Abstract
Branch migration of Holliday junctions is an important step of genetic recombination and DNA repair. In Escherichia coli, this process is driven by the RuvAB complex acting as a molecular motor. Using magnetic tweezers, we studied the RuvAB-directed migration of individual Holliday junctions formed between two ≈6-kb DNA molecules of identical sequence, and we measured the migration rate at 37°C and 1 mM ATP. We directly demonstrate that RuvAB is a highly processive DNA motor protein that is able to drive continuous and unidirectional branch migration of Holliday junctions at a well defined average speed over several kilobases through homologous sequences. We observed directional inversions of the migration at the DNA molecule boundaries leading to forth-and-back migration of the branch point and allowing us to measure the migration rate in the presence of negative or positive loads. The average migration rate at zero load was found to be ≈43 bp/sec. Furthermore, the load dependence of the migration rate is small, within the force range of –3.4 pN (hindering force) to +3.4 pN (assisting force).
Homologous recombination is a fundamental and highly conserved mechanism of genetic content exchange between two homologous DNA molecules and is an essential cellular process required to generate genetic diversity and maintain genome stability. A central intermediate of the homologous recombination is the Holliday junction that connects two recombining DNAs in a four-way branched DNA structure. Branch migration of a Holliday junction, when one DNA strand is progressively exchanged for another, extends the heteroduplex DNA and determines the amount of genetic information transferred between the two DNA molecules (1–3).
In Escherichia coli, RuvA, RuvB, and RuvC proteins process the Holliday-junction intermediate toward the formation of two recombinant DNA molecules (4–6). The three Ruv proteins are thought to form two types of complex with the Holliday junction (7): a RuvAB complex that promotes branch migration (8–12) and a RuvABC complex in which RuvC scans the DNA sequence during RuvAB-mediated branch migration and resolves the Holliday junction preferentially at the consensus sequence 5′-(A/T)TT↓(G/C)-3′ (13–15).
Holliday junctions are specifically bound by RuvA tetramers that unfold the junction in a square planar conformation, energetically favorable to branch-point migration (8, 16–20). RuvB is targeted to the Holliday junction by specific interactions with RuvA and has been reported to assemble symmetrically into hexameric rings around opposite DNA arms of the RuvA–junction complex (21–23). RuvAB promotes branch-point migration in an ATP-dependent manner: the DNA is pumped out of the RuvA–junction assembly by and throughout the RuvB hexamers (24). Therefore, orientation assembly of the RuvAB complex determines the direction of migration.
Despite recent advances in the comprehension of how RuvA and RuvB process the Holliday junction, dynamical aspects of the mechanism are poorly known. Here we present direct measurements of the RuvAB complex activity on Holliday junctions at the single-molecule level. The study explores the dynamical properties of the process and provides direct measurements of the enzyme kinetics.
Materials and Methods
Proteins and DNA Constructs. RuvA and RuvB were expressed and purified as described (12, 25). Protein concentrations were determined by the Bradford method (Bio-Rad) by using BSA as a standard and expressed as moles of monomers.
Two DNA constructs named “multi-ss” and “multiswivel” were used in this work. Both follow the same general scheme (Fig. 1A): a central cruciform core made of oligonucleotides is flanked on the one hand by a pair of short heterologous DNA arms ended by hairpin structures and on the other hand by two long homologous DNA arms with either biotin- or digoxigenin (DIG)-multilabeled ends. Both constructions have the same sequence but differ in their end attachments. Each long homologous DNA arm is obtained from a vector containing the 4.6-kb KpnI–HindIII fragment of λ DNA inserted into the KpnI- and HindIII-digested pBluescript KS(+) vector (Stratagene), resulting in a 7.5-kb pk-λ vector (generous gift of D. Bensimon, Ecole Normale Supérieure). The central cross with the hairpin ends is formed by six oligonucleotides (Eurogentec, Brussels): HOLL1, 5′-ACCATGCTCGTGATTACGAGATATCGATGCATGCGAATTCGAGCTCGGTAC; HOLL2, 5′-AGCTGTACCGAGCTCGA AT TCGCATGCATCGATATA ATACGTGAGGCCTA; HOLL3, 5′-GATCCTAGGCCTCACGTATTATATCGATGCATGCGAATTCGAGCTCGACGC; HOLL4, 5′-AGCTGCGTCGAGCTCGAATTCGCATGCATCGATATCTCGTAATCACGAGCA; Hairpin1, 5′-GGATCGAAGCGAGCGAAAGCTCGCTTC; and Hairpin2, 5′-TGGTGAAGCGAGCGAAAGCTCGCTTC.
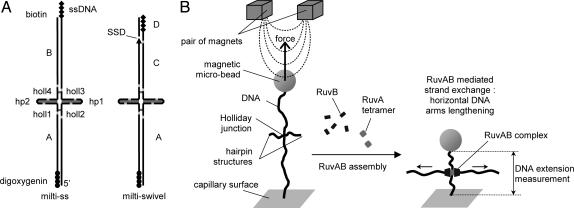
Fig. 1.
Configurations. (A) Schemes of the two DNA constructs used (multi-ss and multiswivel) with their subcomponents. Points of ligation are shown in light gray, and sequence heterologies are shown in dark gray. Everywhere else, the sequence is palindromic to enable homologous strand exchange. In the multi-ss construct, no rotational constraint can occur, because the multilabeled biotin end is single-stranded. In the multiswivel construct, a single-strand discontinuity (SSD, arrow) relaxes the rotational constraint. ssDNA, single-stranded DNA. (B) Experimental configuration. A DNA molecule containing a Holliday junction is tethered between the capillary surface and a magnetic microbead (see Materials and Methods). Initially, the branch point is restricted inside a 27-bp region of homologous sequence by 3 bp of heterology that impede spontaneous branch migration (26, 27) but not RuvAB-mediated branch migration. A shortening of the tethering length caused by a forward RuvAB-mediated migration is illustrated.
For both DNA constructs, the long (5.9-kb) DNA arms are designed to give multiple (hence strong) anchoring of the construction while preserving a rotational degree of freedom (see below). For multi-ss, this is obtained by using a single-stranded multilabeled end. For multiswivel, a single-strand discontinuity is incorporated. Precautions have been taken to avoid UV damaging of the DNA constructs during the purification steps.
Multi-ss Construction. A fragment. Three micrograms of vector pk-λ was digested with BsaI (NEB, Beverly, MA) and dephosphorylated with shrimp alkaline phosphatase (United States Biochemical) followed by heat inactivation of the phosphatase. For labeling, the DNA solution was complemented at 50 mM NaCl final concentration and incubated with 0.6 μl of T4 polymerase (NEB) without dNTP added (37°C, 10 min) such that exonucleolytic digestion occurred at the 3′ extremities. Then, nucleotides were added (final concentration, 60 μM dNTP/12 μM DIG-dUTP) such that the 3′-recessed extremities were filled in (37°C, 1 h) with a fraction of modified nucleotides. The DNA was then purified (Wizard SV gel and PCR, Promega) and digested by HindIII, which was then heat-inactivated. The annealed oligonucleotide pair HOLL1 and HOLL2 then were ligated to the vector DNA (molar ratio, 100:1) by T4 ligase (NEB) followed by heat inactivation of the ligase. The resulting preparation was electrophoresed on a 1% agarose gel, and the large fragment was recovered from the gel with precautions to avoid UV irradiation of the DNA: the gel was run with side lanes incorporating a small fraction of the preparation. Those side lanes were cut and separately stained with ethidium bromide, and the position of the band was marked under UV. The band in the gel then was excised based on the estimation of its position obtained from the stained side lanes and purified to obtain the A fragment.
B fragment. Three micrograms of vector pk-λ was digested with SacI (NEB), followed by heat inactivation of the endonuclease, and purified. To label the B fragment of the construction, we used terminal deoxynucleotidyl transferase. In a final volume of 15 μl, 100 mM sodium cacodylate (pH 7.2), 0.2 mM mercaptoethanol, 2 mM CoCl2, complemented with 0.5 μl of 10 mM dTTP, 0.5 μl of 1 mM DIG-dUTP (Amersham Pharmacia), and 3 units of terminal deoxynucleotidyl transferase (Amersham Pharmacia) per microgram of DNA were incubated for 2 h at 37°C. Then, as for the A fragment, the DNA was purified, digested with HindIII, and ligated with the annealed oligonucleotide pair HOLL3 and HOLL4. Finally, the large fragment was extracted while avoiding UV irradiation to give the B fragment.
A-B assembly. In two separate tubes, the A [respectively (resp) B] fragment and the hairpin oligonucleotide Hairpin2 (resp Hairpin1) (molar ratio, 100:1) were ligated. The two tubes then were mixed, and further ligation was performed. The ligase then was heat-inactivated, and the preparation was used without further purification. The appropriate dilution was determined empirically to obtain a few beads tethered within the sample.
Multiswivel Construction. D fragment. Six micrograms of vector pBluescript KS(+) was digested with BsaI and dephosphorylated with shrimp alkaline phosphatase followed by heat inactivation of the phosphatase. Labeling was performed by using the same protocol as for the A fragment with the same concentration ratio between DNA and T4 DNA polymerase. DNA purification and digestion by Acc65, followed by heat inactivation of Acc65 and DNA purification, gave the D fragment.
C fragment. Three micrograms of vector pk-λ was digested by BsaI and Acc65, dephosphorylated by adding shrimp alkaline phosphatase, and purified and digested by HindIII followed by heat inactivation of HindIII. Ligation at the HindIII ends with the annealed oligonucleotide pair HOLL3 and HOLL4, followed by extraction avoiding UV irradiation, gave the C fragment.
A-C-D assembly. In two separate tubes, the A (resp C) fragment and the hairpin oligonucleotide Hairpin2 (resp Hairpin1) (molar ratio, 100:1) were ligated. The two tubes (equimolar concentrations) were then mixed, and an excess of fragment D (10:1) was added for additional ligation. The ligase then was heat-inactivated, and this preparation was used without additional purification.
In this construction, there is a single-strand discontinuity at the connection between C and D, because C has been dephosphorylated before the ligation with D.
Magnetic Tweezers. The magnetic tweezers were built by using an inverted microscope configuration adapted from a setup developed by Croquette and coworkers (28). The sample was contained in a flow chamber composed of a rectangular crosssection capillary tube (inside dimensions, 0.1 × 1 mm; length, 45 mm). Silicone tubes connected one extremity of the capillary to a small funnel for sample injection and the other extremity to a syringe pump. The capillary was coated with polyclonal antibodies from sheep directed against DIG (Roche Diagnostics). Single DNA molecules, end-labeled with DIG and biotin (see above), were tethered between the capillary surface and streptavidin-coated magnetic microbeads (2.8-μm-diameter M270 or 1-μm-diameter MyOne, Dynal, Oslo) (Fig. 1). A pair of NeFeB magnets (4 × 4 × 4 mm, separated by 1 mm) was positioned above the sample and motor-controlled to be either vertically translated or rotated along the vertical axis to impose a determined stretching force and a known supercoiling to the tethered DNA molecules. Images of the sample were obtained by using a ×100 oil objective (Leica C-Plan; numerical aperture, 1.25) and grabbed with a charge-coupled device camera (model CV-M30, JAI, Copenhagen) connected to a personal computer. The microscope was enclosed in a box regulated at a temperature of 37°C.
Measurements. Lateral and vertical bead positions were measured by real-time image analysis (29) at 60-Hz frequency with, respectively, ≈5 and ≈20 nm accuracy. In brief, a parallel illumination of the sample creates around the beads a pattern of diffraction rings that depends on the distance of the bead to the focal plane of the objective, i.e., on the vertical position of the bead. Before each experiment, we recorded a calibration profile of the bead of interest: the objective was moved stepwise along the vertical direction, and at each step the corresponding radial profile of the diffraction pattern was recorded. During the experiment, the instantaneous radial profile of the bead obtained from each video frame was compared in real time with the calibration profile to determine the vertical position of the bead. Mechanical drifts of the microscope were corrected by simultaneously tracking a bead stuck on the surface of the capillary and serving as reference.
The force exerted on the tethered DNA molecule by the magnetic gradient field was calculated by analyzing the Brownian movement of the bead and using the equipartition theorem F = kBT l/〈δx2〉, where l is the molecule extension, T is the temperature, kB is the Boltzmann constant, and 〈δx2〉 is the lateral fluctuations of the bead.
Branch-Migration Assay (Experimental Procedure). Sample preparation. To limit nonspecific interactions and aggregation between the beads, anchoring of the DNA molecules was performed without magnesium ions by using a low ionic strength buffer [25 mM Tris-acetate/5 mM EDTA/0.1% (wt/vol) BSA/0.01% (wt/vol) NaN3]. Holliday junctions were first incubated with an excess of magnetic microbeads for 5 min to obtain bead–DNA attachment. Then, the mixture was injected into the flow chamber and incubated for 6–10 min for the DIG-labeled DNA ends to anchor to the capillary surface. Finally, the flow chamber was gently rinsed (using the syringe pump) with 200 μl of the same buffer to evacuate the free beads in excess.
Then, before injection of the proteins, the flow chamber containing the tethered DNA molecules was rinsed gently with 70 μl of the reaction buffer chosen for RuvAB [25 mM Trisacetate, pH 8/10 mM magnesium acetate/100 mM NaCl/1 mM ATP/20 mM phosphocreatine/0.5 mM EDTA/1 mM dTT/0.1% (wt/vol) BSA/0.01% (wt/vol) NaN3], in which 10% (vol/vol) glycerol was added. Addition of 10% glycerol at this step was essential to avoid artifacts in bead-position measurements: RuvA and RuvB proteins were stored in a 50% (vol/vol) glycerol buffer, resulting in a 10% final glycerol concentration in the buffer containing the proteins. Because the glycerol modifies the optical index of the buffer, the diffraction patterns around the beads were slightly different in the presence or absence of glycerol. Because calibration profiles were recorded before protein injection, preparation of the sample by rinsing with a buffer containing 10% glycerol, and thus the same glycerol condition as in the presence of the proteins, prevented measurements from artifacts that otherwise could give rise to position-determination errors that could take the appearance of sudden changes in the migration rate up to ≈40% at the micrometer scale (see Fig. 4 and Supporting Text, which are published as supporting information on the PNAS web site).
At this stage, a tethered bead was chosen, and a “supercoiling test” was performed: the magnets were rotated by +100 turns while monitoring the tethering length at ≈0.5 pN to check that neither plectonemes nor braids formed, indicating that the bead was tethered by a single nonsupercoilable DNA molecule. Finally, for various vertical positions of the magnets, we determined the corresponding stretching force.
RuvAB-mediated branch-migration measurement. After injection in the flow chamber of 10 μl of reaction buffer containing the indicated RuvA and RuvB monomer concentrations mixed with 12.5 units/ml creatine phosphokinase, the height of the tethered bead was monitored continuously as a function of time. The RuvAB-mediated branch migration induced the transfer of DNA length between the vertical and the horizontal DNA arms of the Holliday junction (Fig. 1B). We measured the activity of RuvAB by monitoring in real time the shortening (or lengthening) of the tethering length. By convention we called it forward (resp backward) migration when the tethering length decreased (resp increased). If no migration occurred for a significantly long time or if the anchoring broke, a new tethered bead was chosen and the migration activity was monitored again after the above steps were repeated: recording of the calibration profile, supercoiling test of the tethered molecule, and calibration of the force.
Data Analysis. To determine the contour length of the vertical DNA arms (and thus the branch-point position), the tethered bead height (which measures the vertical DNA arms extension) was divided by the relative extension of double-stranded DNA calculated from force-extension calibration curves measured in the same conditions as for the RuvAB experiment except that no protein was present. The distributions for the branch-migration rates then were obtained by linear fitting the migration runs by using a sliding time window (size as indicated) translated by half its size at each step.
Results
Cyclic Activity. After protein injection, migration can occur in two opposite directions depending on the orientation assembly of the RuvAB complex on the junction. Nevertheless, dissociation of the junction was impeded at (i) the “low-extension” boundary by multiattachments of the vertical DNA arm ends to the capillary and to the bead surface that restrict the branch-point movement and (ii) the “high-extension” boundary by the hairpin structures at the horizontal DNA arm extremities. This configuration resulted in a cyclic activity of the migration process in which the RuvAB complex proceeded to forth-and-back migrations between the molecule extremities (see Fig. 2 Left). However, at the high-extension boundary, we could not tell whether the cruciform structure was maintained with the branch point blocked before or inside the heterologous region or whether the cruciform structure was absorbed completely by a total backward migration. The latter event would result in the formation of a transient denaturation bubble and an estimated length change of ≈22 nm, which might be easily hidden in the Brownian noise.
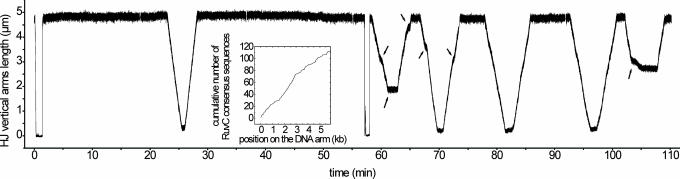
Fig. 2.
Influence of RuvB concentration. The experiment was performed by using a multi-ss construction. All along this experiment, the same DNA molecule was followed, and a constant load force of 0.73 pN was applied. In the first part of the experiment (from 0 to 58 min), 100 nM RuvA and 150 nM RuvB were injected in the sample (see the downfall near 0 min). In the second part (from 58 to 110 min), 100 nM RuvA and 670 nM RuvB were injected (see the downfall near 58 min). Whereas in the presence of 150 nM RuvB only one back-and-forth migration occurs during ≈1 h and with a nearly constant speed, in the presence of 670 nM RuvB the frequency of back-and-forth migrations is clearly higher and the migrations are less regular, because one can perceive migration inversions (arrows) and more frequent variations in speed (arrows). (Inset) Repartition of the RuvC consensus sequence 5′-(A/T)TT(G/C)-3′ along the vertical DNA arm sequence (a high slope corresponds to a high density of RuvC consensus sequences).
Influence of RuvA and RuvB Concentrations, Processivity, and Unidirectionality. Preliminary experiments were done to determine the optimal RuvA and RuvB concentrations to be injected in our assay. Fig. 2 shows an experiment performed with a multi-ss construct and illustrates the effect of increasing the RuvB concentration from 150 (Fig. 2 Left) to 670 nM (Fig. 2 Right) in the presence of 100 nM RuvA. At 150 nM RuvB (Fig. 2 Left), the enzyme processed branch migration continuously, at a constant speed along the 6 kb of the DNA arms, and unidirectionally until the migration process was stopped and reversed at the molecule extremity. Short pauses or irregularities in the motion were sometimes observed (none appear on this recording). At 670 nM RuvB (Fig. 2 Right), mainly two differences could be seen compared with the activity at 150 nM RuvB. First, the migrating complex was destabilized, because we could see motion reversal before reaching the construction boundaries. Also, irregularities in the migration speed (Fig. 2 Right, arrows) were seen at a higher frequency of occurrence. Second, when the branch point had reached the high-extension boundary, the waiting time for the migration to reverse was decreased (i.e., the migration-initiation efficiency was increased). Clearly, when the branch point reached the low-extension boundary, the time for the migration to reverse was shorter than at the high-extension boundary; this was a general feature of all our experiments and may reflect the fact that, at the low-extension boundary, there was more DNA arm length left than at the high-extension boundary to facilitate the loading of RuvB hexamers. At lower concentrations of RuvB (<100 nM), seldom migration activity was detected (data not shown), indicating a dramatic drop in the migration-initiation efficiency.
The influence of RuvA concentration was also tested in the presence of 150–300 nM RuvB monomers. Varying the RuvA concentration to 10, 100, and 700 nM had no perceptible effect (data not shown). Moreover, no migration activity was detected at RuvB monomer concentrations of 0.3, 0.67, 1.5, and 4 μM when experiments were performed in the absence of RuvA (data not shown).
However, with this multi-ss construct, the migration-initiation frequency was found to dramatically decrease after ≈1 h. We therefore designed another Holliday-junction molecule: the multiswivel construct, similar to the multi-ss construct for its central core but differing by its end attachment. No “aging” of the sample was observed with it, and the initiation efficiency at the high-extension boundary was similar compared with the multi-ss construct. However, the initiation efficiency at the low-extension boundary (i.e., Holliday junction in the close vicinity of the single-strand discontinuity, which appears to constitute a block for the branch-point migration) was reduced very substantially with respect to multi-ss construct (data not shown). We have no explanation for the aging of the sample with the multi-ss construct, but it seems to be linked to the presence of the single-stranded tail.
In the following experiments, the multiswivel construct was used, avoiding the aging of the sample, and concentrations of 100 nM RuvA and 200 nM RuvB were used to avoid the perturbing effects of high RuvB concentrations and maintain a reasonable migration-initiation efficiency.
Branch-Migration Rate and Influence of the Force. From the experimental configuration, a 1-bp translocation of the branch point along the sequence led to a 2-bp reduction in the tethering length. Therefore, the branch-migration rate and the rate of the enzyme were taken here by definition to be half the rate of variation of the total tethering DNA contour length, which corresponded to the sum of the length of the two opposed vertical arms.
Thanks to the cyclic activity of RuvAB, and by using the multiswivel construct, we could explore the effect of both negative and positive loads on the complex kinetics. In the forward motion (resp backward), the force was hindering (resp assisting) the migration, and thus the sign of the load was taken as negative (resp positive). The level of force explored covered the range of ±0.7 to ±3.4 pN. Examples of cumulative forward- and backward-migration runs are represented in Fig. 3A. Distributions of branch-migration rates for the corresponding levels of force are represented in Fig. 3B. They were calculated as indicated in Materials and Methods. The time window was chosen so as to find a compromise between the following constraints: (i) the Brownian noise should not dominate the signal (which implies the use of the largest time window) and (ii) the treatment should be sensitive to variations in the complex kinetics (which implies the use of the smallest time window). We chose a time window of 10 sec for the lowest force level of 0.7 pN and used a value of 5 sec for all the points at higher forces.
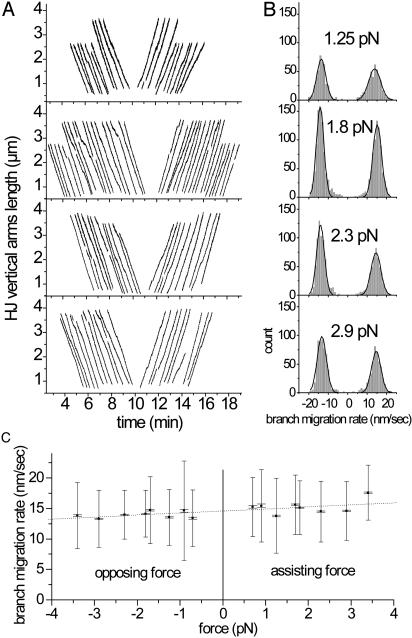
Fig. 3.
RuvAB-mediated branch-migration rate and effect of the applied load. In all experiments, the multiswivel construct was used, and concentrations of RuvA and RuvB were 100 and 200 nM, respectively. (A) Example of migration runs (grouped for presentation convenience). Because the maximum extension may vary from molecule to molecule, runs do not have the same length. Runs usually come in pairs of forward and backward migration, resulting in the apparent symmetry of the figure. Short breaks appearing in some abutted runs correspond to either short computer-generated interruptions of the recording for data storage or clear pauses that were manually removed for the velocity determination. (B) Branch-migration rate distributions (in nm/sec) for four levels of force (1.25, 1.8, 2.3, and 2.9 pN) obtained as indicated in Materials and Methods with a time window of 5 sec. Positive and negative peaks correspond to forward and backward migrations, respectively. Histograms were fitted with Gaussian distributions. (C) Branch-migration rate vs. load. Sign convention: negative loads correspond to forces that hinder the migration. Rates were obtained by the Gaussian fits in B. Two types of error bars are represented. The larger bars correspond to the width of the Gaussian fit, and the smaller bars correspond to the standard errors of the mean obtained from the Gaussian fits in B. The experimental error on the force applied to a given molecule was estimated to be approximately ±10% (position on horizontal axis) and is not represented. Experiments with force within ±5% were grouped together. The error of the mean obtained by the fit does not take into account the uncertainty of the relative extension factor of the molecule: because the uncertainty of the determination of force was estimated to ±10%, this induces a typical uncertainty ranging here from ±2% at 1 pN, lowering under ±1% above 2 pN.
Fig. 3C represents the migration velocity vs. force data obtained for all levels of force explored. Two types of error bars are represented. The larger bars correspond to the width of the Gaussian fit shown in Fig. 3B, and the smaller bars correspond to the standard errors of the mean obtained from the same Gaussian fit. The effective error for the determination of the average velocity is clearly at an intermediate level between the estimated standard deviation and the estimated error of the mean. The absence of a clear overlap of the error of the mean between neighboring points indicates that the effective error of the mean is larger than calculated for each point. We identified several possible (not mutually exclusive) candidate explanations for the “missing” source(s) of error: (i) when we calculated the “error of the mean,” it was assumed that the data followed Gaussian statistics and the calculation derived from the properties of a Gaussian model; (ii) on the velocity vs. force curve, it is not excluded that the enzyme-to-enzyme variability was still apparent as a variation from point to point; and (iii) the error of the mean obtained by the fit did not take into account the uncertainty on the relative extension factor of the molecule (see the Fig. 3 legend).
The data points for the mean velocity in Fig. 3C are within ±10% of a slightly increasing line, and the RuvAB-mediated branch-migration rate is only weakly force-sensitive: the rate increases slightly or decreases slightly depending on whether the load assists or hinders, respectively, the migration. The estimation of the rate at zero force was obtained by a linear fit to the data (Fig. 3C, dashed line) and was found to be ≈14.6 nm/sec, corresponding to ≈43 bp/sec of branch-migration rate with an error estimate on the average of ±10%.
Discussion
Influence of RuvB Concentration. Influence of the RuvB concentration on the migration process, in the presence of 100 nM RuvA, has been studied and showed mainly two effects: a perturbation of the RuvAB complex activity at high RuvB concentration and an influence on the migration-initiation frequency. First, the perturbation of the RuvAB complex activity at high RuvB concentrations may reflect interactions between RuvB and the migrating complex. Such interactions then could lead to the formation on the branch point of nonefficient overloaded RuvAB complexes eventually containing additional RuvB monomers binding the RuvAB complex or where RuvB hexamers are positioned on adjacent DNA arms (14), the simultaneous activity of which would result in nonproductive work. Alternatively, the destabilization can result from interaction of the migrating complex with RuvB hexamers or RuvAB complexes assembled on the duplex DNA arms. Second, at high RuvB concentration the migration cycle frequency is increased significantly. This effect is mainly caused by a decrease of the waiting time needed for the branch migration to restart after a total backward migration when the branch migration is arrested at the high-extension boundary. This observation shows that, at this extremity, the RuvB hexamer assembly is one of the limiting steps of the reorganization of the RuvAB complex for the migration inversion, and thus starting a new forward migration proceeds by the incorporation of new RuvB molecules (and not by a conservation and rearrangement of the components of the complex, which should be independent of the RuvB concentration). At this position, the reorganization of the complex can thus occur in two possible ways: (i) by an initial dissociation followed by a reassembly of the RuvAB complex or (ii) by the binding of new RuvB on the alternate arms of the junction, forming a transient overloaded complex (14) followed by the dissociation of the former RuvB.
When <150 nM RuvB was injected, we observed very few migrations, which is in agreement with previous studies (30, 31) in which global RuvAB branch-migration activity or ATP hydrolysis were observed to be significantly reduced at RuvB concentration below ≈0.2 μM. This observation may reflect the need for a minimum RuvB monomer concentration for RuvB hexamers to assemble cooperatively.
Influence of RuvA Concentration. In our experiments, Holliday-junction migration has not been observed in the absence of RuvA even at high RuvB concentrations. Noticing that our buffer contained 100 mM NaCl, this result is in agreement with the inhibitory effect of NaCl on the RuvB-only migration activity (30). The initiation frequency was not modulated by the RuvA concentration, indicating that the RuvA–Holliday-junction dissociation-reassembly process may not be the limiting event in an RuvAB–Holliday-junction complex reorganization and/or that the RuvA–Holliday-junction complex is highly stable. One also deduces that an excess of free RuvA tetramers does not sequester RuvB, indicating that the RuvAB complex assembles cooperatively with (or is stabilized by) the Holliday junction. Finally, the absence of perturbing effects on RuvAB migration at high RuvA concentration agrees with the high specificity of RuvA for the Holliday junction [>1,000-fold higher than for duplex DNA (32)] and indicates that any putative interaction of RuvA with linear double-stranded DNA is not stable enough and/or tight enough to impede or block the migration of a RuvAB–Holliday-junction complex.
Processivity and Regularity of the Migration Process. At the RuvA and RuvB concentrations subsequently chosen (100 and 200 nM, respectively), the migration process was quite regular, with some occasional perturbations. The present work demonstrates that RuvAB is highly processive. Blocking of the RuvAB complex, and its rearrangement to restart the migration in the other direction, essentially occur at the extremities of the construction. First, this finding indicates that the bidirectionality of RuvAB observed here does not reflect a capacity of the complex to reverse the migration direction by a spontaneous conformational change, but rather it is induced by some constraints to the migration. Second, it indicates that, in our experiment, the DNA molecule length was the most limiting factor to the processivity of the complex, and we thus obtain a lower bound for the enzyme processivity.
We do not know whether multiple hexamers of RuvB bind to the junction itself. In the condition used, in which the RuvB concentration was not too high, it is probable that two hexamers assemble, although we cannot exclude the possibility of only one hexamer assembling on the junction. Also, the formation of RuvAB complexes along double-stranded DNA had been suggested from experiments with DNA containing no Holliday junction, in which the RuvA/B ATPase activity was found to be stimulated by DNA (9, 21, 31, 32). With the concentration chosen above, the presence of those putative complexes was undetected in our experiments (we initially expected that they might possibly lead to velocity variations near the extremities of the construction if they could be pushed like beads on a string by the junction migration).
Motion of the enzyme was found to be processive through DNA sequences that contain multiple RuvC consensus sequences. Fig. 2 Inset represents the repartition of the RuvC consensus sequences along the DNA arms of the junction. On average, a consensus sequence appears every 64 bases (i.e., approximately every second at branch-migration rates of ≈43 bp/sec), which is below the experimental resolution of velocity changes. However, the repartition density of the RuvC consensus sequences vary substantially from one region to another. If these sequences would induce pauses of RuvAB, we should observe reproducible sequence-dependent variations of the migration rate from one region to another. No such reproducible feature could be seen (compare Figs. 2 and 3 and data not shown), which indicates a sequence independence of the RuvAB migration activity. Hence, this observation strengthens the model of RuvC acting in an RuvABC complex rather than the assumption that RuvAB dissociates from the Holliday junction or pauses at the RuvC consensus sequences to allow efficient cleavage by RuvC.
Velocity of the RuvAB Molecular Motor. The RuvAB-mediated branch-migration rate was measured as a function of the force applied to the tethered Holliday junction. The migration rate was found to be only weakly sensitive to the applied force in the range covered. The average velocity slightly increases (resp slightly decreases) if the force assists (resp hinders) the branch migration. The value determined here for the rate at zero load is ≈43 bp/sec. Tsaneva et al. (33) measured the migration rate on an α-structure intermediate from a gapped circular and a linear DNA of ≈5 kb each. At the beginning of the time course, the Holliday junction was approximately in the middle of the linear fragment so that the arms had an initial length of 2.5 kb. Because it took 2.5–5 min to obtain the products, the branch-point migration velocity was 10–20 bp/sec. Thus, there is a factor ranging from 2 to 4 between the two experimental determinations: 43 bp/sec compared with 10–20 bp/sec. Because this previous study was performed in the absence of salt, we performed control experiments in which no salt was added to the reaction buffer. Within the 10% uncertainty, the same migration rates were obtained as in the presence of salt (data not shown), ruling out this origin for the discrepancy. Therefore, we interpret the difference with Tsaneva et al. (33) by the following: In the present experiment, the migration velocity is obtained directly from active RuvAB complexes. In the bulk experiment, it is not clear whether the molecules (in their majority) taken into account for the rate measurement already have an enzyme loaded and are actively moving the junction right from the beginning after the initial mixing with the enzyme. Reinforcing this argument is the fact that we observed a lag time to wait until a migration starts, probably reflecting the time needed for an active complex to assemble. This waiting time was commonly larger than the time taken to perform the full-length translocation (equivalent to runoff).
In conclusion, this single-molecule study of RuvAB-mediated Holliday-junction migration allows a direct in vitro measurement of the extremely important process of RuvAB-driven genetic-material exchange between two homologous DNA strands and provides information on the processivity and velocity of RuvAB activity.
Acknowledgments
This work was funded by Centre National de la Recherche Scientifique, Ministère de la Recherche, and Paris-6 and Paris-7 Universities.
Abbreviation: DIG, digoxigenin.
References
- 1.Holliday, R. (1964) Genet. Res. 5, 282–304. [Google Scholar]
- 2.Dressler, D. & Potter, H. (1982) Annu. Rev. Biochem. 51, 727–761. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski, S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D. & Rehrauer, W. M. (1994) Microbiol. Rev. 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.West, S. C. (1997) Annu. Rev. Genet. 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 5.Shinagawa, H. & Iwasaki, H. (1996) Trends Biochem. Sci. 21, 107–111. [PubMed] [Google Scholar]
- 6.West, S. C. (1996) J. Bacteriol. 178, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.West, S. C. (1998) Cell 94, 699–701. [DOI] [PubMed] [Google Scholar]
- 8.Parsons, C. A., Stasiak, A., Bennett, R. J. & West, S. C. (1995) Nature 374, 375–378. [DOI] [PubMed] [Google Scholar]
- 9.Shiba, T., Iwasaki, H., Nakata, A. & Shinagawa, H. (1991) Proc. Natl. Acad. Sci. USA 88, 8445–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons, C. A., Tsaneva, I., Lloyd, R. G. & West, S. C. (1992) Proc. Natl. Acad. Sci. USA 89, 5452–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki, H., Takahagi, M., Nakata, A. & Shinagawa, H. (1992) Genes Dev. 6, 2214–2220. [DOI] [PubMed] [Google Scholar]
- 12.Tsaneva, I. R., Illing, G., Lloyd, R. G. & West, S. C. (1992) Mol. Gen. Genet. 235, 1–10. [DOI] [PubMed] [Google Scholar]
- 13.Shah, R., Bennett, R. J. & West, S. C. (1994) Cell 79, 853–864. [DOI] [PubMed] [Google Scholar]
- 14.van Gool, A. J., Hajibagheri, N. M. A., Stasiak, A. & West, S. C. (1999) Genes Dev. 13, 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Gool, A. J., Shah, R., Mezard, C. & West, S. C. (1998) EMBO J. 17, 1838–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hargreaves, D., Rice, D. W., Sedelnikova, S. E., Artymiuk, P. J., Lloyd, R. G. & Rafferty, J. B. (1998) Nat. Struct. Biol. 5, 441–446. [DOI] [PubMed] [Google Scholar]
- 17.Ariyoshi, M., Nishino, T., Iwasaki, H., Shinagawa, H. & Morikawa, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roe, S. M., Barlow, T., Brown, T., Oram, M., Keeley, A., Tsaneva, I. R. & Pearl, L. H. (1998) Mol. Cell 2, 361–372. [DOI] [PubMed] [Google Scholar]
- 19.Panyutin, I. G. & Hsieh, P. (1994) Proc. Natl. Acad. Sci. USA 91, 2021–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panyutin, I. G., Biswas, I. & Hsieh, P. (1995) EMBO J. 14, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, A. H. & West, S. C. (1994) J. Mol. Biol. 243, 208–215. [DOI] [PubMed] [Google Scholar]
- 22.Stasiak, A., Tsaneva, I. R., West, S. C., Benson, C. J. B., Yu, X. & Egelman, E. H. (1994) Proc. Natl. Acad. Sci. USA 91, 7618–7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, X., West, S. C. & Egelman, E. H. (1997) J. Mol. Biol. 266, 217–222. [DOI] [PubMed] [Google Scholar]
- 24.Hiom, K. & West, S. C. (1995) Cell 80, 787–793. [DOI] [PubMed] [Google Scholar]
- 25.Grigoriev, M. & Hsieh, P. (1998) Mol. Cell 2, 373–381. [DOI] [PubMed] [Google Scholar]
- 26.Biswas, I., Yamamoto, A. & Hsieh, P. (1998) J. Mol. Biol. 279, 795–806. [DOI] [PubMed] [Google Scholar]
- 27.Panyutin, I. G. & Hsieh, P. (1993) J. Mol. Biol. 230, 413–424. [DOI] [PubMed] [Google Scholar]
- 28.Strick, T. R., Allemand, J. F., Bensimon, D., Bensimon, A. & Croquette, V. (1996) Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- 29.Gosse, C. & Croquette, V. (2002) Biophys. J. 82, 3314–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, A. H. & West, S. C. (1996) J. Biol. Chem. 271, 19497–19502. [DOI] [PubMed] [Google Scholar]
- 31.Marrione, P. E. & Cox, M. M. (1996) Biochemistry 35, 11228–11238. [DOI] [PubMed] [Google Scholar]
- 32.Dickman, M. J., Ingleston, S. M., Sedelnikova, S. E., Rafferty, J. B., Lloyd, R. G., Grasby, J. A. & Hornby, D. P. (2002) Euro. J. Biochem. 269, 5492–5501. [DOI] [PubMed] [Google Scholar]
- 33.Tsaneva, I. R., Muller, B. & West, S. C. (1992) Cell 69, 1171–1180. [DOI] [PubMed] [Google Scholar]