters with 5-fold symmetry at the 12 vertices of the particles. Mutation to K125 changes significantly the local charge distribution (negative) for these regions and as a consequence, C15 with this adaptation apparently gains the ability to bind to HS (or heparin) in addition to CDHR3. The enhanced cell attachment property carries over to HeLa cells which do not express CDHR3 (i.e. HeLa-H1), but it was readily abrogated by the presence of free heparin or HS added as an inhibitor, or by pretreatment of cells with heparinase. This adaptation conferred an advantage only in HeLa cells, because C15a did not show an enhanced growth profile in primary bronchial epithelial cells. Moreover, directed mutation of the same VP1 surface residue in C2 and C41 genome contexts did not change their corresponding HeLa cell binding attributes. All these viruses maintained the property of CDHR3 binding as their primary mechanism of cell attachment, but only C15a could now also increase its binding titer through the use of cell-displayed proteoglycans.
Analogous receptor specificity adjustments have been documented for multiple other RV and the related alphaviruses (Sa-Carvalho et al., 1997; Klimstra et al., 1998; Smit et al., 2002). For example, RV-A89 was tested serially in ICAM-1 deficient HEp-2 cells, or by alternating passage in HEp-2 and HeLa cells (Reischl et al., 2001; Vlasak et al., 2005). As with C15a, the HEp-2-adapted variant of A89 acquired the use of HS proteoglycans as alternative functional cellular receptors (Vlasak et al., 2005). An A54 clinical isolate has also been described which uses proteoglycans natively and infects human rhabdomyosarcoma (RD) cells lacking ICAM-1 expression. However infections via this mechanism are less efficient than those with ICAM-1 (Khan et al., 2007).
The second C15a adaptive mutation altered a highly conserved residue in the 3A protein. Regardless of whether engineered C2, C15 or C41 viruses were attached to HeLa cells by CDHR3 or heparin-dependent mechanisms, a 3A E41K-containing sequence in that genome induced up to a log higher progeny yields (24 h p.i.) than any virus lacking this mutation. Picornaviruses replicate their RNA on reorganized cellular membrane structures designated “replication organelles.” The virus-induced lipid composition of these regions differs significantly from normal cellular membranes (Belov, 2014; van der Linden et al., 2015). For enteroviruses, including the RV, non-structural proteins 2BC and 3A display hydrophobic domains that are somehow involved in this process of internal membrane rearrangement, presumably through interactions with a number of host cell proteins. These include Golgi-specific brefeldin A-resistance guanine nucleotide exchange factor 1 (GBF1), phosphatidylinositol 4-kinase type III β (PI4KIIIβ) and the Golgi adaptor protein acyl-CoA-binding domain-containing protein 3 (ACBD3) (Wessels et al., 2006; van der Linden et al., 2015). It has been demonstrated that transient expression of an A16 3A protein by transfection, can lead to disruption of the Golgi structure and inhibition of cellular protein secretion (Mousnier et al., 2014). Replication of this virus depends on GBF1 and PI4KIIIβ but not on ACBD3, and moreover, PI4KIIIβ recruitment to viral replication sites is mediated by the 3A protein (Dorobantu et al., 2015). Certain single-point mutations in the 3A protein of B14 and some other related picornaviruses, likewise confer resistance to some antiviral compounds (e.g. enviroxime) via bypassing their replication dependency on host factors such as PI4KIII (Heinz & Vance, 1995; van der Schaar et al., 2012; Dorobantu et al., 2016).
Thus, the involvement of 2BC or 3A mutations in RV adaptation is very well documented. When strains A1a, A2, A16 and A39 were adapted for efficient growth in mouse cells by serial/alternate passage in mouse and human (HeLa) cells, the results were variants with improved replication yields (Yin & Lomax, 1983; Harris & Racaniello, 2003; Harris & Racaniello, 2005; Rasmussen & Racaniello, 2011). Many of the responsible changes were traced to only a few amino acids in 2BC or 3A proteins, again suggesting a major role for these proteins in optimizing the replication processes in different host or cell types. Interestingly, the adaptive 3A mutations in A1a and A39 experiments were different from each other and from the 3A E41K now identified for C15a. Moreover, in the current study, when A16 was adapted to HeLa cells, it picked up 2C mutations and not the 3A K41 sequence. Overall, this means picornaviruses, including the RV-C, can adapt to their specific host environments by optimizing internal membrane interactions. Apparently, the specifics of this process can vary with the cell type and virus type, as long as the outcome is improved progeny titers. Further testing will be needed to confirm or deny whether 3A E41K can now provide at least one universal mechanism for this, particularly with regard to multiple-strain RV-C growth in HeLa cells. The advance here is that by controlled cell adaptation, laboratory strains of RV-C are now available which exhibit efficient replication in continuous cell lines (e.g. HeLa-E8). Certainly, these reagents will facilitate large-scale, cost-effective virus production for viral structure studies, CDHR3 investigations, and potentially, RV-C antiviral development.
4. Materials and Methods
4.1. Cell culture
Bronchial epithelial tissue samples were obtained from residual surgical specimens and cultured at air-liquid interface (fully-differentiated) as described previously (Schroth et al., 1999; Ashraf et al., 2015). The protocol was approved by the University of Wisconsin-Madison Human Subjects Committee. HeLa-H1 (ATCC# CRL-1958), HeLa-E8 (Bochkov et al., 2015) and WisL (human embryonic lung fibroblast) cells were grown in Eagle’s Minimum Essential Medium (Lonza) supplemented by non-essential amino acids (Gibco) and 10% fetal bovine serum (Gemini).
4.2. Virus sequencing
In these studies, the parental C15 and A16 sequences were according to GenBank accession numbers GU219984 and KX891411, respectively. Total RNA from a sample of the polyclonal C15a (P10) grown in HeLa-E8 monolayers was extracted for RT reactions and primed with random hexamers (Life Technologies) or OligoT-r primer (Table S1). The viral cDNAs were amplified using C15-specific primers (Table S1). A total of 14 genome-comprehensive PCR amplicons were sequenced directly as a population, and also cloned in pGEM-T Easy vectors, where out-growth colonies were sequenced individually (n = 10 clones per each product). Sequence data were assembled and compared using Lasergene v.12 software (DNAStar). Polyclonal HeLa-adapted A16 (P10) was treated similarly, except that population and cDNA clone sequencing focused on only two genome regions: the VP3–VP1 genes (PCR1, primers RV16-NcoI and RV16-BstXI), and the 2C–3C genes (PCR2, primers RV16-EcoRV and RV16-SnaBI) (Table S1).
4.3. Viral genome cDNAs with adaptive mutations
Full-length cDNA materials encoding C15, C2, C41 and A16 (pR16.11) infectious genomes have been described (Lee & Wang, 2003; Bochkov et al., 2011; Nakagome et al., 2014). pR16.939 encodes an RV-A16 clinical isolate that was cloned and provided by Dr. Wai-Ming Lee (Biological Mimetic Inc). The adaptive point mutations in C15, C2 and C41 infectious clones were engineered by conventional site-directed mutagenesis using the appropriate flanking primers with unique restriction enzyme sites and internal overlapping primers with mutated bases (Table S1) in a two-step PCR method. Adaptive mutations in the A16 clone (pR16.939) were made by linking the purified PCR2 product (Fig.6B) and pR16.939 clone digested with EcoRV and SnaBI using NEBuilder HiFi DNA Assembly Master Mix (NEB). A total of four cDNA clones containing individual mutations (M1, M4) or various linked combinations (M2, M3) were selected. All plasmid DNAs were verified in the regions of interest by sequencing, and then purified by Plasmid Maxi kits (Qiagen) before use in RNA synthesis reactions with T7 polymerase (Promega).
4.4. Viruses and infection
Recombinant rhinoviruses were produced by transfecting full-length T7 RNA transcripts synthesized in vitro from linearized plasmid cDNA, into WisL cells. Virus purification was by ultracentrifugation through a sucrose cushion as previously described (Bochkov et al., 2011; Nakagome et al., 2014; Griggs et al., 2015). Cells grown in 12- or 24-well plates (monolayers) or in Transwell polycarbonate inserts (0.4 μm pore size, Corning; for differentiated cultures of PBE cells) were (typically) inoculated with virus at 4 × 108 vRNA copies per well (unless another dose is indicated) or ALI insert followed by incubation for 2–24 hours at 34°C. At harvest at 2 h p.i. (cell-associated input virus to estimate binding), the monolayers were washed (3× with PBS) to remove any unattached inoculum, before lysis with RLT buffer (Qiagen), whereas at 24 h p.i. (replication) 100 μl of culture medium and whole cell lysate samples were collected to estimate total virus progeny yields. Viral load (vRNA copy number) was determined by RT-qPCR according to standardized RNA preparations. Total RNA was extracted from harvested cells and media samples using RNeasy Mini kits (Qiagen). The RT-qPCR used Power SYBR Green PCR mix (Life Technologies) and RV-specific primers as previously described (Bochkov et al., 2011).
4.5. Cellular cytotoxicity
Lactate dehydrogenase (LDH) concentrations in cell culture medium were measured at 24, 48, or 72 h after infection (4 × 108 vRNA copies per well in a 12-well plate) by using CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega) according to the manufacturer’s instructions. The absorbance (OD490) of the culture medium (background) was subtracted from all values of the experimental wells to calculate LDH release as an indicator of cellular cytotoxicity.
4.6. Inhibition of virus binding
Virus samples were preincubated (30 min, 34°C) with heparin (1–2 mg/ml, Sigma), HS (1 mg/ml, Sigma) or medium alone (control), before inoculation of HeLa cells grown in 12-well plates for 1 h (30 min at room temp and 30 min at 34°C). The cells were then washed with PBS (3×), harvested in RLT buffer (Qiagen) and tested by RT-qPCR for the input viral RNA signals. HeLa-E8 cells (12 well-plates) were also pretreated (37°C, 2 h) with the indicated dose (U) of heparinase I (Sigma) before inoculation with C15a virus (30 min at room temp, 30 min at 34°C) and tested by RT-qPCR as described above.
4.7. Plaque assay
The procedure was done as described previously (Sherry & Rueckert, 1985; Wang et al., 1998) with some modifications. HeLa-E8 cells monolayers were prepared by plating 1.2 × 106 cells per well in 6-well plates and then incubation at 37°C overnight. Cells were infected with 10-fold serial dilutions of C15a virus for 1 h at room temp. The infected monolayers were overlaid, first with 1.5 ml of 0.8% agarose (Promega) in medium P6 (Sherry & Rueckert, 1985), and then (after the agarose solidified) with 1.5 ml of medium P6 containing 4 mM L-glutamine, 4 mM oxaloacetate, 2 mM pyruvate, and 11.2 mM D-glucose. Plaques were allowed to develop at 34°C for 96 h and then visualized by crystal violet staining.
4.8. Statistical analysis
We used Student’s t-test to analyze viral binding and replication data (SigmaPlot 11.0, Systat Software). Significance was defined at p < 0.05. Results from 3–5 independent experiments were expressed as means ± standard deviation (s.d.).
Supplementary Material
Highlights.
We adapted RV-C15 clinical isolate to efficient growth in HeLa-E8 cells.
Serial passaging resulted in stronger cytopathic effects and increased virus binding to cells and progeny yields.
We identified two key mutations which increased virus binding (VP1 T125K) and replication (3A E41K), respectively.
The findings enabled large-scale cost-effective C15 production by infection and the testing of RV-C infectivity by plaque assay.
Acknowledgments
This work was supported by the following NIH grants: UM1 AI114271, U19 AI104317 and P01 HL070831.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. Journal of Medical Virology. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S, Brockman-Schneider R, Gern JE. Propagation of rhinovirus-C strains in human airway epithelial cells differentiated at air-liquid interface. Methods Mol Biol. 2015;1221:63–70. doi: 10.1007/978-1-4939-1571-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA. Modulation of lipid synthesis and trafficking pathways by picornaviruses. Curr Opin Virol. 2014;9:19–23. doi: 10.1016/j.coviro.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, Le Souef PN. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggett SB, Gern JE. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brebion A, Mirand A, Regagnon C, Archimbaud C, Chambon M, Peigue-Lafeuille H, Henquell C. Evaluation of real-time RT-PCR Rhino&EV/Cc r-gene((R)) (bioMerieux) kit versions 1 and 2 for rhinovirus detection. J Clin Virol. 2015;62:110–113. doi: 10.1016/j.jcv.2014.10.019. [DOI] [PubMed] [Google Scholar]
- Calvo C, Casas I, Garcia-Garcia ML, Pozo F, Reyes N, Cruz N, Garcia-Cuenllas L, Perez-Brena P. Role of rhinovirus C respiratory infections in sick and healthy children in Spain. Pediatr Infect Dis J. 2010;29:717–720. doi: 10.1097/INF.0b013e3181d7a708. [DOI] [PubMed] [Google Scholar]
- Conant RM, Hamparian VV. Rhinoviruses: basis for a numbering system. 1 HeLa cells for propagation and serologic procedures. J Immunol. 1968;100:107–113. [PubMed] [Google Scholar]
- Cox DW, Bizzintino J, Ferrari G, Khoo SK, Zhang G, Whelan S, Lee WM, Bochkov YA, Geelhoed GC, Goldblatt J, Gern JE, Laing IA, Le Souef PN. Human rhinovirus species C infection in young children with acute wheeze is associated with increased acute respiratory hospital admissions. Am J Respir Crit Care Med. 2013;188:1358–1364. doi: 10.1164/rccm.201303-0498OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorobantu CM, Ford-Siltz LA, Sittig SP, Lanke KH, Belov GA, van Kuppeveld FJ, van der Schaar HM. GBF1- and ACBD3-independent recruitment of PI4KIIIß to replication sites by rhinovirus 3A proteins. J Virol. 2015;89:1913–1918. doi: 10.1128/JVI.02830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorobantu CM, Albulescu L, Lyoo H, van Kampen M, De Francesco R, Lohmann V, Harak C, van der Schaar HM, Strating JRPM, Gorbalenya AE, van Kuppeveld FJM. Mutations in Encephalomyocarditis Virus 3A Protein Uncouple the Dependency of Genome Replication on Host Factors Phosphatidylinositol 4-Kinase III+¦ and Oxysterol-Binding Protein. mSphere. 2016:1. doi: 10.1128/mSphere.00068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale SB, Alcazar M, Wilson T, Smith M, Zuckerman M, Lauinger IL, Tong CY, Broughton S, Rafferty GF, Johnston SL, Greenough A. Respiratory outcome of prematurely born infants following human rhinovirus A and C infections. Eur J Pediatr. 2014;173:913–919. doi: 10.1007/s00431-014-2262-1. [DOI] [PubMed] [Google Scholar]
- Fawkner-Corbett DW, Khoo SK, Duarte MC, Bezerra P, Bochkov YA, Gern JE, Le Souef PN, McNamara PS, Rose K, Fonceca AM, Hopkins M, Britto M, Cuevas LE, Correia JB. Rhinovirus-C detection in children presenting with acute respiratory infection to hospital in Brazil. Journal of Medical Virology. 2015 doi: 10.1002/jmv.24300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs TF, Bochkov YA, Nakagome K, Palmenberg AC, Gern JE. Production, purification, and capsid stability of rhinovirus C types. J Virol Methods. 2015;217:18–23. doi: 10.1016/j.jviromet.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR, Racaniello VR. Changes in rhinovirus protein 2C allow efficient replication in mouse cells. J Virol. 2003;77:4773–4780. doi: 10.1128/JVI.77.8.4773-4780.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR, Racaniello VR. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J Virol. 2005;79:5363–5373. doi: 10.1128/JVI.79.9.5363-5373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG. Rhinovirus and the lower respiratory tract. Rev Med Virol. 2004;14:17–31. doi: 10.1002/rmv.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz BA, Vance LM. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J Virol. 1995;69:4189–4197. doi: 10.1128/jvi.69.7.4189-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. The nucleotide sequence of human rhinovirus 1B: molecular relationships within the rhinovirus genus. J Gen Virol. 1988;69( Pt 1):49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- Israelsson S, Gullberg M, Jonsson N, Roivainen M, Edman K, Lindberg AM. Studies of Echovirus 5 interactions with the cell surface: heparan sulfate mediates attachment to the host cell. Virus Res. 2010;151:170–176. doi: 10.1016/j.virusres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Khan AG, Pichler J, Rosemann A, Blaas D. Human rhinovirus type 54 infection via heparan sulfate is less efficient and strictly dependent on low endosomal pH. J Virol. 2007;81:4625–4632. doi: 10.1128/JVI.02160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SS, Smith TJ, Chapman MS, Rossmann MC, Pevear DC, Dutko FJ, Felock PJ, Diana GD, McKinlay MA. Crystal structure of human rhinovirus serotype 1A (HRV1A) J Mol Biol. 1989;210:91–111. doi: 10.1016/0022-2836(89)90293-3. [DOI] [PubMed] [Google Scholar]
- Klimstra WB, Ryman KD, Johnston RE. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, Dean A, St George K, Briese T, Lipkin WI. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Lemanske RF, Jr, Evans MD, Vang F, Pappas T, Gangnon R, Jackson DJ, Gern JE. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Wang W. Human rhinovirus type 16: mutant V1210A requires capsid-binding drug for assembly of pentamers to form virions during morphogenesis. J Virol. 2003;77:6235–6244. doi: 10.1128/JVI.77.11.6235-6244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WM, Wang W, Rueckert RR. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes. 1995;9:177–181. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- Liu Y, Hill MG, Klose T, Chen Z, Watters K, Bochkov YA, Jiang W, Palmenberg AC, Rossmann MG. Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc Natl Acad Sci USA. 2016;113:8997–9002. doi: 10.1073/pnas.1606595113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sheng J, Baggen J, Meng G, Xiao C, Thibaut HJ, van Kuppeveld FJ, Rossmann MG. Sialic acid-dependent cell entry of human enterovirus D68. Nat Commun. 2015;6:8865. doi: 10.1038/ncomms9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre CL, Knowles NJ, Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J Gen Virol. 2013;94:1791–1806. doi: 10.1099/vir.0.053686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeish NJ, Witteveldt J, Clasper L, McIntyre C, McWilliam Leitch EC, Hardie A, Bennett S, Gunson R, Carman WF, Feeney SA, Coyle PV, Vipond B, Muir P, Benschop K, Wolthers K, Waris M, Osterback R, Johannessen I, Templeton K, Harvala H, Simmonds P. Development and assay of RNA transcripts of enterovirus species A to D, rhinovirus species a to C, and human parechovirus: assessment of assay sensitivity and specificity of real-time screening and typing methods. J Clin Microbiol. 2012;50:2910–2917. doi: 10.1128/JCM.01172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousnier A, Swieboda D, Pinto A, Guedan A, Rogers AV, Walton R, Johnston SL, Solari R. Human rhinovirus 16 causes Golgi apparatus fragmentation without blocking protein secretion. J Virol. 2014;88:11671–11685. doi: 10.1128/JVI.01170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, Gern JE. Effects of rhinovirus species on viral replication and cytokine production. J Allergy Clin Immunol. 2014;134:332–341. doi: 10.1016/j.jaci.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AL, Racaniello VR. Selection of rhinovirus 1A variants adapted for growth in mouse lung epithelial cells. Virology. 2011;420:82–88. doi: 10.1016/j.virol.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl A, Reithmayer M, Winsauer G, Moser R, Gosler I, Blaas D. Viral evolution toward change in receptor usage: adaptation of a major group human rhinovirus to grow in ICAM-1-negative cells. J Virol. 2001;75:9312–9319. doi: 10.1128/JVI.75.19.9312-9319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa-Carvalho D, Rieder E, Baxt B, Rodarte R, Tanuri A, Mason PW. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J Virol. 1997;71:5115–5123. doi: 10.1128/jvi.71.7.5115-5123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler M, Yerly S, Vieille G, Docquier M, Turin L, Kaiser L, Tapparel C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J Clin Microbiol. 2012;50:2868–2872. doi: 10.1128/JCM.06752-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth MK, Grimm E, Frindt P, Galagan DM, Konno SI, Love R, Gern JE. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol. 1999;20:1220–1228. doi: 10.1165/ajrcmb.20.6.3261. [DOI] [PubMed] [Google Scholar]
- Sherry B, Rueckert R. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J Virol. 1985;53:137–143. doi: 10.1128/jvi.53.1.137-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010;91:2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JM, Waarts BL, Kimata K, Klimstra WB, Bittman R, Wilschut J. Adaptation of alphaviruses to heparan sulfate: interaction of Sindbis and Semliki forest viruses with liposomes containing lipid-conjugated heparin. J Virol. 2002;76:10128–10137. doi: 10.1128/JVI.76.20.10128-10137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway G, Hughes PJ, Mountford RC, Minor PD, Almond JW. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss DM, Glustrom LW, Wuttke DS. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J Mol Biol. 2003;330:225–234. doi: 10.1016/s0022-2836(03)00577-1. [DOI] [PubMed] [Google Scholar]
- Tan CW, Poh CL, Sam IC, Chan YF. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol. 2013;87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden L, Wolthers KC, van Kuppeveld FJ. Replication and Inhibitors of Enteroviruses and Parechoviruses. Viruses. 2015;7:4529–4562. doi: 10.3390/v7082832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Purstinger G, de VE, de Haan CA, Neyts J, van Kuppeveld FJ. Coxsackievirus mutants that can bypass host factor PI4KIIIbeta and the need for high levels of PI4P lipids for replication. Cell Res. 2012;22:1576–1592. doi: 10.1038/cr.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasak M, Goesler I, Blaas D. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J Virol. 2005;79:5963–5970. doi: 10.1128/JVI.79.10.5963-5970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lee WM, Mosser AG, Rueckert RR. WIN 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J Virol. 1998;72:1210–1218. doi: 10.1128/jvi.72.2.1210-1218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E, Duijsings D, Lanke KH, van Dooren SH, Jackson CL, Melchers WJ, van Kuppeveld FJ. Effects of picornavirus 3A Proteins on Protein Transport and GBF1-dependent COP-I recruitment. J Virol. 2006;80:11852–11860. doi: 10.1128/JVI.01225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin FH, Lomax NB. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J Virol. 1983;48:410–418. doi: 10.1128/jvi.48.2.410-418.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zautner AE, Korner U, Henke A, Badorff C, Schmidtke M. Heparan sulfates and coxsackievirus-adenovirus receptor: each one mediates coxsackievirus B3 PD infection. J Virol. 2003;77:10071–10077. doi: 10.1128/JVI.77.18.10071-10077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.