Abstract
Two viral proteins, HIV-1 protease and HIV-1 integrase, have been targeted for inhibitor design to prevent assembly and maturation of HIV-1 virions. The enzymatic mechanism of these proteins involves side-chain groups that serve as general acids or bases. Furthermore, catalytic activity requires that water be removed from the microenvironment surrounding the chemical reaction site or be constrained to serve as an activated nucleophile. Here, we identify previously unrecognized structural features that promote water removal from polar catalytic regions. Packing defects in the form of hydrogen bonds that are insufficiently dehydrated intramolecularly, named “dehydrons,” are strategically placed in the structure to induce an anhydrous enzymatic pathway. Dehydrons become electrostatically enhanced and stabilized upon further desolvation. Thus, packing defects act synergistically with the polar active groups to enhance the enzymatic electrostatics. However, because dehydrons are sticky, they constitute targets for inhibitor design. We noticed that inhibitors attach to polar surfaces by further desolvating dehydrons, thus blocking the active sites or the sites involved in harnessing the substrate. The dehydrons are thus required for functional reasons, making them suitable targets. The differences in success when targeting HIV-1 protease, feline immunodeficiency virus protease, and HIV-1 integrase are rationalized in terms of the dehydron distribution, revealing possible improvements in the targeting strategy. Principles of design optimization are proposed to create an inhibitor that can be neutralized only at the expense of the loss of catalytic function. The possibility of using drugs that wrap dehydrons to block protein–protein associations is also discussed.
The removal of water molecules surrounding backbone and side-chain hydrogen bonds is required to guarantee the structural integrity of soluble proteins (1–7) and also places constraints on the allowed conformational changes along folding pathways (8, 9). Backbone and side-chain hydrogen bonds typically prevail provided that nonpolar groups are clustered around them. This “wrapping” (1, 7) provides an anhydrous microenvironment that makes it thermodynamically unfavorable to expose the backbone amide and carbonyl and side-chain polar groups in the nonbonded state. Thus, soluble protein structure prevails by keeping its hydrogen bonds “dry in water.” However, the hydrogen bonds that are intramolecularly underdehydrated, or overexposed to the solvent, named “dehydrons” (2, 3), constitute structural markers for protein reactivity. This property was demonstrated experimentally (10) as well as statistically by examination of protein–protein interfaces and supramolecular protein assemblies (1, 2). Dehydrons are inherently sticky (10), a property that finds an energetic and a thermodynamic basis: The partial charges of the polar backbone and side-chain groups are descreened as surrounding water is removed, and, in turn, water removal destabilizes the nonbonded state (or equivalently stabilizes the bonded state) by preventing hydration of the polar groups.
Many enzymatic reactions involving nucleophilic attack on scissile bonds become more efficient when surrounding water can be removed to enhance the electrostatic interactions. Occasionally, especially in hydrolysis, a few water molecules must be selectively confined to participate in the reaction. Because dehydrons promote the removal of surrounding water, it is expected that they could play a significant role in shaping the microenvironments at the active site. We explore this aspect in this study, especially in connection with designing inhibitors of catalytic function or protein–protein associations.
Many enzymes involve polar side-chain groups that can serve as general acids and bases as they interact with the substrate in a concerted or multistep fashion. The aspartyl proteinase HIV-1 protease (11–13) and the HIV-1 integrase (14–16) are examples of such enzymes. These proteins have been targeted in inhibitor drug design geared at preventing the full assembly and maturation of HIV-1 virions (17, 18) in AIDS therapy. Partial water exclusion from the microenvironment around the chemical reaction site, whether it is involved in hydrolysis, transphosphoesterification, proton donor-acceptor chemistry, etc., is important to ensure the efficiency of the enzymatic mechanism. In this regard, surface nonpolar groups flanking the active polar groups (see figure 1 of ref. 1) might become useful. However, when the groups interacting with the substrate are themselves polar and no nearby hydrophobic patches assist the enzymatic activity by inducing water removal, an alternative structural feature, the dehydron, could become a primary contributor to the shaping of the functional microenvironment. Being a severely underdehydrated hydrogen bond, the dehydron favors removal of surrounding water without itself engaging nonpolar groups; thus, dehydrons are commonly found in sparsely structured polar surface regions such as those representing enzymatically active sites.
This finding implies that dehydrons may act concurrently and synergistically with polar catalytic groups at the active sites by inducing the descreening of the charges. At the same time, because the dehydron is sticky, it is expected that it would represent a specific and efficient target for inhibitor drug design, as the evidence presented here reveals. This previously uncharacterized structural marker may aid drug design and improve its efficiency.
Methods
To identify the dehydrons in a domain fold, multidomain chain, or complex in one-chain or multiple-chain Protein Data Bank (PDB) entries, we use the program hb desolvator (a rudimentary earlier version may be found at http://sosnick.uchicago.edu/aifoldlab/YAPView/YAPView.html), which identifies dehydrons in a PDB structure according to the following premises. The extent of intramolecular hydrogen-bond desolvation in a monomeric structure is evaluated quantitatively by determining the number of nonpolar groups within a desolvation domain. This domain is defined as two intersecting balls of fixed radius centered at the α-carbons of the hydrogen-bonded residues (2). If we examine complexes or multimers, the count includes nonpolar groups from the monomer as well as those from its binding partner(s). The number of nonpolar groups within the desolvation domain of a hydrogen bond assesses in a simplified manner the local dielectric environment or, equivalently, the charge screening of the paired polar groups (2, 8).
The statistics of hydrogen-bond wrapping vary according to the desolvation radius adopted, but the tails of the distribution invariably single out the same dehydrons in a given structure over a 5.8- to 6.3-Å range in the adopted desolvation radius. For this study, the value 6 Å was chosen. Residues contributing to the wrapping of a backbone hydrogen bond must contain side-chain nonpolar groups at least up to the γ-position to favor or promote the clustering of desolvating nonpolar groups around the bond. This requirement excludes from the counting the “poor backbone wrappers,” G, A, D, N, S, and T (19).
In most stable protein folds (≈92% of PDB entries), at least two-thirds of the backbone hydrogen bonds are wrapped on average by r = 27.1 ± 7.5 nonpolar groups (or 14.0 ± 3.7 if we count only side-chain groups and exclude those from the hydrogen-bonded residue pair itself). Thus, dehydrons are the hydrogen bonds in the tails of the distribution, i.e., with 12 or fewer nonpolar groups in their desolvation domains (their r value is no greater than the mean – 2 Gaussian dispersions).
The microenvironments around side-chain hydrogen bonds on the protein surface are far more difficult to assess precisely because of the relatively large local B factors of side-chain carbon atoms for polar surface residues in the PDB (typically 2- to 3-fold larger than those for α-carbons). These larger B factors introduce a significant uncertainty to the evaluation of the side-chain hydrogen-bond microenvironment. For example, a surface lysine with a B factor of 60 in the α-carbon introduces an uncertainty of 3 in the wrapping r value of an adjacent side-chain hydrogen bond. This uncertainty notwithstanding, the criterion for identifying side-chain dehydrons adopted here is the same as that used for backbone hydrogen bonds, with the caveat that the reliability of the assertion diminishes as it becomes modelspecific.
The dehydron, then, is identified as an insufficiently wrapped hydrogen bond. A more rigorous characterization takes into account the sensitivity of the hydrogen-bond coulombic energy to the removal of surrounding water or a decrease in solvent polarizability (2). Thus, our criterion introduces a sufficient condition for the existence of a dehydron. Although being a satisfactory approximation, this criterion may miss the specific case in which the number of nonpolar wrappers exceeds 12, but they are unevenly distributed in space around the hydrogen bond.
Results
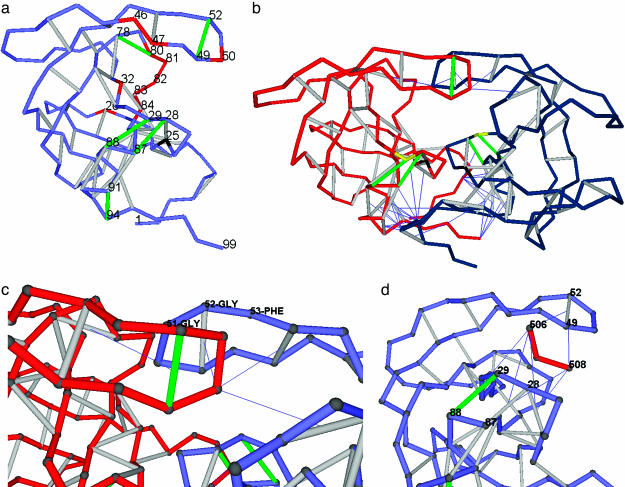
Packing Defects as Drug Targets in HIV-1 Protease. The HIV-1 protease, an aspartyl proteinase, is an obligatory homodimer in its active functional form (11–13), with two equivalent D25 residues acting in opposite ways as general acid and base to catalyze the hydrolysis of the substrate peptide bond. A thermodynamic factor favoring water exclusion becomes essential to ensuring that the substrate is properly anchored and manipulated and that the electrostatics of the nucleophilic attack are properly enhanced along the enzymatic pathway. Thus, it is not surprising to find a cluster of hydrophobic residues, L23, L24, and I84, surrounding the catalytic site D25 (compare with figure 1 of ref. 1). However, the substrate polypeptide must be anchored by the active residues D29 and D30 and dragged along with the aid of a gating mechanism that makes use of the flap 46–55 region (Fig. 1a). This pivoting of the substrate bears resemblance to previously discovered modes of substrate anchoring around the active site of α-chymotrypsin (20). In the acid protease, no hydrophobic patch is present either at the rim of the catalytic pocket or at the flap. Furthermore, because the active residues are themselves polar, removal of surrounding water cannot be promoted by their sole presence. The distribution of dehydrons in the protein (Fig. 1a) reveals how packing defects are strategically placed in the structure to favor the exclusion of surrounding water where needed for the enzymatic activity.
Fig. 1.
Distribution of dehydrons in HIV-1 protease monomer and dimer, flap asymmetry in the wrapping, and the inhibitor as a wrapper of packing defects. (a) Distribution of dehydrons in a monomeric HIV-1 protease unit. The backbone is represented as a light blue chain made up of virtual bonds joining α-carbons. Well wrapped backbone hydrogen bonds (see Methods) are indicated as gray segments joining α-carbons, and dehydrons are marked in green. The catalytically active D25 is shown in black, and the most significant residues undergoing site mutation associated with drug resistance or substrate specificity are shown in red. The dehydrons, together with their functional roles, are as follows: (G49, G52), flap flexibility, dimerization inducer; (G78-T80), induces dehydration of catalytic core; (A28, R87), stickiness of substrate-harnessing region and dimerization inducer; (D29, N88), stickiness of substrate-harnessing region; and (T91, G94), dimerization inducer. (b) Homodimeric HIV-1 protease (PDB entry 1A30) adopting the same virtual-bond backbone representation as in a, except that one monomer is shown in red and the other is shown in blue. Only intermolecular wrapping is shown. Thus, the line joining the α-carbon of a residue in a monomer and the center of a backbone hydrogen bond on the other monomer indicates penetration upon dimerization of at least one nonpolar group of the residue side chain into the desolvation domain of the hydrogen bond. Some dehydrons become well wrapped hydrogen bonds upon dimerization. The remaining dehydrons dictate the pathway of the peptide-chain substrate through the HIV-1 protease. The substrate-anchoring residue D29 is marked in yellow, and the catalytic D25 is marked in black, as above. (c) Detail revealing the broken symmetry upon induced fit in HIV-1 dimerization. The flap angle defined by α-carbons at positions G51, G52, and F53 is distorted in one of the monomers to better wrap the flap dehydron of the other intermolecularly. (d) The tripeptide inhibitor EDL (red, residues 506–508) wrapping the dehydrons (G49, G52), (A28, R87), and (D29, N88) in one monomer of HIV-1 protease within the dimer (PDB entry 1A30). Only (D29, N88) remains a dehydron (green) even upon further wrapping provided by the inhibitor.
The backbone dehydrons in the HIV-1 protease monomer are as follows: (G49, G52), (G78, T80), (A28, R87); (D29, N88), and (T91, G94). In addition, one side-chain dehydron engaging D29 and N88 may be predicted with the highest confidence because its wrapping is <12 (desolvating groups) even when taking into account the uncertainty introduced by the large B factors for solvent-exposed side chains (see Methods). Note that, with the sole exception of R87, all dehydrons involve residues known to be poor wrappers (G, A, D, N, S, or T) (19).
The dehydrons (G49, G52), (G78, T80), and (T91, G94) are determinants of the HIV-1 protease dimerization (1), as revealed by comparing Fig. 1 a and b. This association arises because the nonpolar side-chain groups of a monomer contribute to further and favorably desolvate the dehydrons of the other monomer. Thus, the dehydrons become well wrapped hydrogen bonds upon dimerization. This intermolecular wrapping is depicted in Fig. 1b by thin lines joining the α-carbons of the wrapper residues with the center of the hydrogen bonds, which are being intermolecularly dehydrated. Thus, one functional role of these dehydrons arises because HIV-1 must dimerize to become active.
The importance of underdesolvated hydrogen bonds in driving protein–protein associations is illustrated as follows (1). The nonpolar groups of one binding partner contribute to dehydrate the intramolecularly underdehydrated hydrogen bonds of the other partner, and, in so doing, they electrostatically enhance and stabilize such hydrogen bonds. If examined by using standard arguments, the dimer interface can be seen to involve seemingly inconsistent residue mismatches, such as W6 of one monomer in the proximity of T91 and G94 of the other monomer. However, W6 is actually dehydrating the (T91, G94) dehydron intermolecularly, and, in so doing, it is adding stability to the monomer. Similarly, I50 in one monomer dehydrates the dehydrons (G49, G52) and (G78, T80) of the other monomer intermolecularly.
It should be noted that there is a symmetry-breaking-induced fit upon dimerization (Fig. 1c), as clearly determined by computing the wrapping of PDB entry 1A30 for the HIV-1 protease. This distortion takes place in one monomer as its flap wraps the (G49, G52) dehydron of the other monomer. Thus, one flap remains slightly more flexible in the dimer, as needed for processing, whereas the other becomes slightly rigidified. The reported B factors in 1A30 reflect this observation; thus, the B factors for G52 are 11% larger in chain B than in chain A, and similar discrepancies are found for G49 and I50.
The flexibility of the 46–55 flap requires that its backbone hydrogen bond be partially exposed to water. Thus, the underwrapping of the dehydron (G49, G52) appears as a necessary design feature. However, the feature that confers flexibility also confers stickiness (10) to the flap, as mechanistically needed to drag the substrate peptide chain along the catalytic site. This stickiness suggests a strategy for inhibitor design by wrapping the packing defects.
The functional role of the protease dehydrons can be clearly identified as follows: (G49, G52) confers flap flexibility and is a dimerization inducer (Fig. 1 a and b); (G78-T80) promotes dehydration at the catalytic core (Fig. 1a); (A28, R87) provides stickiness to the substrate-harnessing region and acts as a dimerization inducer (Fig. 1 a and b); (D29, N88) provides stickiness to the substrate-harnessing region (Fig. 1a); and (T91, G94) is a dimerization inducer (Fig. 1 a and b). It should be noted that the pair (D29, N88) is engaged in two dehydrons, one engaging the backbone and the other engaging the side-chain polar groups. Being placed right on the protein surface, the latter side-chain dehydron substantially enhances the stickiness required to anchor the substrate. The spatial orientation of the peptidic substrate chain can be clearly induced by the prevailing dehydrons in the dimeric structure, as shown in Fig. 1b. Note that both (D29, N88) dehydrons prevail in the dimer. It is also worth noting that all residues whose site mutation impacts drug resistance and substrate specificity (marked in red in Fig. 1a) (16) are actually wrappers of the dehydrons in HIV-1 protease, and, thus, amino acid substitution at such sites modulates the sensitivity of the protein to removal of surrounding water (2, 21).
As shown previously, dehydrons constitute highly specific sites for protein–ligand and protein–protein association (1, 2). The fact that they occur precisely at the active-site region for substrate harnessing and at the flap region in the HIV-1 protease makes this molecule an ideal target for inhibitor design. A previously uncharacterized underlying inhibitor strategy is suggested by the thermodynamically and energetically favorable wrapping of packing defects. Thus, the inhibiting EDL-tripeptide (22) wraps all four catalytically important dehydrons in HIV-1 protease: (G49, G52), (A28, R87), and the backbone and side-chain (D29, N88). This intermolecular desolvation is shown in Fig. 1d. In this case, the functionality of the dehydrons as inducers of local water removal or by conferring flexibility to the flap is precisely the property that enables effective drug design. All four dehydrons occur at catalytically active sites, and, because they are inherently sticky (10) and possess intersecting desolvation domains (see Methods), such sites can be blocked simultaneously with a single inhibitor, as shown in Fig. 1d.
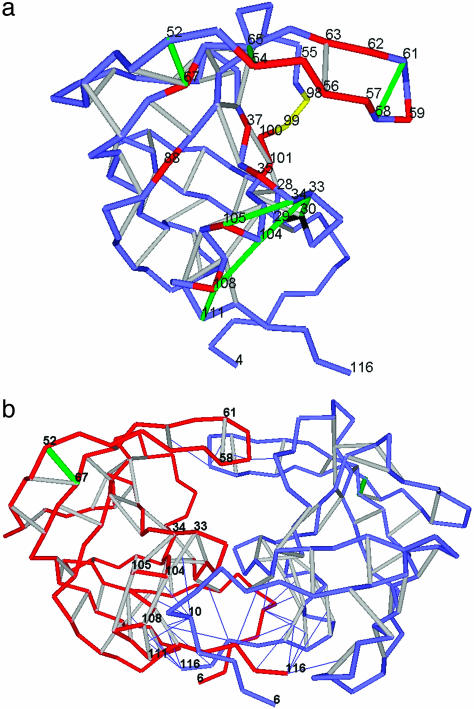
Impairing Feline Immunodeficiency Virus (FIV) Protease by Wrapping Its Unfavorable Protein–Water Interfaces. Given their lower specificity and affinity, drug inhibitors of the homologous FIV protease have not been nearly as successful as those for HIV-1 protease (17, 23). The dehydrons in the monomeric state (PDB entry 4FIV) all engage backbone polar groups and are displayed in Fig. 2a. They are as follows: (G58, G61), (G52, N67), and (Q54, G65) in the flap region; (D30, A33) and (A33, R104) in the catalytic region; (D34, D105) in the substrate-harnessing region; and (R104, I108) and (I108, N111) serving as dimerization promoters. With the exception of dehydron (G52, N67), all dehydrons can be clearly assigned functional roles in strategically inducing water exclusion. (G58, G61) and (Q54, G65) confer flexibility to the flap; (D30, A33) acts jointly with the nonpolar residues L28, L29, and L101 to ensure the dehydration of the catalytic site D30 (marked in black in Fig. 2a); (D34, D105) and (A33, R104) favor water removal as they harness the substrate polypeptide chain; and all dehydrons except (G52, N67) act as anchors in the dimerization of the protease (Fig. 2b). All residue sites assumed to affect substrate and inhibitor specificity (17) are marked in red in Fig. 2a if they are engaged in the wrapping of a dehydron and in yellow if not. With the exception of I98 and Q99, which represent catalytically active sites (17), amino acid substitution in the remaining residues marked in red in Fig. 2a affect the enzymatic activity by modulating the sensitivity of the active site to water removal, as inferred from the distribution of dehydrons along the flap and catalytic rim (Fig. 2a).
Fig. 2.
Packing defects in monomeric and dimeric (functional) FIV protease. (a) Dehydron distribution in FIV protease. The convention of Fig. 1 is followed. Mutation sites that affect substrate and inhibitor specificity (17) are marked in red if the substitution affects the wrapping of a dehydron and in yellow otherwise. (b) Dehydron distribution in the active dimeric FIV protease (PDB entry 3FIV). The intermolecular wrapping of dehydrons by L10 is highlighted. One monomeric chain (A) is depicted in red, and the other chain is depicted in blue. The color convention is the same as in a for dehydrons and the catalytic residue.
The dimer active form of the FIV protease, with its equivalent catalytic D30 residues playing opposing roles as general acid and base in the nucleophilic hydrolyzing attack, contains only one backbone dehydron, (G52, N67). This dehydron is of no direct relevance to enzymatic activity, unless some hitherto-unknown allosteric pathway is invoked. The highly exposed nonpolar residue L10 acts as the major intermolecular wrapper, as shown in Fig. 2b. The lack of dehydrons around the active site and flap in the FIV protease dimer poses a major difficulty in designing specific inhibitors, because the thermodynamically unfavorable protein–water interface is restricted to the localized region with exposed nonpolar residues L28, L29, and L101 around the catalytic D30 (Fig. 2a). No dehydron on the substrate-harnessing track or on the flap contributes to the purported inhibitor binding or pivoting, and, thus, the restricted nature of the binding hot spot reduces the inhibitor specificity.
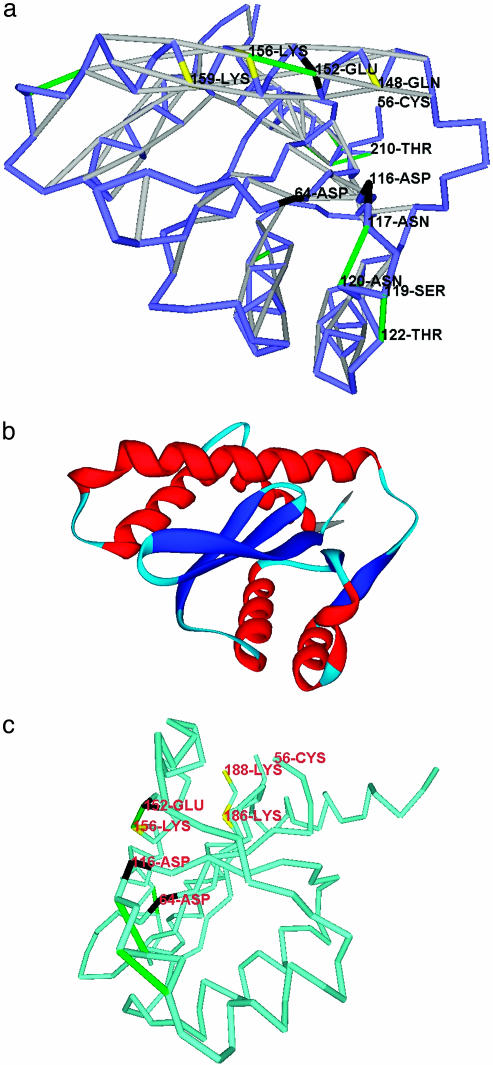
Previously Uncharacterized Drug Epitopes for HIV-1 Integrase. HIV-1 integrase (PDB entry 1B9F), the monomerically active enzyme that integrates viral DNA with the host-cell DNA, is an important inhibitor target in antiviral therapy because it has no equivalent counterpart in the human host cell. Its catalytic residues D64, D116, and E152 form a triad of general acids/bases (16, 18), which have been targeted by drug designers with low or moderate success. The integrase inhibitor 5CITEP and similar drugs (16) bind longitudinally along the major groove of the integrase, parallel to the major 146–164 helix (Fig. 3 a and b). The functionally active residues along this groove (Q148, E152, K156, and K159) are essentially polar. Thus, the removal of water surrounding this region is not “conventionally” induced by the presence of hydrophobic patches on the protein surface and can be promoted only by the presence of the backbone (E152, K156)-dehydron and the side-chain (Q148, E152)-dehydron. The backbone (E152, K156)-dehydron actually serves as the epitope for inhibitor binding (16, 18, 24). The strategic location of this dehydron fully justifies the docking mode of the inhibitor. The additional side-chain dehydron (Q148, E152) can be predicted with high confidence (see Methods) and, because of its central location and orientation along the major groove, also could have been targeted to contribute to the inhibitor docking.
Fig. 3.
Packing defects in the stable fold of HIV-1 integrase. (a) Dehydron distribution for HIV-1 integrase (PDB entry 1B9F). The catalytic residues are marked in black, and the other active residues are marked in yellow. Dehydron (E152, K156), conventionally marked in green, constitutes a major epitope anchoring inhibitor drugs that dock along the major groove region parallel to the 146–164 helix. (b) Ribbon rendering of the HIV-1 integrase as a visual aid. Red, helices; blue, α-strands; light blue, turns and loopy regions. (c) The HIV-1 integrase positioned as in figure 4 of ref. 23, with the dehydrons (N117, N120) and (S119, T122) (a) defining the anchoring track for the viral DNA and the (E152, K156) defining the track for the host-cell DNA. The residue color convention is that of a, and the virtual-bond chain is displayed in lighter blue for visualization purposes.
The distribution of dehydrons in the integrase clearly marks two tracks along which water removal is favored. Although the host DNA is docked along the (E152, K156) dehydron, the viral DNA is anchored by two backbone dehydrons, (N117, N120) and (S119, T122), which, with seven nonpolar wrapping groups each, constitute the most underwrapped backbone hydrogen bonds in the PDB. A third, side-chain dehydron engaging the pair (N117, N120) reinforces the harnessing of the viral DNA.
The two backbone dehydrons (N117, N120) and (S119, T122) and the predicted side-chain dehydron (N117, N120) have not been targeted by any drug inhibitor thus far, although their functional role seems to be paramount to properly orient and attach the viral DNA (compare with figure 4 of ref. 24), as Fig. 3c shows. Thus, rather than merely targeting the (E152, K156) dehydron, we propose an extended inhibitor-binding epitope defined by the two dehydrons (E152, K156) and (S119, T122) and the backbone and side-chain dehydrons involving the pair (N117, N120) around the catalytic triad. This increase in the pivoting binding surface for the inhibitor beyond the (E152, K156) dehydron should ensure a higher specificity and affinity. The need to wrap multiple dehydrons points to the need for macromolecular, rather than small-molecule, inhibitors.
However, a single molecule that could wrap all four dehydrons simultaneously is likely to be problematic in regard to in vivo absorption and transport because of its size and molecular weight. Thus, the (S119, T122) dehydron and the two (N117, N120) dehydrons should be regarded as alternative targets for inhibitor design, suggesting a combination drug treatment including standard inhibitors wrapping the (E152, K156) dehydron and a separate wrapping drug for the remaining dehydrons.
Discussion
This paper addresses the problem of elucidating how protein enzymes may specifically induce the removal of water around catalytically competent polar residues by strategically positioning deficiently packed electrostatic interactions. Surrounding the catalytic polar residue with nonpolar residues to create a thermodynamically unfavorable protein–water interface (see figure 1 of ref. 1) seems an obvious possibility to selectively promote water exclusion because such organizations descreen the catalytic site. In this way, the presence of vicinal nonpolar groups enhances the intermolecular electrostatic interactions. However, when several spatially adjacent polar groups are clustered to shape the enzymatic site, the presence of nearby nonpolar groups becomes a destabilizing factor, and, thus, an alternative factor is required to functionalize the polar groups. This factor is the dehydron, an intramolecular solvent-exposed hydrogen bond. The dehydron is electrostatically enhanced and stabilized by promoting water removal and, thus, acts synergistically with the enzyme intermolecular electrostatics. Therefore, dehydrons induce the descreening of the side-chain charges, as required for their effective participation in enzymatic activity.
The two HIV-1 and FIV viral enzymes investigated reveal different problems faced by inhibitor designers in trying to block catalytic activity. First, we have noticed that dehydrons can be regarded as structural markers for protein–protein and protein–ligand association. This role arises because dehydrons become electrostatically enhanced and stabilized upon further removal of surrounding water. Thus, dehydrons are sticky or, equivalently, promote dehydration, much like nonpolar groups. These markers have been overlooked in current approaches to inhibitor design, although, as this paper shows, the effective inhibitors partially wrap or “correct” the intramolecular packing defects of the protein enzymes. Second, the distribution of dehydrons on the surface of the viral proteins considered differs radically, thus posing different and hitherto unforeseeable difficulties to the drug designer.
Thus, the functionality of dehydrons as modulators of the solvent environment (dehydrators) for the enzymatic activity also makes them good candidates for drug targeting. This functionality arises because dehydrons have been evolutionary placed at the active site of the enzyme catalysts to guarantee enzymatic efficiency by promoting the formation of an anhydrous environment. Thus, effective inhibitors may attach to polar surfaces by wrapping dehydrons.
This perspective clearly leads to a strategy to identify hot spots for inhibitor binding based on the location of functional packing defects in protein enzymes. The results reveal how to improve the inhibitor binding specificity and affinity for the HIV-1 integrase by focusing on heretofore overlooked packing defects along the viral DNA track.
This work dealt with the discovery of previously uncharacterized markers for protein–ligand association in the form of dehydration-promoting packing defects. Because of their inherent stickiness, the same markers have been shown to be important factors driving protein–protein associations, reconciling seemingly inconsistent hydrophobic polar residue mismatches on interfaces. Thus, the same physicochemical principles that guided this study could be applied to justify the adoption of dehydrons as specific targets to block protein–protein interactions. This mode of antagonism constitutes a challenge in drug discovery (25) because it is difficult to design a small molecule that competes with a protein in binding another protein. However, this structural marker for protein association might represent a relatively small target worthy of exploration. The same electrostatic considerations single out underdehydrated or partially exposed intramolecular salt bridges as possible hot spots for the antagonism of protein–protein associations (25): Underdehydrated salt bridges promote removal of surrounding water and, thus, should be sticky.
The assessment of a hydrogen-bond microenvironment and the identification of dehydrons require a simple calculation based on the atomic coordinates in the PDB structure. The results of this analysis facilitate the a priori prediction of binding sites because dehydrons promote the exclusion of surrounding water (1, 2, 10). However, dehydrons have been shown to be evolutionarily conserved and therefore functionally relevant structural elements (3). The combination of these two properties has clear implications for drug design. Mutations fostering drug resistance are unlikely to affect the dehydron pattern of the protein because such an alteration would impair the protein reactivity, and, thus, the mutation is unlikely to be inheritable. In fact, none of the 27 mutations contributing to drug resistance in HIV-1 protease (17) change the dehydron pattern of this protein. These observations suggest that the inhibitors could be effectively engineered to block protein interactions even in highly mutating targets if they are designed to wrap the evolutionarily conserved dehydrons.
It has been shown that inhibitors of the catalytic function or reactivity associated with a protein fold effective across species are not likely to elicit drug resistance by amino acid mutation (26). Thus, because dehydrons are highly conserved and promoters of interactions (3), it makes sense that “universal inhibitors” (i.e., those effective across species and not eliciting drug resistance) should be designed precisely to wrap such dehydrons. Such inhibitors are expected to be rigid in the region that wraps dehydrons and conformationally flexible or adaptable elsewhere, as required to accommodate amino acid variability (27, 28) in the target protein.
The “universal inhibition” may be effective across species only if the dehydrons are conserved in the functional states of the proteins, which are not necessarily monomeric. Thus, the catalytically relevant substrate-anchoring dehydrons (28, 87) and (29, 88) in HIV-1 protease are conserved (3) in the monomeric FIV protease, becoming the dehydrons (33, 104) and (34, 105), respectively (Figs. 1a and 2a). However, they are not present in the dimeric FIV protease, i.e., in the functional form of this protein (Figs. 1b and 2b), which has evolved to better wrap such bonds. For this reason, we expect that an HIV-1 protease inhibitor that wraps the substrate-anchoring dehydrons in HIV-1 protease will not be as effective with FIV protease.
The same physicochemical principles that serve to guide inhibitor design could be applied to build more efficient enzymes. Thus, the selective amino acid substitution of a good wrapping residue for a poorer wrapper may result in the creation of a dehydron that can promote water removal at an enzymatic site or enhance the anchoring of the substrate, thereby improving enzymatic efficiency.
Acknowledgments
A.F. thanks R. Stephen Berry for insightful discussions. A.F. acknowledges support from the Indiana Genomics Initiative and an unrestricted grant from Eli Lilly. H.A.S. was supported by National Institutes of Health Grant GM-14312 and National Science Foundation Grant MCB00-03722.
Abbreviations: FIV, feline immunodeficiency virus; PDB, Protein Data Bank.
References
- 1.Fernández, A. & Scheraga, H. A. (2003) Proc. Natl. Acad. Sci. USA 100, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernández, A. & Scott, R. L. (2003) Biophys. J. 85, 1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández, A. (2004) J. Mol. Biol. 337, 477–483. [DOI] [PubMed] [Google Scholar]
- 4.Nemethy, G. & Scheraga, H. A. (1962) J. Chem. Phys. 36, 3382–3400. [Google Scholar]
- 5.Nemethy, G. & Scheraga, H. A. (1962) J. Chem. Phys. 36, 3401–3417. [Google Scholar]
- 6.Nemethy, G. & Scheraga, H. A. (1962) J. Phys. Chem. 66, 1773–1789. [Google Scholar]
- 7.Fernández, A. & Berry, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández, A., Sosnick, T. & Colubri, A. (2002) J. Mol. Biol. 321, 659–675. [DOI] [PubMed] [Google Scholar]
- 9.Fernández, A., Kardos, J. & Goto, Y. (2003) FEBS Lett. 536, 187–192. [DOI] [PubMed] [Google Scholar]
- 10.Fernández, A. & Scott, R. L. (2003) Phys. Rev. Lett. Vol. 91 (July 1). Available at http://link.aps.org/abstract/PRL/v91/e018102. Accessed July 12, 2004.
- 11.Navia, M. A., Fitzgerald, M. D. P., McKeever, B. M., Leu, C. T., Heimbach, J. C., Herber, W. K., Sigal, I. S., Darke, P. L. & Springer, J. P. (1989) Nature 337, 615–620. [DOI] [PubMed] [Google Scholar]
- 12.Wlodawer, A., Miller, M., Jaskolski, M., Sathyanarayana, B. K., Baldwin, E., Weber, I. T., Selk, L. M., Clawson, L., Schneider, J. & Kent, S. B. H. (1989) Science 245, 616–621. [DOI] [PubMed] [Google Scholar]
- 13.Brik, A. & Wong, C. H. (2003) Org. Biomol. Chem. 1, 5–14. [DOI] [PubMed] [Google Scholar]
- 14.Engelman, A., Bushman, F. D. & Craigie, R. (1993) EMBO J. 12, 3269–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyda, F., Hickman, A. B., Jenkins, T. M., Engelman, A., Craigie, R. & Davies, D. R. (1994) Science 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- 16.Goldgur, Y., Craigie, R., Cohen, G. H., Fujiwara, T., Yoshinaga, T., Fujishita, T., Sugimoto, H., Endo, T., Murai, H. & Davies, D. R. (1999) Proc. Natl. Acad. Sci. USA 96, 13040–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, Y. C., Beck, Z., Morris, G. M., Olson, A. J. & Elder, J. H. (2003) J. Virol. 77, 6589–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schames, J. R., Henchman, R. H., Siegel, J. S., Sotriffer, C. A., Ni, H. & McCammon, J. A. (2004) J. Med. Chem. 47, 1879–1881. [DOI] [PubMed] [Google Scholar]
- 19.Fernández, A., Scott, L. R. & Scheraga, H. A. (2003) J. Phys. Chem. B 107, 9929–9932. [Google Scholar]
- 20.Platzer, K. E. B., Momany, F. A. & Scheraga, H. A. (1972) Int. J. Pept. Protein Res. 4, 201–219. [DOI] [PubMed] [Google Scholar]
- 21.Fernández, A., Kardos, J., Scott, L. R., Goto, Y. & Berry, R. S. (2003) Proc. Natl. Acad. Sci. USA 100, 6446–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis, J. M., Dyda, F., Nashed, N. T., Kimmel, A. R. & Davies, D. R. (1998) Biochemistry 37, 2105–2110. [DOI] [PubMed] [Google Scholar]
- 23.Schnolzer, M., Rackwitz, H. R., Gustchina, A., Laco, G., Wlodawer, A., Elder, J. H. & Kent, S. B. (1996) Virology 224, 268–275. [DOI] [PubMed] [Google Scholar]
- 24.Adesokan, A. A., Roberts, V., Lee, K.-W., Lins, R. & Briggs, J. (2004) J. Med. Chem. 47, 821–828. [DOI] [PubMed] [Google Scholar]
- 25.Arkin, M. R. & Wells, J. A. (2004) Nat. Rev. Drug Discovery 3, 301–317. [DOI] [PubMed] [Google Scholar]
- 26.Kervinen, J., Lubkowski, J., Zdanov, A., Bhatt, D., Dunn, B. M., Hui, K. Y., Powell, D. J., Kay, J., Wlodawer, A. & Gustchina, A. (1998) Protein Sci. 7, 2314–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das, K., Clark, A. D., Lewi, P. J., Heeres, J., De Jonge, M. R., Koymans, L. M., Vinkers, H. M., Daeyaert, F., Ludovici, D. W., Kukla, M. J., et al. (2004) J. Med. Chem. 47, 2550–2560. [DOI] [PubMed] [Google Scholar]
- 28.Wlodawer, A., Gustchina, A., Reshetnikova, L., Lubkowski, J., Zdanov, A., Hui, K. Y., Angleton, E. L., Farmerie, W. G., Goodenow, M. M. & Bhatt, D. (1995) Nat. Struct. Biol. 2, 480–488. [DOI] [PubMed] [Google Scholar]