Abstract
Transthyretin, β2-microglobulin, lysozyme, and the prion protein are four of the best-characterized proteins implicated in amyloid disease. Upon partial acid denaturation, these proteins undergo conformational change into an amyloidogenic intermediate that can self-assemble into amyloid fibrils. Many experiments have shown that pH-mediated changes in structure are required for the formation of the amyloidogeneic intermediate, but it has proved impossible to characterize these conformational changes at high resolution using experimental means. To probe these conformational changes at atomic resolution, we have performed molecular dynamics simulations of these proteins at neutral and low pH. In low-pH simulations of all four proteins, we observe the formation of α-pleated sheet secondary structure, which was first proposed by L. Pauling and R. B. Corey [(1951) Proc. Natl. Acad. Sci. USA 37, 251–256]. In all β-sheet proteins, transthyretin and β2-microglobulin, α-pleated sheet structure formed over the strands that are highly protected in hydrogen-exchange experiments probing amyloidogenic conditions. In lysozyme and the prion protein, α-sheets formed in the specific regions of the protein implicated in the amyloidogenic conversion. We propose that the formation of α-pleated sheet structure may be a common conformational transition in amyloidosis.
Amyloid disease involves the conversion of a protein or peptide from its soluble native state into insoluble amyloid fibrils. There are many different human amyloid diseases linked to a specific precursor protein or peptide (1–3), the most well known being Alzheimer's disease. The most aggressive forms of amyloid disease tend to be linked to pathogenic mutations that increase the buildup of amyloid in affected tissues (4). Transthyretin (TTR), β2-microglobulin (β2m), lysozyme, and the prion protein (PrP) are four of the experimentally best-characterized proteins implicated in amyloid disease (Fig. 1). TTR deposits cause senile systemic amyloidosis and familial amyloid polyneuropathy (5); dialysis and hereditary renal amyloidosis is associated with β2m (6); lysozyme has been implicated in autosomal dominant hereditary amyloidosis (7); and PrP has been linked to transmissible spongiform encephalopathies, including Creutzfeld–Jakob disease and bovine spongiform encephalopathy (8). In addition, many nondisease-related proteins (9) and homopolymers such as poly(l-glutamine) and poly(l-lysine) can form amyloid and protein gels depending on solution conditions (10, 11).
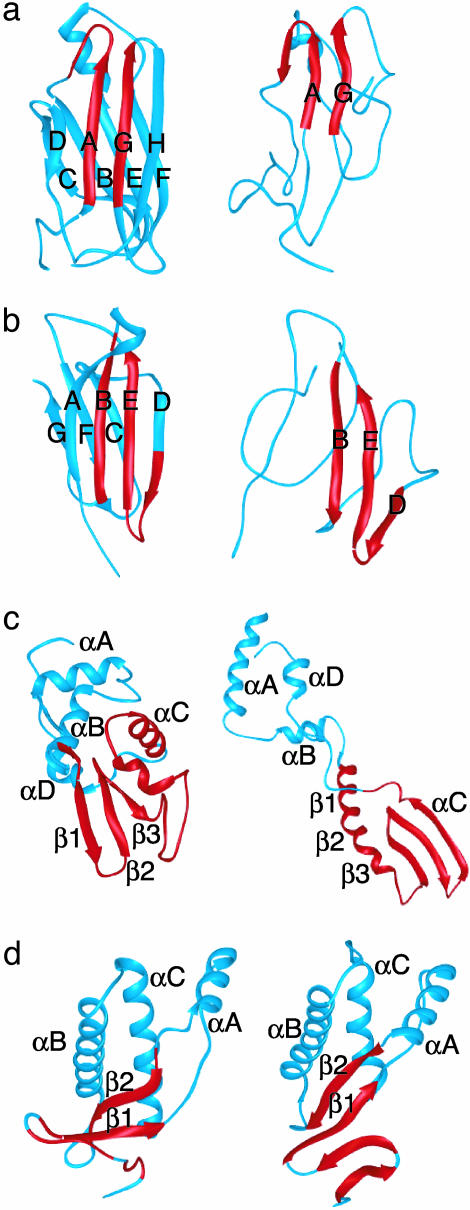
Fig. 1.
MD-generated α-sheet intermediate structures for four amyloidogenic proteins: transthyretin (a), β2m (b), D67H lysozyme (c), and bovine PrP (d). (Left) The native structure. (Right) α-Sheet intermediates from unfolding simulations. Regions shown in red convert to α-extended chain conformations.
Given that many different sequences can form amyloid fibrils of similar architecture, there may be some common structural elements of the prefibrillar amyloidogenic intermediate. It has been shown by synchrotron x-ray fiber diffraction that the final structure of insoluble amyloid fibrils is composed of cross β-pleated sheet secondary structure (12). Therefore, it has been widely held that the formation of amyloid fibrils involves a transition to β-sheet secondary structure in the amyloidogenic intermediate. However, the mechanism of self-assembly at the atomic level remains elusive. Another twist is that soluble oligomeric intermediates, not the insoluble well-ordered fibrils, are responsible for cellular toxicity (13). An antibody was recently identified that is specific for these soluble oligomeric intermediates, but not the insoluble fibrils nor the soluble precursor protein (14). Furthermore, this antibody inhibits toxicity associated with the intermediates, implying a common mechanism of toxicity (14). As this antibody is specific for the soluble oligomeric species of many amyloidogenic proteins and peptides [Aβ42, α-synuclein, islet amyloid polypeptide, poly(l-glutamine), lysozyme, human insulin, and PrP peptide 106–126], they may have a common structure. Based on these observations, the authors (14) proposed that the oligomer-specific antibody recognizes a unique conformation of the polypeptide backbone that is independent of amino acid side chains. Another similar antibody was found to bind specifically to insoluble amyloid fibrils [TTR, islet amyloid polypeptide, β2m, and poly(l-glutamine)] but not to the soluble precursor proteins (15). The authors of this study (15) suggest that if the antibody recognizes a unique polypeptide backbone conformation it must be different from the edge strand β-sheet conformation of TTR, because the antibody does not bind to the native state (15). Based on molecular dynamics (MD) simulations, we propose an intermediate secondary structure in the amyloidogenic transition, the α-pleated sheet. The α-sheet is a possible backbone conformation that may explain these observations.
Pauling and Corey proposed the α-pleated sheet structure, or the “polar pleated sheet” (16, 17), before the parallel and antiparallel β-pleated sheet structures (18). α-Pleated sheet secondary structure is formed by regular hydrogen bonding between adjacent strands in the “α-extended chain” conformation. Rather than being formed by average repeating (φ,ψ) angles, as with the α-helix and β-strand, the α-extended chain conformation is defined by an alternation of residues in the helical, αR, and the αL conformations. A feature of this sheet is the alignment of peptide NH groups on one side of the sheet and carbonyl groups on the other. The α- and β-pleated sheets have the same meridonal repeat distance (4.75 Å) and the same hydrogen-bond distance (2.3 Å). Pauling and Corey (16) provided the coordinates of three α-pleated sheets: one that was flat, one with a 7° rotation, and another with a 20° rotation. They concluded that β-pleated sheets would be less stable flat and more stable with a slight rotation between sheets (18). β-Sheets usually exhibit a 15° rotation between strands, both in protein structures and the cross β-structure of highly ordered amyloid fibrils (12). Flat sheets (1.0–2.5°) have recently been observed in amyloid fibrils by using 3D reconstructions from electron crystallography (19, 20). As α-pleated sheet secondary structure is rare in protein crystal structures, it has not received much attention since its original proposal by Pauling and Corey. Here, we report the formation of α-sheet and α-extended chain conformations in MD simulations of four amyloidogenic proteins (TTR, lysozyme, β2m, and PrP) under amyloidogenic conditions (low pH).
Methods
All unfolding simulations were performed at low pH (Asp, Glu, and His residues protonated) to simulate amyloidogenic conditions. Simulations were performed at high temperature (498 K) to accelerate the folding process. High temperature does not appear to affect the overall pathway of unfolding, but it does accelerate the process (21). All four of the proteins investigated show similar conformational changes at room or physiological temperature at low pH, and the behavior of PrP at low temperature has been described in depth (22, 23).
Six MD unfolding simulations of TTR [Protein Data Bank (PDB) ID code 1tta] (24) were performed at 498 K for 3 ns each. For clarity, we will focus on a single, representative simulation for comparison with the experimental hydrogen-exchange data (25). Details of all six TTR unfolding simulations, as well as 310-K simulations, will be presented elsewhere. Six simulations of β2m (PDB ID code 1lds) (26) were also performed at 498 K for 3 ns each. We focus on a representative simulation for comparison with the experimental hydrogen-exchange data (27). In the simulations of β2m, the native disulfide bond was left intact, as it is known to remain oxidized during the formation of fibrils (28). A single unfolding simulation was performed at 498 K for both WT lysozyme (PDB ID code 1lz1) (29) and the disease-causing variant D67H (PDB ID code 1lyy) (30) for 10 ns each. In the simulations of lysozyme, the four native disulfide bonds were intact, as they remain oxidized during the formation of fibrils (30). A single MD unfolding simulation at 498 K was performed for bovine PrP (PDB ID code 1dx1) (31) (including residues 110–220) for 10 ns. For the simulation of PrP, the native disulfide bond was intact, as it, too, remains oxidized in PrPSc and is necessary for infectivity (32, 33). Control simulations at 298 or 310 K at neutral pH were also performed for each protein.
As another control, simulations of poly(l-Lys) were performed at 276 and 323 K for 10 ns each. Both simulations of a 40-mer started from the ideal poly(Pro) II helix (PII) conformation defined by (φ,ψ) = (–79°, +149°).
All simulations were performed with the program encad (34). The protocols and force field have been described (35, 36). Structures were saved every 0.2 ps for analysis.
Results and Discussion
TTR. In multiple unfolding simulations of TTR (Fig. 1), the DAGH-sheet underwent a transition from β-sheet to α-sheet structure, predominantly over the A and G strands. The average local secondary structure of the native and intermediate states is shown in Fig. 2. The β-sheet → α-sheet transition occurred sequentially through individual transitions of backbone (φ,ψ) angles over 200 ps, rather than all at once in a concerted manner. The transition occurred through the PII conformation (Fig. 3), which is discussed in more depth below. A “peptide-plane flip” like this involves rotation of the peptide plane changing the (φ,ψ) angle of residues i and i+1, with only a minor change in the orientation of the side chains (37). This domino-like effect began on the AG strands and then moved to the H strand, with little change in side-chain packing. In protein crystal structures this transition was predicted in a recent study investigating possible peptide-plane flips, but the most common was the interconversion of the type I and type II β-turn (37).
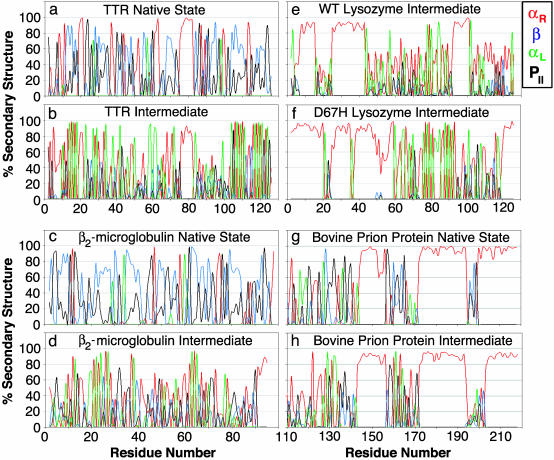
Fig. 2.
Average local secondary structure by residue. (a) Average over the first nanoseconds of the native simulation of TTR at 310 K. (b) Average over the α-sheet unfolding intermediate of TTR. (c) β2m at 310 K. (d) α-Sheet intermediate of β2m. (e) α-Sheet intermediate of WT lysozyme. (f) α-Sheet intermediate of D67H lysozyme. (g) Bovine PrP at 298 K. (h) α-Sheet intermediate of bovine PrP. A residue was classified in a particular conformation if its (φ,ψ) angles were within ± 30° of the average values that follow: αR = (45°, 92°); αL = (–87°, –49°); β-structure (both parallel and antiparallel) = (–165° ≤ φ ≤ –83°) and (89° ≤ ψ ≤ 169°); and PII = (–79°, 149°). Using these definitions there is some overlap between β and PII.
Fig. 3.
Peptide plane flip in the transition from β-to α-sheet structure in TTR. Blue points represent the distribution of (φ,ψ) angles for the entire chain in the TTR α-sheet intermediate. Red dots are the angles sampled for Lys-15 on the A strand during the transition from theβ to theαR conformation. Green dots are for the adjacent residue, Val-16, during the same transition as it converts from β to αL. These residues pass through the right-handed PII and the left-handed PII conformations, respectively. Black dots represent average (φ,ψ) values for: βpar (–119, 113), βanti (–139, 135), αR-helix (–57, –47), αR-extended (–87, –49), αL-helix (47, 57), αL-extended (45, 92), PII(R) (–149, 79), and PII(L) (–79, 149).
α-Pleated sheet secondary structure is formed by regular hydrogen bonding between adjacent strands in the α-extended chain conformation. Rather than being formed by average repeating (φ,ψ) angles, as with the α-helix and β-strand, the α-extended chain conformation is defined by an alternation of residues in the αR and the αL conformations. The (φ,ψ) angles each residue sampled was used to calculate average (φ,ψ) angles over the α-pleated sheet structures obtained in 13 independent TTR simulations at both 310 and 498 K: αL = (45 ± 8°, 92 ± 28°) and αR = (–87 ± 7°, –49 ± 4°) (Fig. 3). The average (φ,ψ) angles do not correspond exactly to ideal values for the helical αL or αR conformations. According to the program procheck (38), our αL conformation is in the “additionally allowed” region and our αR conformation is in the “most favored” region. These angles are not in “disallowed” regions of the Ramachandran plot; in fact, they are in regions that are populated by many common polypeptide conformations. A search of the nonredundant PDB (39) was performed for examples of peptide segments (at least four residues) that exhibit the characteristic alternation of residues in the αL and the αR conformations. The search required that all four residues be within ±45° of ideal α-sheet (φ,ψ) angles: [(φ,ψ) = (45, 90°)] and [(φ,ψ) = (–90°, –45°)], and >40 unique PDB entries were found to have this conformation (data not shown). Another recent study of protein structures in the PDB has shown that a short alternation of residues in the αR and the αL conformations can create anion and cation binding site “nests” in native protein crystal structures (40, 41).
For all six simulations, unfolding intermediates were identified by using projections of the simulation into Cα rms deviation space, where clusters represent periods of time where there is little change in the structure (42). The unfolding intermediates were compared to the experimental hydrogen-exchange data at pH 4.5 (25). An amide hydrogen is predicted to exchange with solvent based on the percentage of simulation time that it hydrogen bonds to water. The single intermediate that best represented the experimental data is shown in Fig. 1a. This intermediate correctly predicts the behavior of 89 of 117 amides used as probes: it correctly predicts 13/14 amides that remain protected at low pH (residues 12–22, 25, 107, and 111) and 10/13 that exchange at pH 4.5 but are strongly protected at pH 5.75 (residues 23, 30, 31–35, 42, 69, 71, 72, 93, and 95). α-Sheet structure provides a unique explanation for the high protection observed in the A and G strands via both the orientation of amide groups in the α-sheets formed between the AG strands and the high frequency of α-sheet structure between the AG strands in the ensemble of intermediates at low pH.
β2m. In the unfolding simulation of β2m, the ABED-sheet underwent a transition from β-to α-sheet structure over a period of 0.1–0.2 ns. Very stable α-sheet structure formed over the B, E, and D strands (Fig. 1). Again, the β-sheet → α-sheet transition occurred sequentially through individual transitions of backbone angles. The α-sheet hydrogen bonding between the B and E strands in this simulation was long-lived. The α-extended chain conformation was also observed with high frequency in either the F or C strands, but there was less hydrogen bonding between these strands. The average local secondary structure of the native and intermediate states is shown in Fig. 2.
The conformational properties of the amyloidogenic intermediate of β2m has been studied experimentally by hydrogen exchange (27), in which 11 residues were strongly protected against urea denaturation at pH 3.6. The simulated amyloidogenic intermediate in best agreement with the data correctly predicts 46/57 amides and is shown in Fig. 1. This intermediate correctly predicts 10/11 amides (residues 22, 23, 26, 61, 64–68, 77, and 88) that remain strongly protected and 36/46 that exchange. Similar to the TTR hydrogen exchange data for the A strand, the α-pleated sheet structure provides an explanation of the high protection observed for residues 61–68 on the E strand of β2m. In the α-pleated sheet, the amide hydrogens from the E strand are strongly protected by hydrogen bonding between the B strand. In all of our unfolding simulations of β2m, the A and G strands are unstructured and predicted to exchange with solvent, in agreement with hydrogen-exchange data for the monomer. Hydrogen exchange has also been investigated in the intact β2m amyloid fibril (43), which shows that residues 20–87 exhibit much greater protection than the A and G strands, suggesting that they are not incorporated into the fibril. Proteolysis of ex vivo β2m amyloid fibrils also agree with this observation and indicate that proteolytic cleavage occurs between residues 1–20 and 87–99, leaving an intact core comprised of residues 20–87 (44).
Human Lysozyme. Hydrogen-exchange experiments monitored by NMR and MS indicate that the D67H mutation alters the stability of the β-domain and the C-helix of lysozyme (45). The rate of hydrogen exchange was >10 times that of WT in the region of residues 39–100 (45). Our unfolding simulations of WT lysozyme compared to the D67H variant are in good agreement with this observation. The D67H simulation predicts much less protection in the region of 39–100 (data not shown). The C-helix was moderately stable in the WT simulations, but was significantly unfolded for the D67H variant (Fig. 1c). In both simulations, there was a complex equilibrium in the unfolded conformations of residues 39–100, giving rise to short segments of α-helix, β-strands, PII, and α-extended chain conformations (Fig. 2). The intact disulfides between residues 65–81 and 77–95 provided a significant constraint to the available conformations in this region. In the unfolding simulation of WT lysozyme, a small α-pleated sheet intermediate formed between residues 65 and 95, although the C-helix did not fully unfolded.
There was much more formation of α-pleated sheet in D67H when the C-helix was unfolded (Fig. 2). α-Sheet formed first between residues 65 and 95, similar to the WT α-sheet (residues 82–87, 74–80, and 65–70). In the D67H α-sheet intermediate there was extensive formation of α-extended chain structure (Fig. 1c). The segments in the region of residues 39–62 were in dynamic, disordered α-extended chain conformations. In these regions, regular α-extended chain structures would be interrupted by residues in the PII or β, or α-helical conformation (the helical conformer is shown in Fig. 1c). In the WT simulation, there was a large population of α-helical conformations over residues 39–60 over the 10-ns average. The formation of these helical segments in residues 39–60 may occur in the unfolded conformational ensemble for the WT protein. Cooperative formation of α-helical structure in the WT protein in unfolded states may protect the protein from amyloid formation, and exchange between α-helix and α-sheet was observed in D67H. A recent study of WT lysozyme with Raman optical activity spectroscopy detected a large population of PII structure in the disordered, unfolded conformations of residues 39–100 under amyloidogenic conditions of 57°C and pH 2 (46), in agreement with our simulations. Fourier transform IR spectroscopy of D67H under physiological amyloidogenic conditions revealed a predominance of β-structure and loss of some helical structure relative to the native state (30).
Bovine PrP. Prion diseases are caused by the conversion of the normal cellular form of the protein, PrPC, to the misfolded protease-resistant isoform known as PrPSc (8). PrPC undergoes a pH-mediated conformational change in the region of pH (4.4–6), with a loss of helix and a gain in β-structure (47). Both CD and Fourier transform IR spectroscopy studies have shown that PrPC is highly helical, whereas PrPSc contains a large amount of β-structure (48, 49). Acid-induced unfolding intermediates have been observed for the recombinant human fragment of residues 90–231 (47). The N-terminal region of the protein has been implicated in conversion to PrPSc, as residues 90–120 are antigenically accessible in PrPC but are masked in PrPSc (50–52). Hydrogen-exchange studies have shown that for the recombinant human protein the most protected amide hydrogens are on the B- and C-helix in the vicinity of the disulfide bond (53). We have previously described MD simulations of the hamster, bovine, and human forms of PrP (22, 23, 54), and we have reported conversion of the 109–129 region into disordered β-structure at low pH.
In the unfolding simulation of bovine PrP, disordered α-pleated sheet structure formed in an early unfolding intermediate (0.3–0.6 ns) (Fig. 1d). This disordered sheet was defined by residues 112–117, 119–123, 126–132, and 158–164. The region of residues 109–129 in the bovine prion was similar to lysozyme (residues 39–100), in that there is a complex equilibrium between short segments of α-helix, β-strands, PII, and α-extended chain conformations (Fig. 2). The largest population of repeating α-extended chain structure was found in residues 126–129 (Fig. 2h). The longest and most persistent segment of the PII segment was within residues 135–139, perhaps because residue 137 is a proline. Amyloidogenic mutations in the N-terminal region of the protein may perturb the equilibrium between segments of PII and α-extended structure. It is interesting that Pro → Leu mutations at residues 102 and 105 cause disease in humans, suggesting that the Pro-associated PII structure may be protective.
Poly(l-Lysine). Vibrational Raman optical activity (ROA) spectroscopy of a β-sheet-rich, reduced isoform of PrP shows that it contains flat sheets not β-helix (11). The amide III region of the ROA spectra was similar to the unusually flat β-sheet of the protein Con A (55), but the amide I and amide II regions are different from normal β-sheet proteins (11). The flat sheets in PrP were also found in the ROA spectra of poly(l-Lys) at pH 11 and 50°C, where β-sheet structure is the dominant conformation. If the ionic strength of a solution of poly(l-Lys) is high enough, it will also form protein gels and amyloid fibrils (11).
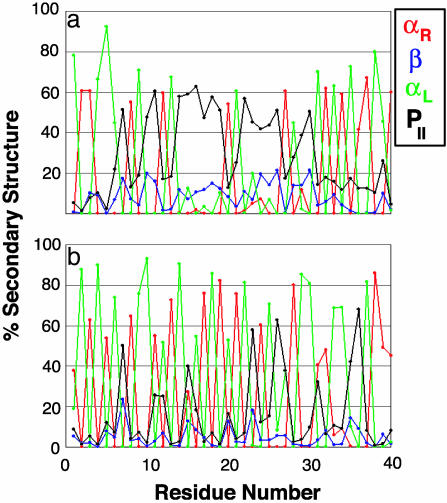
At high pH, there is a temperature-dependent α-helix → β-sheet transition in poly(l-Lys). At low temperatures and neutral pH, it populates PII conformations (56). Here, we have performed two simulations of a 40-mer of poly(l-Lys) starting from the ideal PII conformation, one at 276 K, where α-helix and PII are more favorable, and one at 323 K, where disordered structure and β-sheet are more favorable. A comparison of these two simulations demonstrates the temperature dependence of the equilibrium between the PII, β-strand, and α-extended chain conformation (Fig. 4). At 276 K there is much more repeating PII structure as expected (56). At 323 K there is less PII structure, accompanied by a complex conformational equilibrium between PII and α-extended chain structure, and as shown for TTR (Fig. 3), PII is an intermediate in the conversion of β-sheet → α-sheet. At 323 K there is much more disorder in the chain under conditions where sheet is preferred, and the conformational equilibrium is similar to the unfolded conformations in lysozyme (residues 39–100) and PrP (residues 110–142). This effect is not limited to polyLys; the importance of the PII conformation is becoming increasingly appreciated (57–61).
Fig. 4.
Average local secondary structure for poly(l-lysine). (a) At 276 K. (b) At 323 K. Definitions are provided in the legend for Fig. 2.
We hypothesize that flat β-sheets associated with amyloid emanate from segments of α-extended chain through an α-sheet intermediate. The general temperature dependence of the formation of repeating α-extended structure (favored at higher temperature) compared with repeating PII structure (favored at low temperature) may provide another interpretation, in addition to the hydrophobic effect, for the temperature dependence of nonspecific aggregation.
Implications for α-Pleated Sheet Intermediates in Amyloid Disease. The most noticeable characteristic of α-pleated sheet structure is the alignment of the carbonyl and amide groups participating in hydrogen bonds between the strands forming a “polar pleated sheet” (Fig. 5). This particular alignment of peptide groups also nicely explains the hydrogen-exchange protection observed for TTR and β2m, as well as the lack of protection observed for many amides that should be protected if the protection were caused by β-sheet formation. We have also investigated MD-generated structures of normal globular proteins (chymotrypsin inhibitor 2, protein A, barnase, catechol O-methyl transferase, α-lytic protease, barstar, and the engrailed homeodomain) under similar conditions. In contrast to the amyloidogenic proteins presented here, these normal proteins do not form extensive extended α-sheet structure, although occasional isolated stretches of ≤4 residues form (data not presented). The unique alignment of peptide bonds in α-sheet suggests a mechanism for self-assembly into amyloid.
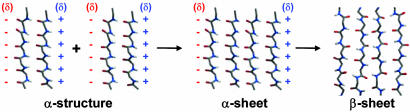
Fig. 5.
α-Sheet intermediate model for self-assembly into amyloid. A main-chain model for an α-sheet is shown with partial charges on the interface (red for negative, blue for positive charges).
Arnsdorf and coworkers (62) proposed that proteins build up a molecular dipole upon encountering low pH conditions, giving rise to attractive forces for linear self-assembly during amyloidosis. The α-pleated sheet structure is a polypeptide backbone conformation that may explain this general dipole model for linear self-assembly. Fig. 5 shows how the partial charges from the peptide backbone create two complementary charged interfaces of opposite charge. As the molecular dipole builds up, individual peptide groups undergo peptide plane flips and align with the overall molecular dipole to form α-pleated sheet. In contrast to β-structure, the peptide groups in α-sheet structure share a similar orientation, as with the α-helix. As described by Worcester (63) and Pauling (64), the diamagnetic anisotropy of the planar peptide groups in an α-helix contributes to a greater total magnetic anisotropy for α-helices compared with β-sheets. The diamagnetic anisotropy (63, 64) of the peptide groups in the α-sheet structure may explain why amyloid fibrils grow more efficiently and in an ordered orientation in the presence of a magnetic field (65, 66). Both amyloid fibrils (65, 66) and α-helices (63, 64) orient with the helical axis parallel to the magnetic field.
We propose that as an amyloidogenic protein unfolds under mildly acidic conditions α-sheet structure facilitates self-association into amyloid protofibrils. Once the soluble protofibrils are formed via an α-sheet intermediate, the transition from α-sheet to flat β-sheet becomes increasingly more favorable as the protofibrils undergo a transition from the cytotoxic soluble phase to insoluble more highly ordered amyloid fibrils (Fig. 5). Several recent solid-state NMR studies suggest that these amyloid fibrils are composed of β-sheet (67, 68). Yet the α-sheet conformation in TTR also reproduces the experimental chemical shifts and atomic distances determined by solid-state NMR of an TTR peptide amyloid (R.S.A., D.O.V.A., and V.D., unpublished work). In addition, the α-pleated sheet appears to be the only backbone conformation that can explain the differential binding of antibodies to soluble oligomeric protofibrils, soluble native structures, and insoluble amyloid fibrils (15).
Although individual amino acids have different propensities for α-sheet structure, the main-chain configuration presented is very similar. The experimental observation that specific peptide fragments are more amyloidogenic than others suggests that there is a sequence propensity for amyloid formation. For TTR, β2m, lysozyme, and PrP, we have observed the formation of α-sheet structure over regions corresponding to the most amyloidogenic peptide fragments. The most amyloidogenic peptide fragments from TTR have been mapped to the A strand (residues 10–19) (69, 70) and G strand (residues 105–115) (71), although peptides derived from the α-helix and the DE loop are also amyloidogenic under certain conditions (72). Amyloidogenic peptide fragments of β2m have been mapped to the B strand, residues 21–31 (73), residues 20–41 (74), and the C-terminal 28 residues (75), which also show some tendency toward α-sheet structure (Fig. 2). The most amyloidogenic fragment of lysozyme (residues 49–64) (76) is from the β-domain, which has been specifically implicated in amyloidogenic conversion. For PrP, amyloidogenic peptide fragments have been mapped to residues 106–147 [residues 106–126 (77); 106–147 (78); 118–135 (79); and 113–120 (80)]; this region is also implicated in conversion to PrPSc. Consideration of α-sheet structure may provide an improved understanding of the sequence propensity for amyloid formation.
Acknowledgments
We thank Drs. David Eisenberg and Neville Kallenbach for helpful comments. We are grateful for financial support from National Institutes of Health Grants R01 GM 50789 (to V.D.) and Training Grants T32 GM 08268 (to R.S.A.) and T32 GM07750 (to M.L.D.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β2m, β2-microglobulin; MD, molecular dynamics; PII, poly(Pro) II helix; PrP, prion protein; TTR, transthyretin; PDB, Protein Data Bank.
References
- 1.Carrell, R. W. & Gooptu, B. (1998) Curr. Opin. Struct. Biol. 6, 799–909. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, J. (1998) Curr. Opin. Struct. Biol. 8, 101–106. [DOI] [PubMed] [Google Scholar]
- 3.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J. S., Taddei, N, Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416, 507–511. [DOI] [PubMed] [Google Scholar]
- 4.Chiti, F., Stefani, M., Taddei, N., Ramponi, G. & Dobson, C. M. (2003) Nature 424, 805–808. [DOI] [PubMed] [Google Scholar]
- 5.Saraiva, M. J. M. (2001) Hum. Mutat. 17, 493–503. [DOI] [PubMed] [Google Scholar]
- 6.Inoue, S., Kuroiwa, M., Ohashi, K., Hara, M. & Kisilevsky, R. (1997) Kidney Int. 52, 1543–1549. [DOI] [PubMed] [Google Scholar]
- 7.Pepys, M. B., Hawkins, P. N., Booth, D. R., Vigushin, D. M., Tennent, G. A., Soutar, A. K., Totty, N., Nguyen, O., Blake, C. C. F., Terry, C. J., et al. (1993) Nature 362, 553–557. [DOI] [PubMed] [Google Scholar]
- 8.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiti, F., Webster, P., Taddei, N., Clark, A., Stefani, M., Ramponi, G. & Dobson, C. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perutz, M. F. (1999) Trends Biochem. Sci. 24, 58–63. [DOI] [PubMed] [Google Scholar]
- 11.McColl, I. H., Blanch, E. W., Gill, A. C., Rhie A. G. O., Ritchie, M. A., Hecht, L., Nielsen, K. & Barron, L. D. (2003) J. Am. Chem. Soc. 125, 10019–10026. [DOI] [PubMed] [Google Scholar]
- 12.Blake, C. & Serpell, L. (1996) Structure (London) 4, 989–998. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 14.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486–489. [DOI] [PubMed] [Google Scholar]
- 15.O'Nuallain, B. & Wetzel, R. (2002) Proc. Natl. Acad. Sci. USA 99, 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauling, L. & Corey, R. B. (1951) Proc. Natl. Acad. Sci. USA 37, 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauling, L. & Corey, R. B. (1951) Proc. Natl. Acad. Sci. USA 37, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauling, L. & Corey, R. B. (1951) Proc. Natl. Acad. Sci. USA 37, 729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez, J. L., Guijarro, J. L., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jimenez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Day, R., Bennion, B. J., Ham, S. & Daggett, V. (2002) J. Mol. Biol. 322, 189–203. [DOI] [PubMed] [Google Scholar]
- 22.Alonso, D. O. V., DeArmond, S. J., Cohen, F. E. & Daggett, V. (2001) Proc. Natl. Acad. Sci. USA 98, 2985–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alonso, D. O. V., An, C. & Daggett, V. (2002) Philos. Trans. R. Soc. London A 360, 1165–1178. [DOI] [PubMed] [Google Scholar]
- 24.Hamilton, J. A., Steinrauf, L. K., Braden, B. C., Liepnieks, J., Benson, M. D., Holmgren, G., Sandgren, O. & Steen, L. (1993) J. Biol. Chem. 268, 2416–2424. [PubMed] [Google Scholar]
- 25.Liu, K., Cho, H. S., Laushuel, H. A., Kelly, J. W. & Wemmer, D. E. (2000) Nat. Struct. Biol. 7, 754–757. [DOI] [PubMed] [Google Scholar]
- 26.Trinh, C. H., Smith, D. P., Kalverda, A. P., Phillips, S. E. V. & Radford, S. E. (2002) Proc. Natl. Acad. Sci. USA 99, 9772–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McParland, V. J., Kalverda, A. P., Homans, S. W. & Radford, S. E. (2002) Nat. Struct. Biol. 9, 326–331. [DOI] [PubMed] [Google Scholar]
- 28.Katou, H., Kanno, T., Hoshino, M., Hagihara, Y., Tanaka, H., Kawai, T., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Protein Sci. 11, 2218–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artymiuk, P. J. & Blake, C. C. F. (1981) J. Mol. Biol. 152, 737–762. [DOI] [PubMed] [Google Scholar]
- 30.Booth, D. R., Sunde, M., Bellotti, V., Robinson, C. V., Hutchinson, W. L., Fraser, P. E., Hawkins, P. N., Dobson, C. M., Radford, S. E., Blake, C. C. F. & Pepys, M. B. (1997) Nature 385, 787–793. [DOI] [PubMed] [Google Scholar]
- 31.Garcia, F. L., Zahn, R., Riek, R. & Wuthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann, L. M. & Caughey, B. (1998) NeuroReport 9, 2457–2461. [DOI] [PubMed] [Google Scholar]
- 33.Turk, E., Teplow, D. B., Hood, L. E. & Prusiner, S. B. (1998) Eur. J. Biochem. 176, 21–30. [DOI] [PubMed] [Google Scholar]
- 34.Levitt, M. (1990) encad: Energy Calculations and Dynamics (Stanford Univ., Palo Alto, CA).
- 35.Levitt, M., Hirshberg, M., Sharon, R. & Daggett, V. (1995) Comp. Phys. Comm. 91, 215–231. [Google Scholar]
- 36.Levitt, M., Hirshberg, M., Sharon, R., Laidig, K. E. & Daggett, V. (1997) J. Phys. Chem. B 101, 5051–5061. [Google Scholar]
- 37.Hayward, S. (2001) Protein Sci. 10, 2219–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 39.Holm, L. & Sander, C. (1998) Bioinformatics 14, 423–429. [DOI] [PubMed] [Google Scholar]
- 40.Watson, J. D. & Milner-White E. J. (2002) J. Mol. Biol. 315, 171–182. [DOI] [PubMed] [Google Scholar]
- 41.Watson, J. D. & Milner-White, E. J. (2002) J. Mol. Biol. 315, 183–191. [DOI] [PubMed] [Google Scholar]
- 42.Li, A. J. & Daggett, V. (1994) Proc. Natl. Acad. Sci. USA, 91, 10430–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332–336. [DOI] [PubMed] [Google Scholar]
- 44.Monti, M., Principe, S., Giorgetti, S., Mangione, P., Merlini, G., Clark, A., Bellotti, V., Amoresano, A. & Pucci, P. (2002) Protein Sci. 11, 2362–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Canet, D., Last, A. M., Tito, P., Sunde, M., Spencer, A., Archer, D. B., Redfield, C., Robinson, C. V. & Dobson, C. M. (2002) Nat. Struct. Biol. 9, 308–315. [DOI] [PubMed] [Google Scholar]
- 46.Blanch, E. W., Morozova-Roche, L. A., Cochran, D. A. E., Doig, A. J., Hecht, L. & Barron, L. D. (2000) J. Mol. Biol. 301, 553–563. [DOI] [PubMed] [Google Scholar]
- 47.Swietnicki, W., Petersen, R., Gambetti, P. & Surewicz, W. K. (1997) J. Biol. Chem. 272, 27517–27520. [DOI] [PubMed] [Google Scholar]
- 48.Caughey, B. W., Dong, A., Bhat, K. S., Ernst, D., Hayes, S. F. & Caughey, W. S. (1991) Biochemistry 30, 7672–7680. [DOI] [PubMed] [Google Scholar]
- 49.Pan, K. M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Huang, Z., Fletterick, R. J., Cohen, F. E. & Prusiner, S. B. (1993) Proc. Natl. Acad. Sci. USA 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williamson, R. A., Peretz, D., Pinilla, C., Ball, H., Bastidas, R. B., Rozenshteyn, R., Houghten, R. A., Prusiner, S. B. & Burton, D. R. (1998) J. Virol. 72, 9413–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peretz, D., Williamson, R. A., Kaneko, K., Vergara, J., Leclerc, E., Schmitt-Ulms, G., Mehlhorn, I. R., Legname, G., Wormald, M. R., Rudd, P. M., et al. (2001) Nature 412, 739–743. [DOI] [PubMed] [Google Scholar]
- 52.White, A. R., Enever, P., Tayebi, M., Mushens, R., Linehan, J., Brandner, S., Anstee, D., Collinge, J. & Hawke, S. (2003) Nature 422, 80–83. [DOI] [PubMed] [Google Scholar]
- 53.Hosszu, L. L. P., Baxter, N. J., Jackson, G. S., Power, A., Clarke, A. R., Waltho, J. P., Crave, C. J. & Collinge, J. (1999) Nat. Struct. Biol. 6, 740–743. [DOI] [PubMed] [Google Scholar]
- 54.DeMarco, M. L. & Daggett, V. (2004) Proc. Natl. Acad. Sci. USA 101, 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deacon, A., Gleichmann, T., Kalb, A. J., Price, H., Raftery, J., Bradbrook, G., Yariv, J. & Helliwell, J. R. (1997) J. Chem. Soc. Faraday. Trans. 93, 4305–4312. [Google Scholar]
- 56.Tiffany, M. L. & Krimm, S. (1972) Biopolymers 11, 2309–2316. [DOI] [PubMed] [Google Scholar]
- 57.Barron, L. D., Blanch, E. W. & Hecht, L. (2002) Adv. Protein Chem. 62, 51–90. [DOI] [PubMed] [Google Scholar]
- 58.Shi, Z. S., Woody, R. W. & Kallenbach, N. R. (2002) Adv. Protein Chem. 62, 163–240. [DOI] [PubMed] [Google Scholar]
- 59.Shi, Z. S., Olson, C. A., Rose, G. D., Baldwin, R. L. & Kallenbach N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keiderling T. A. & Xu, Q. (2002) Adv. Protein Chem. 62, 112–161. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, G., Hecht, L. & Barron, L. D. (1996) Biochemistry 35, 12518–12525. [DOI] [PubMed] [Google Scholar]
- 62.Xu, S. H., Bevis, B. & Arnsdorf, M. F. (2001) Biophys. J. 81, 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Worchester, D. L. (1978) Proc. Natl. Acad. Sci. USA 75, 5475–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pauling, L. (1979) Proc. Natl. Acad. Sci. USA 76, 2293–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fraser, P. E., Duffy, L. K., O'Mally, M. B., Nguyen, J., Inouye, H. & Kirschner, D. A. (1991) J. Neurosci. Res. 28, 474–485. [DOI] [PubMed] [Google Scholar]
- 66.Malinchik, S. B., Inouye, H., Szumowski, K. E. & Kirschner, D. A. (1998) Biophys. J. 74, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antzutkin, O. H., Balbach, J. J., Leapman, R. D., Rizzo, N. W., Reed, J. & Tycko, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tycko, R. (2000) Curr. Opin. Chem. Biol. 4, 500–506. [DOI] [PubMed] [Google Scholar]
- 69.Chamberlain, A. K., MacPhee, C. E., Zurdo, J., Morozova-Roche, L. A., Hill, H. A. O., Dobson, C. M. & Davis, J. J. (2000) Biophys. J. 79, 3282–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacPhee, C. E. & Dobson, C. M. (2000) J. Mol. Biol. 297, 1203–1215. [DOI] [PubMed] [Google Scholar]
- 71.Jaroniec, C. P., MacPhee, C. E., Astrof, N. S., Dobson, C. M. & Griffin, R. G. (2002) Proc. Natl. Acad. Sci. USA 99, 16748–16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gustavsson, A., Engstrom, U. & Westermark, P. (1997) Amyloid Int. J. Exp. Clin. Invest. 4, 1–12. [Google Scholar]
- 73.Hiramatsu, H., Goto, Y., Naiki, H. & Kitagawa, T. (2004) J. Am. Chem. Soc. 126, 3008–3009. [DOI] [PubMed] [Google Scholar]
- 74.Ohhashi, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2004) J. Biol. Chem. 279, 10814–10821. [DOI] [PubMed] [Google Scholar]
- 75.Ivanova, M. I., Gingery, M., Whitson, L. J. & Eisenberg, D. (2003) Biochemistry 42, 13536–13540. [DOI] [PubMed] [Google Scholar]
- 76.Krebs, M. R. H., Wilkins, D. K., Chung, E. W., Pitkeathly, M. C., Chamberlain, A. K., Zurdo, J., Robinson, C. V. & Dobson, C. M. (2000) J. Mol. Biol. 300, 541–549. [DOI] [PubMed] [Google Scholar]
- 77.Forloni, G., Angeretti, N., Chiesa, R., Monzani, E., Salmona, M., Bugiani, O. & Tagliavini, F. (1993) Nature 362, 543–546. [DOI] [PubMed] [Google Scholar]
- 78.Tagliavini, F., Prelli, F., Verga, L., Giaccone, G., Sarma, R., Gorevic, P., Ghetti, B., Passerini, F., Ghibaudi, E., Forloni, G., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 9678–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chabry, J., Ratsimanohatra, C., Sponne, I., Elena, P. P., Vincent, J. P. & Pillot, T. (2003) J. Neurosci. 23, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lundberg, K. M., Stenland, C. J., Cohen, F. E., Prusiner, S. B. & Millhauser, G. L. (1997) Chem. Biol. 4, 345–355. [DOI] [PubMed] [Google Scholar]