Abstract
Tumor-associated neutrophils are increasingly recognized for their ability to promote tumor progression, mediate resistance to therapy, and regulate immunosuppression. Evidence from various murine models has shown that the chemokine receptor CXCR2 attracts neutrophil into tumors and, therefore, represents a tractable therapeutic target. Here, we report prominent expression of a neutrophil gene signature in a subset of human pancreatic adenocarcinoma (PDA). CXCL5 was the most prominently expressed CXCR2 ligand in human PDA and its expression was higher in PDA than in any other common tumor represented in The Cancer Genome Atlas. Using a genetically engineered mouse model of PDA, we found that tumor and stromal cells differentially expressed CXCR2 ligands, with Cxcl5 high in tumor and Cxcl2 high in stroma. Cxcl5 expression was associated with mutant Kras expression and regulated by NF-κB activation. Host CXCR2 inhibition by genetic ablation prevented neutrophil accumulation in pancreatic tumors and led to a T cell–dependent suppression of tumor growth. In the absence of neutrophils, activated and functional T cells infiltrated pancreatic tumors otherwise devoid of effector T cells. Thus, the CXCR2-ligand axis helps establish an immunosuppressive microenvironment in PDA, highlighting the potential utility of targeting this axis as a novel therapy for this deadly disease.
INTRODUCTION
Despite advances using combination chemotherapy, pancreatic adenocarcinoma (PDA) remains one of the most lethal malignancies, with a dismal 5-year survival rate of ~7.7% (http://seer.cancer.gov/statfacts/html/pancreas.html) (1,2). Even with surgery and adjuvant therapy in patients with resectable PDA, the 5-year survival rate is 20% (3). Alarmingly, PDA now kills more Americans than breast cancer and is predicted to become the second leading cause of cancer-related deaths in the United States by 2020 (4). Therefore, expanding our knowledge of mechanisms promoting PDA progression and resistance is urgently needed.
Avoiding host immunity is a key feature of tumor progression (5), and understanding the mechanisms by which cancer cells evade immune destruction is critical for the development of more effective treatment strategies. To describe the immune-tumor interactions in PDA, our lab previously characterized the immune infiltrate during progressive stages of PDA development using the KrasLSL-G12D/+;Trp53LSL-R172H/+;Pdx1-Cre (KPC) murine model (6,7). We found extensive infiltration of immunosuppressive Treg and myeloid cells in both pancreatic intraepithelial neoplasia (PanINs) and PDA. In contrast, infiltration of effector T cells was scant in the tumor microenvironment. These observations suggested that a highly immunosuppressive environment is established even at the earliest stages of tumor development. Although immune checkpoint blockade with anti–PD-1/PD-L1 or anti–CTLA-4 is ineffective in stimulating antitumor immunity, depleting or “re-educating” immunosuppressive myeloid populations has proven to be more effective at eliciting antitumor T-cell responses in models of PDA (8–13).
Tumor-associated neutrophils (TANs) play important roles in cancer development, progression, and resistance to therapy (14–17). A meta-analysis of the literature concluded that TANs are typically pro-tumor and are strongly associated with poorer prognosis in the majority of human cancers (18). TANs often exhibit protumorigenic functions, including promotion of angiogenesis, metastases, and immunosuppression (19–23). However, TANs also can be antitumor in early-stage cancer or when TGFβ is inhibited (24,25). Thus, whether TANs are pro- or antitumor depends in part on the specific cancer type and the stage of the tumor. In the context of PDA, the abundance of TANs is strongly associated with poorer prognosis (26,27). High intratumoral CXCL5, a chemokine for neutrophils, have also been associated with poorer overall survival (28). In the KPC model, systemic depletion of GR1+ myeloid cells, which includes neutrophils, can increase infiltration of effector T cells and inhibit tumor growth (13,29). Therefore, targeting neutrophils may be therapeutic in PDA.
CXCR2 ligands are essential for neutrophil egression from the bone marrow and trafficking toward sites of inflammation (30,31). CXCR2 is also essential for the recruitment of TANs in various cancers (32,33). In the KPC model, CXCR2 blockade by genetic ablation or pharmacologic inhibition reduces the recruitment of MPO+ neutrophils to the PDA tumor microenvironment and potently suppresses metastases (19). CXCR2 inhibition sensitized PDA tumors that were otherwise highly resistant to anti–PD-1D-1 therapy. Here, using both human and mouse data, we build on the above results by detailing the mechanisms involved in CXCR2-ligand activation in PDA and discerning the impact on T-cell responses in the setting of CXCR2 disruption. We show that a subset of human PDA have significant elevation of TAN-related genes. Analysis of gene expression in human PDA revealed a correlation between high expression of CXCR2 ligands and enrichment of neutrophils and NF-κB related pathways. We further showed that the KPC model faithfully recapitulates human disease in the expression profile of CXCR2 ligands, with CXCL5 being the most prominent in both. Using a KPC-derived PDA cell line, we discovered that NF-κB activation can potently induce CXCL5 expression and secretion. We found that host CXCR2 ablation dramatically inhibited TAN accumulation and resulted in a spontaneous, T cell–dependent suppression of tumor growth. Therefore, our data support the hypothesis that the CXCR2-ligand axis is a promising therapeutic target in PDA.
MATERIALS AND METHODS
Analysis of TCGA RNA-seq Data
Normalized RSEM counts (rsem.genes.normalized_results) from primary tumor samples were downloaded from the NIH TCGA Research Network through the GDAC data portal (http://gdac.broadinstitute.org/) and included all available Illumina HiSeq 2000 Level 3 gene-level data as of January 28, 2016. All samples marked as normal tissues were excluded. The following 15 cancer types (TCGA project, n = sample size) were included in our analysis: hormone receptor positive breast cancer (BRCA, n = 823), colorectal adenocarcinoma (COADREAD, n = 379), esophageal carcinoma (ESCA, n = 184), glioblastoma (GBM, n = 153), head and neck squamous cell carcinoma (HNSC, n = 520), kidney renal clear cell carcinoma (KIRC, n = 533), kidney renal papillary carcinoma (KIRP, n = 290), liver hepatocellular carcinoma (LIHC, n = 371), lung adenocarcinoma (LUAD, n = 515), lung squamous carcinoma (LUSC, n = 501), pancreatic ductal adenocarcinoma (PAAD, n = 134), prostate adenocarcinoma (PRAD, n = 497), colorectal adenocarcinoma (COADREAD, n = 382), liver hepatocellular carcinoma (LIHC, n = 373), lung adenocarcinoma (LUAD, n = 517), lung squamous cell carcinoma (LUSC, n = 501), skin cutaneous melanoma (SKCM, n = 103), stomach adenocarcinoma (STAD, n = 415), and thyroid carcinoma (THCA, n = 501). Genes in which less than 30% of samples have a normalized RSEM count of more than 1 were filtered out from subsequent analysis. The normalized RSEM values were then log-transformed using the formula: log2(RSEM values+1).
The neutrophil gene signature was calculated as the log-average of normalized RSEM expression of 31 genes that constitute a previously defined neutrophil-signature (34). To compare expression of CXCL5 and neutrophil-signature across the 15 different cancer cohorts, the log2-normalized RSEM values were transformed to z-scores using the formula: z-score = (X − average(X)) / stdev(X), where X represents the RSEM values. The average and standard deviation were calculated from the expression of X across all samples included in this study.
Unsupervised hierarchal clustering was used to cluster the 134 PDA samples based on the expression of the neutrophil-signature genes or CXCR2 ligands. PDA subtype classifier and canonical gene sets were acquired from the indicated literature and Broad Institute’s Molecular Signature Database (34–37). The “GSVA” package available in R/Bioconductor was used to calculate GSVA signature scores for each gene set (rnaseq=T, mx.diff=T). Differential expression of GSVA signature scores between cluster groups was calculated using the Holm-Sidak multiple comparison test. Adjusted values of ≤ 0.05 were considered significant.
Histological Analysis of Human PDA
Hematoxylin and eosin stained slides from the resected tumors of 12 patients with previously untreated, resectable PDA were prepared per routine at the Department of Pathology, Hospital of the University of Pennsylvania. The percent of cancer epithelium, stroma, and lumen that have significant neutrophil involvement for each sample were then recorded. These studies were approved by the University of Pennsylvania Institutional Review Board.
Mouse PDA Models
All mouse protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. Animals were maintained in a specific pathogen-free facility. KrasLSL-G12D/+;Trp53LSL-R172H/+;RosaLSL-YFP;Pdx1-Cre (KPCY) and KrasLSL-G12D/+;Trp53LSL-R172H/+;Pdx1-Cre (KPC) mice were bred in-house and backcrossed for over ten generations with C57BL/6J mice (6,38). All of these mice were confirmed on the C57BL/6 background at the DartMouse™ Speed Congenic Core Facility at the Giesel School of Medicine at Dartmouth College (11). Four- to six- months old KPC/KPCY mice with palpable tumors or with tumors >100mm3 by ultrasound were used in this study, with their age-matched controls. Cxcr2−/− and Cxcr2+/+ littermates were bred in-house from backcrossed, syngeneic Cxcr2+/− mice purchased from the Jackson Laboratories. These mice were maintained on acidified water (pH ~ 3–4) to minimize opportunistic infections. Genotypes were determined by Transnetyx. C57BL/6J wild-type mice were purchased from Jackson Laboratories. Six to twelve week old mice were used for tumor implantation studies.
Mouse PDA Cell Lines
All murine PDA cell lines, including 4662, were derived from primary pancreatic tumors of KPC mice. Briefly, dissociated cells from primary tumor were plated in a 6-well dish with serum-free DMEM to select for tumor cells. After two weeks, the cells were expanded in DMEM with 10% Fetal Bovine Serum (FBS). Only cells of ten or fewer passages were used for experiments. The pancreatic-lineage origin of all PDA cell lines were validated by PCR for the presence of rearranged KrasLSL-G12D allele (39). Cell lines were tested and authenticated using IMPACT and RADIL. Cell lines used for implantation studies were also tested and confirmed to be Mycoplasma and endotoxin free. All cell lines were maintained at 37°C and 5% CO2 in complete media (cDMEM), which contains Dulbecco’s Modified Eagle Medium (DMEM-GlutaMAX™) (Thermo Scientific) supplemented with 10% fetal bovine serum, 1% l-glutamine, and 50 µg/mL gentamicin (Gibco).
In vitro treatment of 4662 PDA cell line with inhibitors and siRNA
4662 PDA cells were seeded at a density of 1 × 105 cells/mL onto 6-well plates in cDMEM for 24 hours in triplicates. After washing with DMEM, cells were incubated for 24 h with DMEM containing DMSO control, 10 ng/mL mTNFα (R&D), or 10µM U0126 (MEK1/2-i, Cell Signal Technologies), each with or without 20 µM Wedelolactone (IKK1/2-i, Sigma-Aldrich). After 24 h, the supernatant was collected and frozen at −20°C until further analysis. Qiagen’s RNeasy-Plus Mini Kit was used to isolate total RNA from pelleted cells. RNA quality was assessed using a NanoDrop ND-1000. Only samples with a 260/280 value of ≥1.9 were used. First strand cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primer probes for 18S (Applied Biosystems), Csf2 (GM-CSF), Kras, Yap1, Cxcl1, Cxcl2, Cxcl3, Cxcl5, Cxcl7 (PPBP), and Cxcr2 (IDT) were used for qPCR in a ViiA™ 7 Real-Time PCR System. Relative expression was determined after normalizing to 18S expression.
For siRNA inhibition of Yap1 and Kras, 4662 PDA cells were seeded as described above. After washing with DMEM, the cells were transfected with 10 nM of siRNA against Yap1 (Sigma-Aldrich, NM_009534), Kras (Life Technologies, S68935), and a Td-Tomato fluorescent negative control (Sigma-Aldrich, SIC005) using the Lipofectamine® RNAiMAX Transfection Reagent (Life Technologies). After 48 hours of incubation, the supernatant and RNA were collected and analyzed as described above.
In vivo subcutaneous tumor implantation studies
PDA cells were cultured and harvested at 80–90% confluence. After assessing for > 90% viability via hemocytometer with Trypan blue staining, 5 × 105 cells were injected subcutaneously into the right flank. Tumor volume was monitored using digital calipers and calculated as (l × w2)/2, where l is the longest dimension and w is the perpendicular dimension. Mice were euthanized if tumor volume reached > 1 cm3. For T-cell depletion studies, 200 µg of anti-CD4 (BioXCell, GK1.5l) and 200 µg of anti-CD8 (BioXCell, YTS 169.4), or 400 µg of isotype control (BioXCell, LTF-2) were administered intraperitoneally every 3 days for the duration of the study, starting 4 days prior to tumor implantation, an approach validated as previously published (39).
Processing of plasma, tumor supernatant, and single-cell suspension from tissues
Mice were euthanized in a CO2 chamber. Whole blood from cardiac puncture was collected in EDTA-containing Eppendorf tubes. The tubes were centrifuged at 12,000 × g for 10 mins. The resulting supernatants were centrifuged at 14,000 × g for 15 mins to remove remaining debris. Plasma samples were stored at −20°C until further use. Pancreata or subcutaneous tumors were dissected and rinsed with RPMI. To prepare samples for flow cytometric analysis or FACS, tissues were minced with fine scissors (≥200 cuts) and incubated in collagenase IV solution (1 mg/ml in RPMI) for 30–45 minutes at 37°C. The solution was then placed on ice and a 1:4 dilution with ice cold RPMI + 10%FBS was added to stop the reaction. Dissociated cells were passed through a 70µm cell strainer and pelleted. The supernatant was collected and frozen until further use. The pelleted cells were washed with FACS Buffer (PBS + 0.5% BSA + 0.5 mM EDTA). After centrifugation, the cells were resuspended in FACS Buffer, filtered through another 70 µm cell strainer, and kept on ice.
Histopathology and Immunofluorescence of tumors
Fresh pancreatic or subcutaneous tumors were rinsed in PBS and placed in Zinc Formalin Fixative (Sigma-Aldrich) overnight. Then they were dehydrated serially in 70%, 95%, and 100% ethanol. The tissues were then embedded in paraffin, which were processed to generate H&E and unstained sections. To rehydrate for immunofluorescence, unstained sections were serially submerged in Xylene (Sigma), 100% ethanol, 95% ethanol, 70% ethanol, and PBS. Sections were then blocked with 10% donkey serum + 0.1% TritonX-100 in PBS overnight at 4°C; followed by incubation with a goat anti-GFP antibody (Abcam, ab6673; 1:200) and a rat anti-mouse Ly6G antibody (BioXCell, BE0075-1; 1:200) for 1 hour at room temperature. After washing, the sections were incubated with Alexa Fluor 488 donkey anti-goat IgG (Invitrogen, A-11055; 1:200), Alexa Fluor 488 or Alexa Fluor 594 donkey anti-rat IgG (Invitrogen, A-21208 or A-21209; 1:200), and DAPI (Biolegend; 1:1000) for 1 hour at RT in the dark. Sections were then washed and mounted with Aqua-Poly/Mount (Polysciences). A Nikon Eclipse Ti-U fluorescent microscope was used to acquire images at a 64-bit data depth.
Measurement of chemokine concentrations in plasma, tumor and cell culture supernatant
Plasma, tumor and cell culture supernatant CXCL1 and CXCL5 protein levels were measured using the LEGENDplex™ Mouse Proinflammatory Chemokine Panel (13-plex) kit following manufacturer’s protocol. Bead identity and intensity were measured with a FACSCanto and subsequently analyzed with FlowJo.
FACS Sorting and Transcriptional analysis
YFP+ and YFP− populations from KPCY pancreata were sorted using a FACSAria II Flow Cytometer directly into TRIzol Reagent (Invitrogen). Total RNA was isolated following recommendations from the manufacturer. Briefly, cells were homogenized in TRIzol reagent. Chloroform (0.2ml per 1ml of TRIzol Reagent) was then added, vigorously mixed, and incubated for 2–3 mins. After centrifugation at 12,000 × g for 15 mins at 4°C, the aqueous phase was isolated. Glycogen was added to the RNA solution to aid subsequent visualization of RNA pellet. Isopropanol was added to precipitate RNA, which was followed by washing with ethanol. The RNA pellet was resuspended in DNase/RNase-free water. RNA quality was assessed using a NanoDrop ND-1000. First strand cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primer probes for 18S (Applied Biosystems), Cxcl1, Cxcl2, Cxcl3, Cxcl5, Cxcl7, and Cxcr2 (IDT) were used for qPCR in a ViiA™ 7 Real-Time PCR System. Relative expression was determined after normalizing to 18S expression.
Flow Cytometry
Between 5 × 106 and 107 cells were plated per well in a 96 well plate. For ex vivo stimulation, cells were incubated with the Leukocyte Activation Cocktail, with BD GolgiPlug™ (1:500) in RPMI for 5 hours at 37°C before subsequent staining. Viability was assessed using Live/Dead Fixable Aqua Dead Cell Stain Kit. Cells were then washed and stained with labeled antibodies (1:100) against surface markers at 4°C for 30 minutes in FACS Buffer. For intracellular stains, cells were fixed, permeabilized, and stained with labeled intracellular antibodies using the Transcription Factor Staining Buffer Set (eBioscience). After washing, labeled cells were analyzed using a FACSCanto or a LSR II flow cytometer (BD Biosciences). The FlowJo software was used to perform subsequent gating and quantification of cell populations. The gating strategies from several recent publications was used to define immune cell populations (12,15).
Cell labeling was performed with the following fluorescently conjugated mAbs directed against mouse CD31 (MEC 13.3), CD45 (30-F11), CD3ε (145-2C11), CD4 (RM4-5), CD8a (53-6.7), CD44 (IM7), CD62L (MEL-14), CD11a (M17/4), CD11c (N418), CD19 (6D5), F4/80 (BM8), CD11b (M1/70), Ly6C (HK1.4), Ly6G (1A8), IFNγ (XMG1.2), TNFα (MP6-XT22), IL10 (JESS-16E3), IL2 (JES6-5H4), IL17 (eBio17B7), and FOXP3 (FJK-16s) – all acquired from Biolegend, eBiosciences, or BD Biosciences.
Statistical Analysis
Significance in variations between two groups was determined by two-tailed, unpaired t-test. Significance in variations between two groups for many factors was calculated using the Holm-Sidak multiple comparison test. Differences among three (or more) related groups for one factor were analyzed by one-way ANOVA, followed by Tukey’s multiple comparison test. Differences in tumor growth curves and immune populations were analyzed by two-way ANOVA, followed by Dunnett’s multiple comparison test. Statistical analyses were performed using GraphPad Prism (GraphPad). A P value ≤ 0.05 was considered statistically significant.
RESULTS
A subset of human PDA has significant TAN involvement
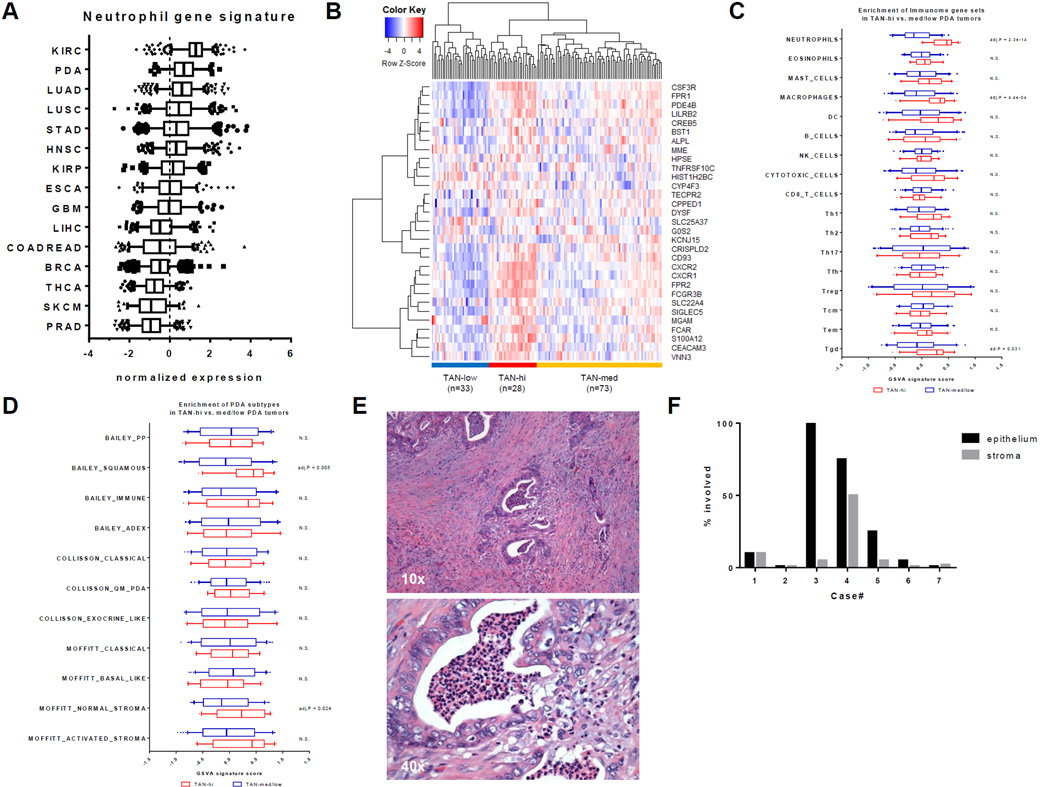
To investigate the extent of neutrophil infiltration in human PDA, we compared the normalized expression of a previously defined neutrophil gene signature in 134 resected PDA compared to 14 other primary cancer cohorts using RNA-sequencing data available from The Cancer Genome Atlas (TCGA) (34,40). PDA ranked second, on average, among these 15 cancers in the expression of the neutrophil gene signature (Fig. 1A). Thus, TANs may be relatively abundant in PDA. The PDA cohort naturally clusters into a TAN-high, a TAN-medium, and a TAN-low group (Fig. 1B). To identify immune populations co-enriched in TAN-high tumors, gene set variation analysis (GSVA) was performed using previously defined “immunome” gene sets that specifically identify immune populations (34). Confirming our cluster definition, the neutrophil-specific gene set was significantly enriched in the TAN-high group (Fig. 1C). Of the 16 other immune populations tested, the macrophage and γδ T-cell gene sets were also significantly enriched in TAN-high compared to TAN-medium and TAN-low tumors (Fig. 1C).
Figure 1. A subset of human PDA have significant TAN involvement.
(A) Normalized expression (z-scores) of the neutrophil gene signature in primary tumors across 15 TCGA cancer cohorts. Boxplot whiskers at 5–95th percentile. Dashed line represents the average expression value. (B) Clustering of 134 human TCGA PDA samples using the 31 genes in the neutrophil signature into TAN-high, TAN-med, and TAN-low groups. (C) Comparison of GSVA signature scores for 17 different immune cell types between TAN-high and TAN-med/low groups. Holm-Sidak multiple comparison test; N.S. = Not Significant. (D) Comparison of GSVA signature scores for PDA subtypes between TAN-high and TAN-med/low groups. Holm-Sidak multiple comparison test; N.S. = Not Significant. (E) H&E stain of a representative, resected human PDA sample (n = 12) showing TAN involvement in the cancer epithelium, stroma, and lumen. (F) Bar graph of the percentage of cancer epithelium or stroma involved in each of the 7 PDA cases with TAN infiltration.
A number of landmark studies have delineated several subtypes of PDA based on genomic and transcriptomic data (35,36,41). Four main subsets of PDA were identified by Bailey et al.: squamous, aberrantly differentiated endocrine exocrine (ADEX), pancreatic progenitor (PP), and immunogenic (Immune) (35). Squamous, ADEX, and PP subtypes are most similar to Collisson et al.’s quasimesenchymal (QM-PDA), exocrine-linked, and classical subtypes, respectively (35,36). PDA tumors could be further divided based on whether they have normal or activated stroma (41). To explore whether neutrophil infiltration is associated with certain PDA subtypes, we used GSVA to compare the enrichment of previously defined PDA subtype gene sets in TAN-high and TAN-medium/low tumors (35,36,41). Our analysis showed that TAN-high tumors have significant enrichment of genes in the normal stroma subtype and the squamous subtype, which has the poorest prognosis (Fig. 1D). However, high TAN involvement was not significantly associated with the immunogenic subtype, which is characterized by infiltration of adaptive immune cells.
To understand the spatial distribution of TANs in human PDA, a cohort of PDA resected tumors from previously untreated patients (n = 12) were examined by standard pathologic examination, in which neutrophils are readily identified on H&E staining. Eight of 12 tumors exhibited a mild to extensive degree of neutrophil infiltration, with a few tumors having significantly more TANs (Table 1). Four tumors were devoid of neutrophils. One tumor had neutrophils only in neoplastic ductal lumens. In the remaining seven samples, neutrophils were also found within the cancer epithelium and stroma (Fig. 1F). Because none of the samples in this cohort was of the rare squamous subtype, comparisons of neutrophil infiltration between different PDA subtypes, as suggested by our analysis of TCGA data (Fig. 1D) could not be made on a pathological basis.
Table 1.
Percentage of cancer epithelium, stroma, and lumen with TAN involvement in a cohort of 12 patients with resectable PDA.
| TAN infiltration | count | percentage | |
|---|---|---|---|
| epithelium | 7/12 | 58.3% | |
| stroma | 7/12 | 58.3% | |
| lumen | 6/12 | 50.0% | |
| overall | 8/12 | 66.7% | |
CXCR2 ligand expression strongly associated with neutrophil and NF-κB pathway in human PDA
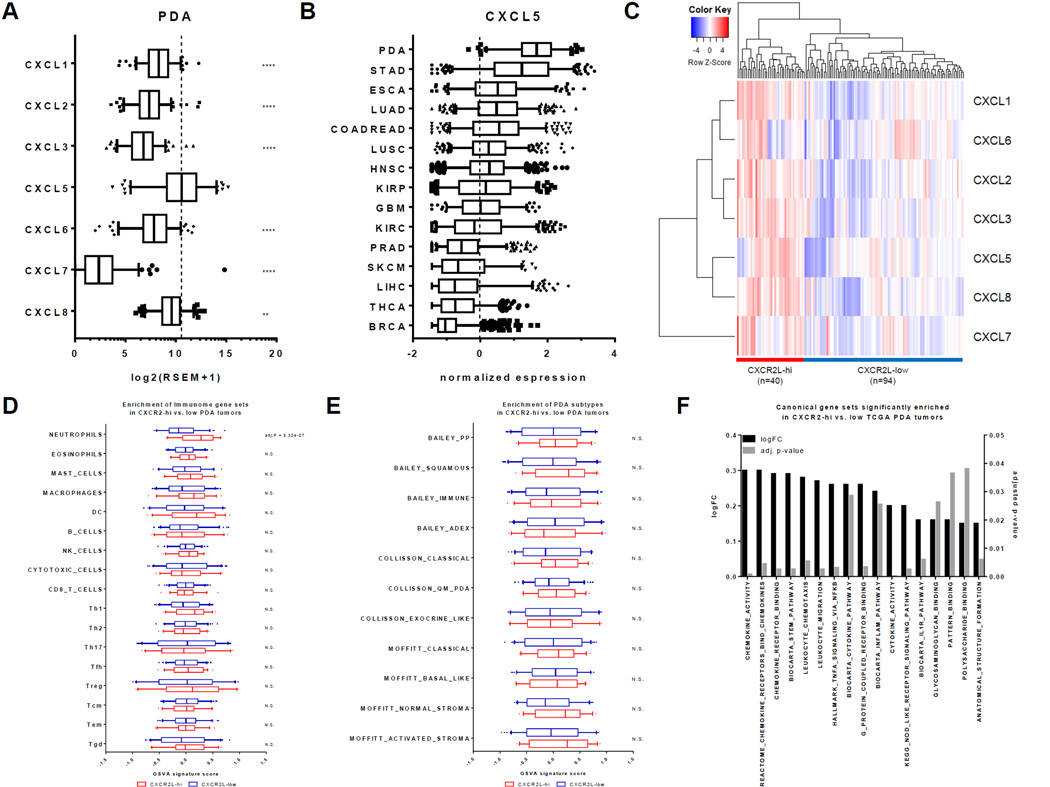
To investigate whether CXCR2 ligands are involved in pancreatic cancer, we compared the expression of CXCR2 ligands (CXCR2Ls) in 134 PDA samples from TCGA. The expression of CXCL5 was not only higher than the other CXCR2Ls, but it was also markedly elevated in PDA compared to the other tumor types (Figs. 2A–B). In general, tumors with higher CXCL5 expression had higher neutrophil gene signature expression, and vice versa (Figs. 1A and 2B). However, even though PDA had the highest CXCL5 expression, it was not the highest in neutrophil gene expression. This may be explained by the role of other CXCR2Ls besides CXCL5 in the recruitment of TANs in other cancers. Although CXCL5 expression was the highest among CXCR2Ls in human PDA, we also noted significant expression of other CXCR2Ls, especially CXCL8. Because these chemokines are likely redundant in their function, all CXCR2Ls were included in the subsequent analysis.
Figure 2. CXCR2 ligand expression is strongly associated with neutrophil and NF-κB pathway gene sets in human PDA.
(A) Distribution of RSEM expression for all CXCR2 ligands in 134 human TCGA PDA tumors. Boxplot whiskers at 5–95th percentile. Dashed line represents the average expression value of CXCL5. **, P ≤ 0.01; ****, P ≤ 0.001 (1-way ANOVA, Dunnett’s multiple comparison test against CXCL5). (B) Normalized expression (z-scores) of CXCL5 in primary tumors across 15 TCGA cancer cohorts. Boxplot whiskers at 5–95th percentile. Dashed line represents the average expression value. (C) Clustering of 134 human TCGA PDA samples using CXCR2L expressions into CXCR2L-high and CXCR2L-low groups. (D) Comparison of GSVA signature scores for 17 different immune cell types between CXCR2L-high and CXCR2L-low groups. Holm-Sidak multiple comparison test; N.S. = Not Significant. (E) Comparison of GSVA signature scores for PDA subtypes between CXCR2L-high and CXCR2L-low groups. Holm-Sidak multiple comparison test; N.S. = Not Significant. (F) Log fold-change of GSVA signature scores and the adjusted p-values of canonical gene sets that are significantly elevated in CXCR2L-high compared to CXCR2L-low groups.
Unsupervised hierarchal clustering divided the PDA cohort into a CXCR2L-high group and a CXCR2L-low group (Fig. 2C). To identify immune populations enriched in CXCR2L-high tumors, GSVA analysis was done using the “immunome” gene sets as above (34). Of the 17 immune populations tested, only the neutrophil gene set showed significant enrichment in CXCR2L-high compared to CXCR2L-low tumors (Fig. 2D). This result supported the hypothesis that CXCR2 ligands are specifically and selectively important for the recruitment of tumor-associated neutrophils (TANs), but not for other immune populations.
Because CXCR2L-high tumors should in theory overlap with TAN-high tumors, we hypothesized that CXCR2L-high tumors would be relatively more enriched in genes related to the squamous subtype. However, GSVA analysis of PDA subtypes showed that CXCR2L-high tumors are not significantly associated with any PDA subtypes, though there was a trend toward enrichment of the squamous subtype with an adjusted P value of 0.13 (Fig. 2E). Therefore, the CXCR2-ligand axis probably plays a role in PDA regardless of subtype.
In addition to immune populations and PDA subtypes, we were also interested in signaling pathways and biological processes that may be enriched in CXCR2L-high tumors. Of the 2,838 Hallmark, Canonical Pathways, and Gene Ontology gene sets that were curated by Broad Institute’s Molecular Signature Database, only 17 gene sets (0.6%) were significantly enriched in CXCR2L-high tumors (Fig. 2F) (37). Confirming our stratification of the PDA cohort into chemokine-high and low groups, the majority of these significant gene sets (8 of 17) involved leukocyte trafficking and chemokine/cytokine-receptor signaling. As may be expected of tumors enriched with neutrophils, pathways involved in innate immune functions, such as pattern recognition and glycosaminoglycan (GAG) binding, were also significantly enriched. Gene sets related to IL1R, NOD-like receptor (NLR), and TNFα signaling were also significantly enriched in CXCR2L-high tumors. Importantly, these inflammatory pathways all converged on NF-κB signaling downstream. These findings suggested that increased NF-κB signaling may be associated with elevated CXCR2 ligand expression in PDA.
KPC tumors have elevated neutrophil infiltration and Cxcl5 expression
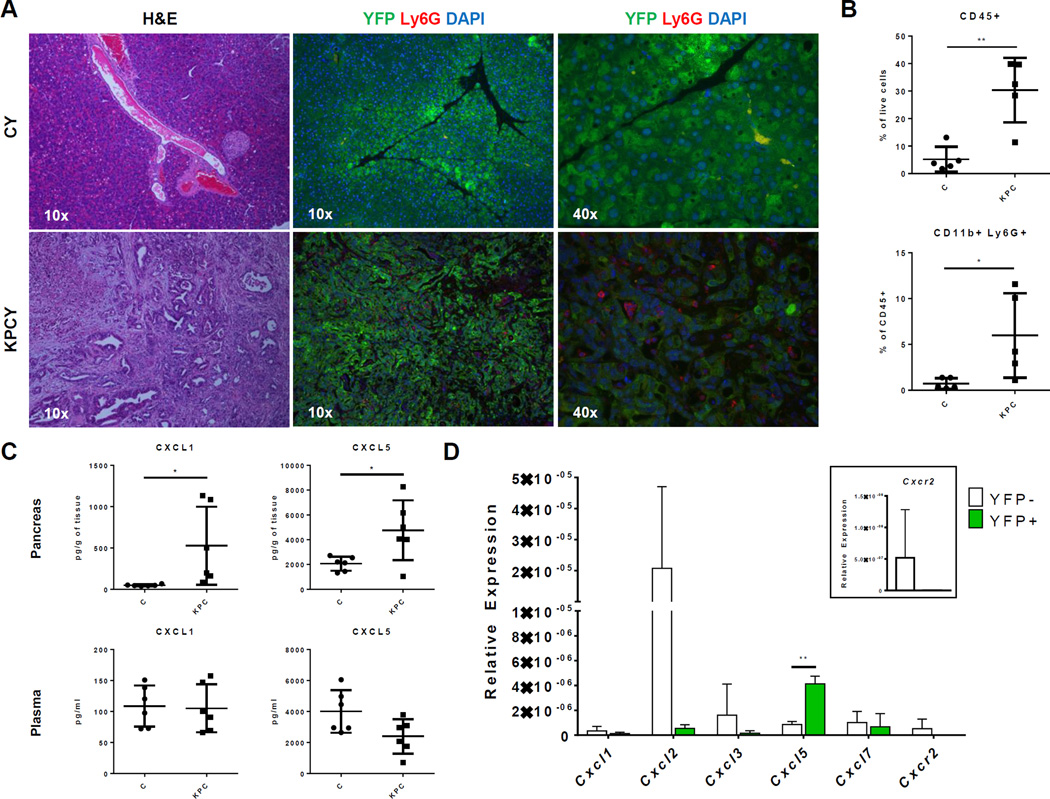
To further elucidate the role of TANs and the CXCR2-ligand axis in PDA, we then studied their involvement in murine models of PDA using KrasLSL-G12D/+;Trp53LSL-R172H/+;Pdx1-Cre (KPC) and KPC with the RosaLSL-YFP allele (KPCY) mice (38). Hematoxylin and eosin (H&E) staining of the pancreatic tumors of KPCY mice showed remarkable histological similarity with human PDA (compare Figs. 1B and 3A). Similar to human disease, immunofluorescent staining of murine neutrophils with an antibody to Ly6G showed extensive infiltration of neutrophils throughout the tumor microenvironment in most KPC tumors (Fig. 3A). In contrast, almost no staining was observed in the pancreata of age-matched Pdx1-Cre (C) control mice. Flow cytometric analysis further confirmed a significant accumulation of CD11b+Ly6G+ neutrophils in KPC tumors compared to control pancreas (Fig. 3B). Therefore, the murine KPC and KPCY models faithfully recapitulated TAN involvement observed in human PDA.
Figure 3. KPC tumors have elevated neutrophils infiltration and Cxcl5 expression.
A–D, An independent cohort of 4–6 months old, tumor-bearing KPC/KPCY and matched controls was used for each of the following figures. Each of these experiments was done only once. (A) Representative H&E (10×) and YFP-Ly6G-DAPI (10× and 40×) stains of slides from the pancreas of 4–6 months old tumor-bearing KPCY mice and their age-matched CY controls (n = 4 per group). (B) Flow cytometric analysis of CD45+ immune cells and CD11b+Ly6G+ neutrophils in the pancreas of tumor-bearing KPC mice (n = 5) compared to age-matched controls (n = 5). Graphs show mean ± s.d. of one experiment. *, P value ≤ 0.05; **, P value ≤ 0.01 (unpaired t-test). (C) Protein quantification of CXCL1 and CXCL5 in the pancreas and plasma of KPC (n = 6) compared to controls (n = 6) mice. Graphs show mean ± s.d. of one experiment. *, P value ≤ 0.05; (unpaired t-test). (D) CXCR2 ligand expression in YFP+ cancer cells compared to YFP− stromal cells in KPCY pancreatic tumors (n = 3). Inset shows Cxcr2 expression on a different scale. Gene expressions were normalized to 18S. Graphs show mean ± s.d. of one experiment. *, P value ≤ 0.05; **, P value ≤ 0.01 (unpaired t-test).
To investigate CXCR2 ligands in murine PDA, we quantified CXCL1 and CXCL5 protein in KPC tumors and normal pancreas (Fig. 3C). The results showed that both CXCL1 and CXCL5 were significantly elevated in pancreatic tumors compared to control pancreas. However, the plasma concentrations of both of these chemokines did not differ significantly between KPC and control mice. This implied steeper chemokine gradients for CXCL1 and CXCL5 toward the pancreas of tumor-bearing KPC compared to control mice. To further address which cell populations were responsible for CXCR2 ligand expression in PDA, we compared the gene expression of these chemokines in YFP+ pancreatic-lineage and YFP− stromal cells in tumor-bearing KPCY mice (Fig. 3D). Similar to human PDA, Cxcl5 was the most highly expressed CXCR2 ligand in YFP+ pancreatic cancer cells in KPCY mice (Fig. 3D). However, YFP− stromal cells in the tumor also expressed many CXCR2 ligands, particularly Cxcl2. Thus, both tumor and stromal cells were involved in the expression of CXCR2 ligands and the recruitment of TANs.
Although not statistically significant, Cxcr2 was expressed primarily by YFP− stromal cells and not by YFP+ pancreatic cancer cells (Fig. 3D, inset). Cxcr2 gene expression was below detectable limits in 8 of 8 KPC PDA cell lines (data not shown). In contrast, all of these cell lines highly expressed Cxcl5 (data not shown). Although both cancer and stromal cells contribute to expression of multiple CXCR2 ligands, expression of Cxcr2 receptor itself was primarily found in stromal populations rather than in cancer cells.
TNFα and Kras/MEK inhibition induce CXCL5 expression in a NF-κB dependent manner
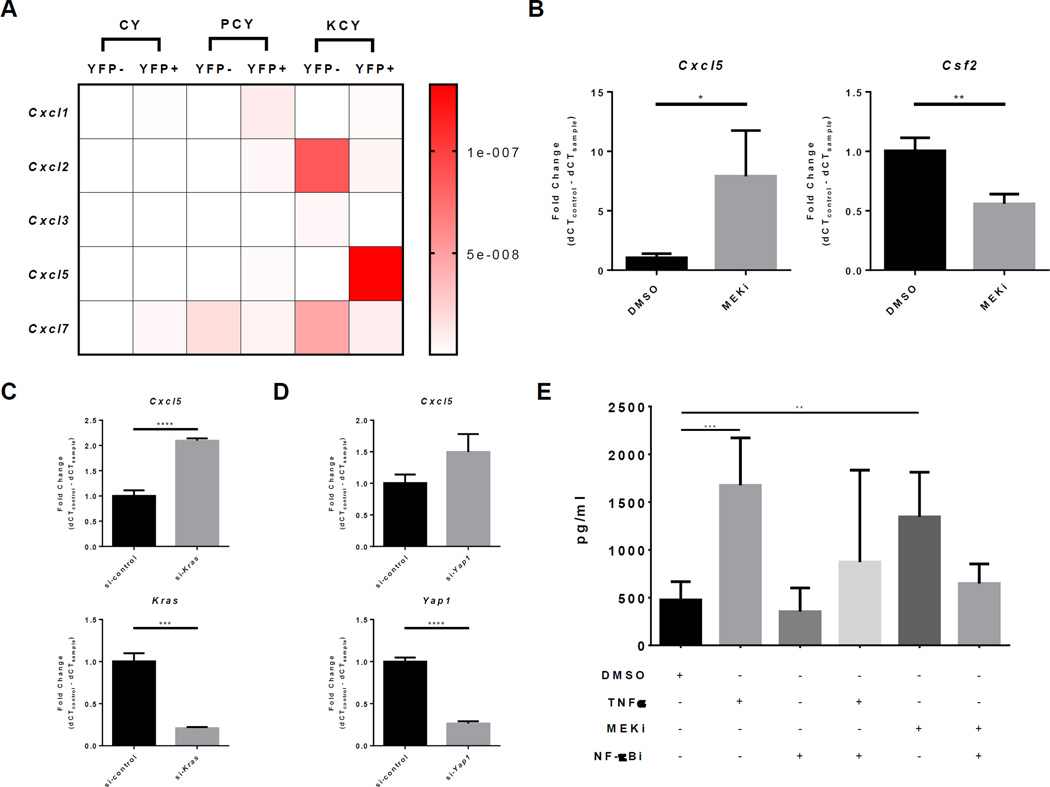
To explore the effects of mutant Kras and mutant Trp53 in regulating CXCR2 ligand expression, we compared the expression of all CXCR2 ligands in YFP+ pancreatic-lineage or YFP− stromal cells in 4–6 months old CY, PCY, and KCY mice (Fig. 4A). Increased expression of Cxcl5 in YFP+ tumor cells and Cxcl2 in YFP− stromal cells were associated with mutant Kras and not mutant Trp53 expression. This data led us to hypothesize that pathways directly downstream or indirectly induced by oncogenic Kras may be responsible for regulating CXCR2 ligand expression. We therefore compared the expression of Cxcl5 via RT-qPCR in 4662 KPC cells treated with U0126 (a MEK1/2 inhibitor) or DMSO. The 4662 cell line is a well-characterized KPC-derived PDA cell line that has been previously described (8,11,42). Inhibition of MEK resulted in a significant increase in Cxcl5 expression compared to control (Fig. 4B). In contrast, MEK inhibition led to a significant reduction in Csf2 (GM-CSF) expression in PDA, which confirmed previously published observations regarding GM-CSF regulation by oncogenic Kras (9). Cxcl5 expression was also significantly increased in Kras siRNA–treated 4662 cells compared to control (Fig. 4C). Thus, KRAS-MEK signaling is not directly responsible for Cxcl5 expression; rather, Cxcl5 expression was likely regulated by pathways that are activated in response to KRAS-inhibition in the setting of oncogenic Kras expression.
Figure 4. TNFα and KRAS/MEK inhibition induce CXCL5 expression in a NF-κB dependent manner.
(A) Heatmap of relative CXCR2 ligand expression by YFP+ pancreatic and YFP− stromal cells in 4–6 months old CY, PCY, and KCY mice (n = 3 per group). (B) Fold change of Cxcl5 and Csf2 (GM-CSF) expression in 4662 PDA cells treated with 10µM U0126 (MEK inhibitor) compared to DMSO. Graphs show mean ± s.d. of 3 independent experiments. *, P ≤ 0.05; **, P ≤ 0.01 (unpaired t-test). (C) Fold change of Cxcl5 and Kras expression in Kras siRNA–treated compared to control siRNA–treated 4662 PDA cells. Graph shows mean ± s.d. of 3 independent experiments. ***, P ≤ 0.001; ****, P ≤ 0.0001 (unpaired t-test). (D) Fold change of Cxcl5 and Yap1 expression in si-Yap1 treated compared to si-control treated 4662 PDA cells. Graph shows mean ± s.d. of 3 independent experiments. ****, P ≤ 0.0001 (unpaired t-test). (E) CXCL5 protein level in the supernatant of 4662 PDA cells treated with the indicated combinations of DMSO control, 10ng/mL TNFα, 10µM U0126 (MEK inhibitor), or 20µM Wedeloactone (NF-κB inhibitor). Graph shows mean ± s.d. of 3 independent experiments. *, P ≤ 0.05; **, P ≤ 0.01 (1-way ANOVA, Holm-Sidak’s multiple comparison test).
An elegant study using an inducible oncogenic Kras model of PDA has shown that YAP1 activation in PDA cancer cells allows escape from oncogenic Kras addiction (43). Recently, the same group showed that YAP1 directly regulates Cxcl5 expression in a murine model of prostate cancer (44). To test the hypothesis that YAP1 directly regulates Cxcl5 expression in PDA, we compared the expression of Cxcl5 in Yap1 siRNA– and control siRNA–treated 4662 cells (Fig. 4D). Unlike the observations in prostate cancer, knock down of Yap1 did not significantly alter Cxcl5 expression in PDA cells in our model.
From our analysis of TCGA PDA data, we noted that tumors with high CXCR2 ligand expression were significantly enriched in expression of genes associated with inflammatory signaling pathways which all converged on NF-κB signaling. Therefore, we hypothesize that NF-κB signaling may be regulating Cxcl5 expression in PDA cells. To test this hypothesis, we compared the amount of secreted CXCL5 in the supernatant of 4662 PDA cells treated with mouse TNFα, which is a potent inducer of NF- κB activity, and those treated with DMSO control (Fig. 4E). In support of our hypothesis, TNFα treatment significantly increased CXCL5 secretion. The TNFα-induced increase in CXCL5 was abrogated upon co-treatment with wedelolactone (NF-κB inhibitor), a selective inhibitor of IKKα/β that does not affect p38 MAPK or AKT activities. Importantly, NF-κB inhibition alone did not significantly alter baseline CXCL5 secretion. These results showed that NF-κB activity can potently induce CXCL5 secretion in PDA cells, but does not seem to affect baseline levels.
We then tested the hypothesis that NF-κB signaling in the setting of KRAS/MEK inhibition is responsible for the increased CXCL5 level. Indeed, NF-κB inhibition completely abrogated the increase in CXCL5 in the presence of MEK inhibition (Fig. 4E). Therefore, NF-κB signaling may be an important pathway induced in response to KRAS/MEK inhibition in PDA cancer cells.
CXCR2 ablation specifically prevents TAN accumulation and inhibits tumor growth
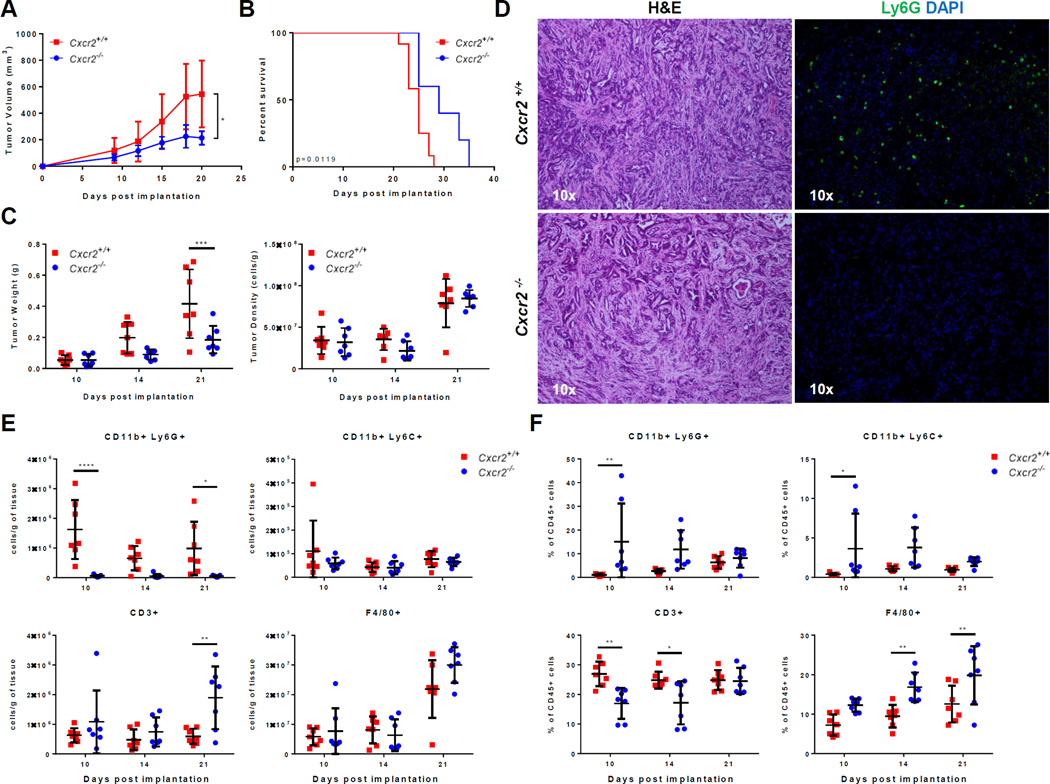
The expression of multiple CXCR2 ligands by different cell populations in the tumor microenvironment suggested significant redundancy in chemotactic signal to recruit TANs in PDA. This complexity made targeting individual CXCR2 ligands difficult and confounded. However, CXCR2 ligands all converged on their binding to CXCR2, which as noted above, was primarily expressed in the stromal population, consistent with previous reports (32,44). To study the role of stromal CXCR2 in PDA, we compared the subcutaneous growth of 4662 KPC tumor cells in syngeneic Cxcr2−/− or Cxcr2+/+ hosts. Tumor growth was significantly delayed in the absence of host Cxcr2 (Fig. 5A). A significant survival benefit was also observed in Cxcr2 knockouts (Fig. 5B). Differences in tumor weight between Cxcr2−/− and Cxcr2+/+ hosts only became significant two to three weeks post implantation (Fig. 5C). The lack of initial differences in growth kinetics and tumor weights showed that the 4662 PDA cell line was able to seed and establish tumors equally well in both Cxcr2−/− and Cxcr2+/+ hosts, suggesting that late factors were likely responsible for the observed differences in growth kinetics.
Figure 5. CXCR2 ablation specifically prevents TAN accumulation and inhibits tumor growth.
A–F, An independent cohort of Cxcr2−/− and Cxcr2+/+ mice was used for Figures A, C, and E–F, another cohort for Figure B, and another cohort for Figure D. Each of these experiments was done once unless otherwise indicated. (A) 4662 PDA tumor growth in Cxcr2−/− compared to Cxcr2+/+ littermates after subcutaneous implantation (n = 7 per group). Graph shows mean ± s.d. of one experiment. *, P ≤ 0.05 on Day 21 (2-way ANOVA, Dunnett’s multiple comparison test). The observed difference in tumor growth until Day 21 was observed in a second, independent experiment. (B) Kaplan-Meier survival analysis of Cxcr2−/− (n = 6) compared to Cxcr2+/+ (n = 10) littermates subcutaneously implanted with 4662 PDA tumors. (P value = 0.0119, log rank test). This result is representative of two independent experiments. (C) Comparison of tumor weights and cell-density in Cxcr2−/− compared to Cxcr2+/+ hosts on Day 10, 14, and 21 (n = 7 per group/day). Graph shows mean ± s.d. of one experiment. ***, P ≤ 0.001 (2-way ANOVA, Sidak’s multiple comparison test). (D) Representative H&E (10×) and Ly6G-DAPI (10×) stain in Cxcr2−/− compared to Cxcr2+/+ controls (n = 8 per group). (E) Flow cytometric measurement of the density of CD11b+Ly6G+ TANs, CD11b+Ly6C+ monocytes, CD3+ T cells, and F4/80+ macrophages in the tumors of Cxcr2−/− and Cxcr2+/+ littermates on Day 10, 14, and 21 (n = 7/day/group). Graph shows mean ± s.d. of one experiment. *, P ≤ 0.05; **, P ≤ 0.01; ***, P<0.001 (2-way ANOVA, Sidak’s multiple comparison test). The observed differences of TANs, monocytes, T cells, and macrophages on Day 21 were repeated in another independent experiment. (F) The percentage of CD11b+Ly6G+ TANs, CD11b+Ly6C+ monocytes, CD3+ T cells, and F4/80+ macrophages in the spleens of Cxcr2−/− and Cxcr2+/+ littermates on Day 10, 14, and 21 (n = 7/day/group). Graph shows mean ± s.d. of one experiment. *, P ≤ 0.05; **, P ≤ 0.01 (2-way ANOVA, Sidak’s multiple comparison test).
To understand if CXCR2 signaling was required for TAN accumulation in this PDA model, we compared the tumor-infiltrating immune populations in Cxcr2−/− and Cxcr2+/+ hosts. Histological analysis with H&E staining showed remarkable similarity between the subcutaneous model and the KPC/KPCY model of PDA (Figs. 3A and 5D). Immunofluorescent staining with anti-Ly6G on tumor sections showed diffuse tumor infiltration of neutrophils in Cxcr2+/+ hosts, similar to the pattern seen in human and KPCY PDA tumors. In contrast, TANs were almost completely absent in the tumors of Cxcr2−/− hosts (Fig. 5D). Using flow cytometry to compare tumor-infiltrating immune populations on days 10, 14, and 21 after implantation, we noted a striking reduction in the density of tumor-infiltrating CD11b+Ly6G+ neutrophils in Cxcr2−/− compared to Cxcr2+/+ hosts, with no difference in the density of F4/80+ macrophages and CD11b+Ly6C+ monocytes (Figs. 5E). An analysis using absolute number rather than density of CD11b+Ly6G+ neutrophils yielded the same conclusion (Supplementary Fig. S1). Consistent with a previously published study, CD11b+Ly6G+ granulocytes were found to be highly abundant in the spleen of Cxcr2−/− hosts at the earliest time point, which suggested that CXCR2 signaling primarily affected trafficking and not the differentiation of granulocytes (Fig. 5F) (31) Importantly, the density of tumor infiltrating CD3+ T cells was significantly elevated in Cxcr2−/− compared to Cxcr2+/+ hosts 21 days post-implantation. Therefore, these data support the hypothesis that CXCR2 signaling is specifically required for the accumulation of TANs, but not for other myeloid populations. Furthermore, lack of TAN accumulation corresponded with increased accumulation of tumor-infiltrating T cells 2–3 weeks post-implantation.
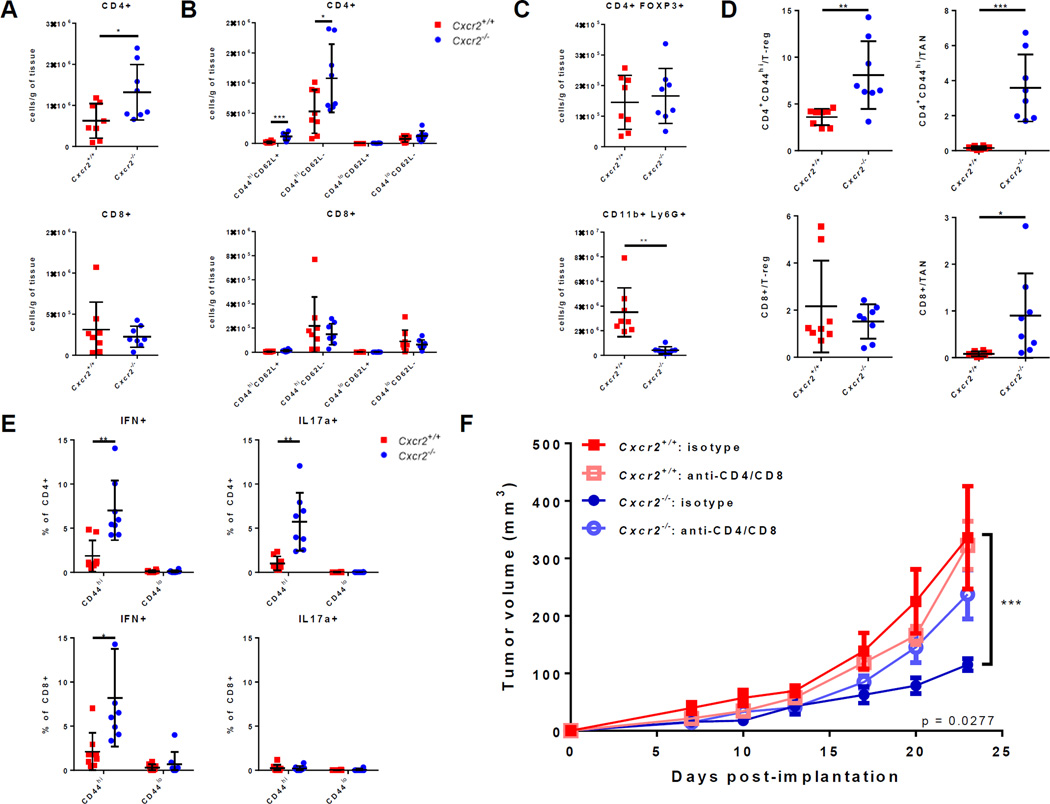
Absence of TANs correlates with increased tumor infiltration and function of activated T cells
Flow cytometric analysis showed no significant difference in the density of CD45−CD31+ endothelial cells (data not shown). This result suggested that TAN modulation in our experiment did not affect tumor angiogenesis. In contrast, we found a significant increase in the infiltration of CD3+ T cells in Cxcr2−/− compared to Cxcr2+/+ hosts (Fig. 5E). Correspondingly, there was also a significant increase in the density of tumor-infiltrating CD4+ T cells in Cxcr2−/− compared to Cxcr2+/+ hosts (Fig. 6A). More detailed analysis showed that activated CD44hiCD62L+ memory and CD44hiCD62L− effector CD4+ T cells were both significantly increased in Cxcr2−/− hosts (Fig. 6B). In contrast, no difference was observed in the density of infiltrating CD4+FOXP3+ Tregs (Fig. 6C). As above, TANs were almost completely absent in this cohort of Cxcr2−/− compared to wild-type hosts (Fig. 6C). Therefore, the absence of TANs corresponded with significant infiltration of activated T cells in the TME.
Figure 6. Absence of TANs leads to increased infiltration and function of activated T cells.
A–E, Cxcr2−/− and Cxcr2+/+ mice were subcutaneously implanted with the 4662 cell line and sacrificed at 4 weeks (n = 8 per group). Graphs show mean ± s.d. of one experiment, which was done only once. *P < 0.05, ** P < 0.01, *** P < 0.001 (unpaired t-test). (A) Densities of tumor-infiltration CD4+ and CD8+ T cells. (B) Densities of CD44hiCD62L+ memory, CD44hiCD62L− effector, and CD44loCD62L+ or CD44loCD62L− naïve CD4+ or CD8+ T cells. (C) Densities of CD4+FOXP3+ Tregs CD11b+Ly6G+ tumor-associated neutrophils. (D) Ratios of the densities of CD4+CD44hi or CD8+ effector T cells to the densities of CD4+FOXP3+ Tregs or CD11b+Ly6G+ TANs. (E) The percentage of CD44hi activated or CD44lo naïve CD4+ or CD8+ T cells that expresses IFNγ or IL17 after ex vivo stimulation with PMA/ionomycin for 5 hours at 37°C. (F) 4662 PDA tumor growth in Cxcr2−/− and Cxcr2+/+ mice treated with 200mg anti-CD4/anti-CD8 depleting antibodies or 200mg isotype every three days intraperitoneally (n ≥ 7 per group). Graph shows mean ± s.e.m. of one experiment. *** P ≤ 0.001 on Day 23 between isotype-treated Cxcr2−/− and Cxcr2+/+ mice (2-way ANOVA with column factor P value = 0.0277, Dunnett’s multiple comparison test). Results are representative of two independent experiments.
To highlight the change in the proportion of effector to suppressive immune populations, ratios of the density of activated CD4+CD44hi T cells to the density of Tregs cells and TANs were calculated and found to be increased (Fig. 6D). The ratio of the density of tumor infiltrating CD8+ T cells to the density of TANs was also significantly elevated. These ratios highlighted the significant increase in the proportion of effector T cells and corresponding decrease in the proportion of suppressive immune cells in the tumors of Cxcr2−/− hosts. We further hypothesized that tumor-infiltrating effector T cells are more functional in Cxcr2−/− than in Cxcr2+/+ hosts. Indeed, ex vivo PMA/ionomycin stimulation induced higher proportions of IFNγ-expressing cells in both CD4+ and CD8+ T-cell populations in Cxcr2−/− compared to Cxcr2+/+ hosts (Fig. 6E). A higher proportion of CD4+ T cells was also induced to express IL17, which indicated more functional Th17 cells. These results supported the hypothesis that T cells were indeed more functional in the absence of TANs.
To test the hypothesis that T cells were responsible for inhibiting tumor growth in Cxcr2−/− mice, we compared tumor growth in Cxcr2−/− and Cxcr2+/+ mice treated with dual depleting antibodies against CD4 and CD8 or isotype control (Fig. 6F). Tumor growth did not differ between control and CD4/8-depleted Cxcr2+/+ mice, which suggested that CD4+ and CD8+ T cells do not naturally impact PDA tumor growth, confirming our recent results (39). There was a significant difference in tumor growth comparing Cxcr2+/+ and Cxcr2−/− mice in the control group, but in contrast, tumor growth in isotype-treated Cxcr2+/+ and CD4/CD8-depleted Cxcr2−/− mice was not significantly different. Thus, depletion of T cells in Cxcr2−/− hosts completely rescued tumor growth. This result strongly supported the hypothesis that CD4+/CD8+ T cells were responsible for inhibiting tumor growth in Cxcr2−/− mice. Altogether, these data show that the CXCR2-ligand axis is required for recruitment of TANs, which regulates T-cell immunity in PDA.
DISCUSSION
Multiple studies have demonstrated the importance of CXCR2 in the recruitment of tumor-promoting and immunosuppressive myeloid cells in various cancers, including PDA (19,32,33,44). It has been shown that CD3+ T-cell infiltration increases upon CXCR2 inhibition in murine PDA (19). Here, we aimed to understand the relevant CXCR2 ligands and mechanisms regulating CXCR2 ligand expression in the TME, as this insight may help advance efforts at clinical translation. Our work adds to previously published studies in several key areas. We report: (1) neutrophils are an important aspect of the human PDA TME particularly in the squamous subtype; (2) human PDA has particularly high CXCL5 expression and other CXCR2 ligands compared to other cancer types, as confirmed in our murine model; (3) CXCL5 expression is correlated to NF-kB signaling pathways in human PDA and, in our mouse model, CXCL5 is strongly induced by NF-kB activation; (4) abrogation of CXCR2 signaling slows tumor growth in mice and triggers an influx of activated and functional CD4+ T cells into the TME; and (5) depletion of T cells completely reverses the antitumor effects of CXCR2 inhibition. Our data adds to important accumulating evidence that the CXCR2-ligand axis is a promising target for the treatment of PDA (19,45,46).
We have previously shown that the PDA tumor microenvironment has elevated frequency of myeloid cells (7,13). Several landmark studies have described novel methods of using predefined gene signatures to derive the relative abundance of immune populations from gene expression data in complex tissues (34,47). Here, similar to comparing relative expression of a single gene between samples, we compare the expression of such previously defined gene sets as a surrogate for the relative abundance of immune cells, pathway activation, and PDA subtypes. Our analysis of human TCGA data confirms that at least a subset of human PDA, especially those of the squamous subtype, also has significantly elevated infiltration of TANs. This is in remarkable congruence with the recent work by Steele et al. showing that absence of TAN accumulation in murine KPC PDA tumors is associated with decreased expression of genes in the squamous subtype compared to controls (19,35). We also report here that TAN-high tumors have elevated expression of macrophages and γδ T-cell related genes. This result supports the recent evidence that γδ T cells can promote TAN accumulation (48) and may relate to new data regarding γδ T cells in PDA (49). TANs have also been shown to highly express CCL chemokines, which can recruit macrophages (16,50). Although it remains to be confirmed, our analysis suggests that such interactions between TANs, macrophages, and γδ T cells may also exist in PDA.
Despite ample evidence of TAN accumulation in PDA, the mechanism leading to increased TAN infiltration is not fully understood and may be due to active recruitment via chemokines or passive response to tissue damage. Recently, Steele et al. demonstrated that Cxcl2 expression is significantly elevated in the KPC mouse model of PDA and showed that CXCR2 inhibition significantly reduces the infiltration of MPO+ neutrophils using IHC (19). In another study also using the KPC model, Seifert et al. showed that CXCL1 is elevated in a RIP1/3-dependent manner and that anti-CXCL1 treatment reduces the infiltration of GR1+CD11b+ cells, which consist of a heterogeneous population including TANs and monocytes (51). Using an implantable model of KPC PDA, we confirmed that CXCR2 regulates the accumulation of TANs. In addition, our data revealed that CXCR2 ablation specifically inhibited the accumulation of neutrophils, without affecting infiltration of other myeloid populations. A role for CXCR1 was not directly studied in our work and cannot be excluded from our results here. Indeed, CXCR1 and CXCR2 have both been shown to be individually sufficient for chemotaxis of human neutrophils in vitro when induced by CXCL8 and CXCL1, respectively (52). However, from the near absence of granulocytes recruited to the tumor in vivo using our Cxcr2 knockout mice, we hypothesize that CXCR2, rather than CXCR1, plays the dominant role in the recruitment of granulocytes in our mouse PDA model. Considering the inherent differences between mouse and human genome, whether this conclusion also applies in human PDA remains unknown and difficult to study in vivo using our immune-competent PDA mouse model. It is possible that CXCR1 inhibition may further decrease granulocyte recruitment when in combination with CXCR2 inhibition.
Although previous studies have shown elevation of various CXCR2 ligands in PDA, only a few have attempted to delineate the source of these chemokines within the tumor microenvironment (19,44,51). Here, an unbiased analysis of all CXCR2 ligands using TCGA RNA-sequence data revealed that CXCL5 and CXCL8 expression were orders of magnitude higher than other CXCR2 ligands in PDA. Furthermore, CXCL5 expression was much higher in PDA compared to other solid tumors. Analysis of all murine CXCR2 ligands in KPCY tumors revealed an abundance of Cxcl2 and Cxcl5 expression, which are primarily expressed by stromal and pancreatic-lineage cells, respectively. Interestingly, Steele et al. also reported an enrichment of Cxcl2 and Cxcl5 expression in the tumor epithelium of KPC tumors compared to WT pancreas (19). Assigning direct orthologous relationships between human and mouse chemokines can sometimes be difficult (53). For instance, the mouse has no homolog of human CXCL8. CXCL8, which binds to both CXCR1 and CXCR2, plays an important role in the recruitment of neutrophils in human cancers (54). Indeed, CXCL8 was the second highest expressed CXCR2 ligand in the PDA samples in TCGA and may also be playing an important role in neutrophil recruitment in human disease. Because CXCL8 is absent in the mouse genome, its role cannot be further explored using our KPC or KPCY mouse models. Furthermore, human CXCL5 and CXCL6 are both very similar to Cxcl5 in the mouse, which also does not have Cxcl6. Given the difference between species, we find the congruence of elevated CXCL5 expression in human and Cxcl5 expression in mouse PDA even more remarkable. CXCL5 protein level is strongly associated with reduced overall survival in a cohort of human PDA (28). Thus, our data highlighted the prominence and uniqueness of CXCL5 expression in PDA.
Because CXCL5 was the most highly and universally expressed CXCR2 ligand, we studied the regulation of its expression in more detail. Here, we discovered that NF-κB activation can potently increase CXCL5 protein level in KPC PDA cells. Again, this was consistent with human TCGA data, in which PDA tumors with high CXCR2 ligand expression are also significantly enriched in the expression of inflammatory pathways involving NF-κB. Although the populations expressing these pathways could not be determined using TCGA data alone, our results from the mouse model suggested that NF-κB activity may be enhanced in the cancer cells themselves. Studies in a mouse model of pancreatic intraepithelial neoplasia (PanIN), show that RELA is activated in the presence of oncogenic KRAS and regulates the expression of Cxcl1 (55). Indeed, RELA/p50 is constitutively activated in almost 70% of pancreatic cancers (56). An elegant study by Ling et al. using pancreas-targeted knockout of IKKβ in Pdx1-Cre;LSL-KrasG12D;Ink4a/ArfF/F mice showed that inactivation of NF-κB signaling completely inhibited PDA development (57). They further demonstrated that KRASG12D-driven AP-1 activation can induce the expression of IL1α, which in turn acted in an autocrine manner and activated NF-κB to induce more IL1α expression in a positive feedforward loop. Contrary to the implications from this study, we found that NF-κB activity actually increased when KRAS or MEK was inhibited. Although the mechanism of this increased activity remains to be determined, we speculate that it may have resulted from enhanced PI3K/AKT signaling, which is upstream of NF-κB activation, upon MEK inhibition (58). Altogether, our data suggested that NF-κB activation is important for inducing CXCL5 expression in PDA.
The frequency of tumor-infiltrating T cells increases significantly in the setting of CXCR2 inhibition (19,44). This observation was also true in our subcutaneous, implantable murine model of PDA. This subcutaneous model faithfully recapitulated the histology, immune infiltration, and even response to therapy of spontaneous KPC pancreatic tumors (8,11,12). Another group used the spontaneous, autochthonous KPC model and reported similar conclusions to those presented in this paper (19). In this study, pharmacologic CXCR2 inhibition suppressed metastasis and prolonged survival in KPC mice. Furthermore, in the context of anti–PD-1 therapy, co-treatment with a CXCR2 inhibitor led to increased infiltration of effector CD4+ and CD8+ T cells. Adding to this study, we observed that the tumor-infiltrating T cells in Cxcr2−/− hosts consisted mostly of activated, effector CD4+ T cells. Although the density of tumor infiltrating CD8+ T cells was not increased in Cxcr2−/− mice, the proportion of IFNγ-producing cells increased among both activated CD4+ and CD8+ T-cell populations. This result supported the hypothesis that TANs are immunosuppressive in PDA.
However, the specific mechanisms of how TANs suppress infiltration of T cells in PDA remain to be elucidated. One possible mechanism is the secretion of arginase 1 (ARG1) by TANs, which depletes l-arginine from the microenvironment and inhibits T-cell proliferation (23). Because granulocytic-myeloid derived suppressor cells (G-MDSCs) are defined in mice as CD11b+Ly6G+Ly6Clo cells that suppresses T-cell proliferation or function, TANs or at least a subset of them in our PDA model, fulfill this definition (15). Future work must be done to further delineate the relationship between TANs and G-MDSCs. Given that PDA is naturally void of effector T cells, the observation that effector T cells could infiltrate in the absence of TANs is particularly exciting. Indeed, CXCR2 inhibition sensitized the otherwise highly resistant KPC PDA to anti–PD-1 therapy, with durable response in a small subset of tumors (19). Therefore, our data support the emerging notion that targeting specific components of the pancreatic tumor microenvironment may be needed in order to sensitize tumors to combination chemotherapy and immune checkpoint inhibitors (8,11,59–61).
Besides their immunosuppressive function, TANs can also promote angiogenesis (21). In fact, reduced blood vessel density was observed in xenografts of a CXCL8 knockdown human PDA cell line compared to controls (62). However, in our model with syngeneic immunocompetent mice, depletion of CD4+ and CD8+ T cells was sufficient to rescue tumor growth in Cxcr2−/− hosts. Furthermore, the density of endothelial cells did not differ significantly between the tumors of Cxcr2+/+ and Cxcr2−/− hosts. These results argued that TANs primarily promote tumor growth via immunosuppressive mechanisms in our model rather than through reduction in angiogenesis.
In summary, we conclude that CXCR2 is required for the recruitment of TANs, which in turn can suppress antitumor T-cell responses. We showed that CXCR2 ligands, particularly CXCL5, are elevated in both human and mouse PDA. Furthermore, expression and secretion of CXCL5 in our mouse model is potently induced by NF-κB activation. Finally, we showed that PDA tumor growth can be inhibited in a T cell–dependent manner in the context of CXCR2 inhibition. Therefore, the CXCR2-ligand axis is emerging as a potential target for the treatment of PDA.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 CA169123 (R.H.V.) and the Parker Institute for Cancer Immunotherapy (R.H.V.). We thank Drs. Katelyn Byrne, David Balli, and Ben Stanger for helpful discussions; the TCGA Research Network for generating the human gene expression data and making it publically available (http://cancergenome.nih.gov/).
Footnotes
Conflict of interest: The authors declare no potential financial conflicts of interest
REFERENCES
- 1.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. New England Journal of Medicine. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 2.Hoff Von DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. New England Journal of Medicine. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Research. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg R. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 7.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA, Vonderheide RH. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Research. 2007;67:9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 8.Winograd R, Byrne KT, Evans RA, Odorizzi PM, Meyer ARL, Bajor DL, et al. Induction of T-cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunology Research. 2015;3:399–411. doi: 10.1158/2326-6066.CIR-14-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–847. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne KT, Vonderheide R. CD40 stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Reports. 2016;15:2719–2732. doi: 10.1016/j.celrep.2016.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beatty GL, Winograd R, Evans RA, Long K, Luque S, Lee JW, et al. Exclusion of T cells from pancreatic carcinomas in mice is regulated by Ly6C low F4/80+ extratumoral macrophages. Gastroenterology. 2015;149:201–210. doi: 10.1053/j.gastro.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayne L, Beatty G, Jhala N, Clark C, Rhim A, Stanger B, et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic vancer. Cancer Cell. 2012;21:822–835. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffelt SB, Wellenstein MD, de Visser KE. Neutrophils in cancer: neutral no more. Nat Rev Cancer. 2016;16:431–446. doi: 10.1038/nrc.2016.52. [DOI] [PubMed] [Google Scholar]
- 15.Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunology. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fridlender Z, Albelda S. Tumor-associated neutrophils: friend or foe? Carcinogenesis. 2012;33(5):949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 18.Shen M, Hu P, Donskov F, Wang G, Liu Q, Du J. Tumor-associated neutrophils as a new prognostic factor in cancer: a systematic review and meta-analysis. PLoS ONE. 2014;9:e98259. doi: 10.1371/journal.pone.0098259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L, et al. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell. 2016;29:832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiegel A, Brooks MW, Houshyar S, Reinhardt F, Ardolino M, Fessler E, et al. Neutrophils suppress intraluminal NK cell-mediated tumor cell clearance and enhance extravasation of disseminated carcinoma cells. Cancer Discovery. 2016;6:630–649. doi: 10.1158/2159-8290.CD-15-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shojaei F, Wu X, Zhong C, Yu L, Liang X-H, Yao J, et al. Bv8 regulates myeloid-cell-dependent tumour angiogenesis. Nature. 2007;450:825–831. doi: 10.1038/nature06348. [DOI] [PubMed] [Google Scholar]
- 22.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Research. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 24.Fridlender ZG, Sun J, Kim S, Kapoor V, Cheng G, Ling L, et al. Polarization of tumor-associated neutrophil phenotype by TGF-β:“N1” versus ‘N2’ TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singhal S, Bhojnagarwala PS, O'Brien S, Moon EK, Garfall AL, Rao AS, et al. Origin and role of a subset of tumor-associated neutrophils with antigen-presenting cell features in early-stage human lung cancer. Cancer Cell. 2016;30:120–135. doi: 10.1016/j.ccell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid MD, Basturk O, Thirabanjasak D, Hruban RH, Klimstra DS, Bagci P, et al. Tumor-infiltrating neutrophils in pancreatic neoplasia. Mod Pathol. 2011;24:1612–1619. doi: 10.1038/modpathol.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W-Q, Liu L, Xu H-X, Wu C-T, Xiang J-F, Xu J, et al. Intratumoral infiltrating immune cells and gene mutations in pancreatic ductal adenocarcinoma. Br J Surg. 2016 doi: 10.1002/bjs.10187. [DOI] [PubMed] [Google Scholar]
- 28.Li A, King J, Moro A, Sugi MD, Dawson DW, Kaplan J, et al. Overexpression of CXCL5 is associated with poor survival in patients with pancreatic cancer. The American Journal of Pathology. 2011;178:1340–1349. doi: 10.1016/j.ajpath.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stromnes IM, Brockenbrough JS, Izeradjene K, Carlson MA, Cuevas C, Simmons RM, et al. Targeted depletion of an MDSC subset unmasks pancreatic ductal adenocarcinoma to adaptive immunity. Gut. 2014;63:1769–1781. doi: 10.1136/gutjnl-2013-306271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eash KJ, Greenbaum AM, Gopalan PK, Link DC. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. Journal of Clinical Investigation. 2010;120:2423–2431. doi: 10.1172/JCI41649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mei J, Liu Y, Dai N, Hoffmann C, Hudock KM, Zhang P, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. Journal of Clinical Investigation. 2012;122:974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Science Translational Medicine. 2014;6:237ra67. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh H, Wang D, Daikoku T, Sun H, Dey S, DuBois R. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Walder M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 36.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature Medicine. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhim A, Mirek E, Aiello N, Maitra A, Bailey J, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans RA, Diamond M, Rech AJ, Chao T, Richardson M, Lin J, et al. Lack of immunoediting in murine pancreatic cancer reversed with neo-antigen. JCI Insight. 2016;1(14):e88328. doi: 10.1172/jci.insight.88328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart JM Network CGAR. The cancer genome atlas pan-cancer analysis project. Nature Genetics. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SGH, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nature Genetics. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo A, Wang L-CS, Scholler J, Monslow J, Avery D, Newick K, et al. Tumor-promoting desmoplasia is disrupted by depleting FAP-expressing stromal cells. Cancer Research. 2015;75:2800–2810. doi: 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W, N N, Nandakumar Nandakumar, Shi Y, Manzano M, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Science Signaling. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discovery. 2016;6:80–95. doi: 10.1158/2159-8290.CD-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ijichi H, Chytil A, Gorska AE, Aakre ME, Bierie B, Tada M, et al. Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves survival in a mouse model of pancreatic ductal adenocarcinoma. Journal of Clinical Investigation. 2011;121:4106–4117. doi: 10.1172/JCI42754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purohit A, Varney M, Rachagani S, Ouellette MM, Batra SK, Singh RK. CXCR2 signaling regulates KRAS (G12D)-induced autocrine growth of pancreatic cancer. Oncotarget. 2016;7:7280–7296. doi: 10.18632/oncotarget.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature Medicine. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coffelt SB, Kersten K, Doornebal CW, Weiden J. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, et al. γδ T cells support pancreatic oncogenesis by restraining αβ T cell activation. Cell. 2016;166:1485–1499. doi: 10.1016/j.cell.2016.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fridlender ZG, Sun J, Mishalian I, Singhal S, Cheng G, Kapoor V, et al. Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLoS ONE. 2012;7:e31524. doi: 10.1371/journal.pone.0031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seifert L, Werba G, Tiwari S, Ly NNG, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hammond ME, Lapointe GR, Feucht PH, Hilt S, Gallegos CA, Gordon CA, et al. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol. 1995;155:1428–1433. [PubMed] [Google Scholar]
- 53.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfaro C, Teijeira A, Oñate C, Pérez G, Sanmamed MF, Andueza MP, et al. Tumor-produced interleukin-8 attracts human myeloid-derived suppressor cells and elicits extrusion of neutrophil extracellulartraps (NETs) Clinical Cancer Res. 2016;22:3924–3936. doi: 10.1158/1078-0432.CCR-15-2463. [DOI] [PubMed] [Google Scholar]
- 55.Lesina M, Wörmann SM, Morton J, Diakopoulos KN, Korneeva O, Wimmer M, et al. RelA regulates CXCL1/CXCR2-dependent oncogene-induced senescence in murine Kras-driven pancreatic carcinogenesis. Journal of Clinical Investigation. 2016;126:2919–2932. doi: 10.1172/JCI86477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang WX, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ. The nuclear factor-kappa B RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clinical Cancer Research. 1999;5:119–127. [PubMed] [Google Scholar]
- 57.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, et al. Kras G12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wee S, Jagani Z, Xiang KX, Loo A, Dorsch M, Yao Y-M, et al. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Research. 2009;69:4286–4293. doi: 10.1158/0008-5472.CAN-08-4765. [DOI] [PubMed] [Google Scholar]
- 59.Nywening TM, Wang-Gillam A, Sanford DE, Belt BA, Panni RZ, Cusworth BM, et al. Targeting tumour-associated macrophages with CCR2 inhibition in combination with FOLFIRINOX in patients with borderline resectable and locally advanced pancreatic cancer: a single-centre, open-label, dose-finding, non-randomised, phase 1b trial. Lancet Oncology. 2016;17:651–662. doi: 10.1016/S1470-2045(16)00078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H, Hegde S, Knolhoff BL, Zhu Y, Herndon JM, Meyer MA, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nature Medicine. 2016;22:851–860. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feig C, Jones JO, Kraman M, Wells RJB, Deonarine A, Chan DS, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci USA. 2013;110:20212–20217. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.