Abstract
To elucidate the biogenetic pathways for the generation of lysosome-related organelles, we have chosen to study the Drosophila eye pigment granules because they are lysosome-related and the fruit fly provides the advantages of a genetic system in which many mutations affect eye color. Here, we report the molecular identification of two classic Drosophila eye-color genes required for pigment granule biogenesis, claret and lightoid; the former encodes a protein containing seven repeats with sequence similarity to those that characterize regulator of chromosome condensation 1 (RCC1, a guanine nucleotide exchange factor for the small GTPase, Ran), and the latter encodes a rab GTPase, Rab-RP1. We demonstrate in transfected cells that Claret, through its RCC1-like domain, interacts preferentially with the nucleotide-free form of Rab-RP1, and this interaction involves Claret's first three RCC1-like repeats that are also critical for Claret's function in pigment granule biogenesis in transgenic rescue experiments. In addition, double-mutant analyses suggest that the gene products of claret and lightoid function in the same pathway, which is different from that of garnet and ruby (which encode the δ- and β-subunit of the tetrameric adaptor protein 3 complex, respectively). Taken together, our results suggest that Claret functions as a guanine nucleotide exchange factor for Lightoid/Rab-RP1 in an adaptor protein 3-independent vesicular trafficking pathway of pigment granule biogenesis.
The Drosophila eye pigment granules are lysosome-related organelles, a group of cell type-specific organelles that also include melanosomes produced in melanocytes and retinal pigment epithelial cells, platelet-dense granules, lytic granules of cytotoxic T lymphocytes, MHC class II compartments of antigen-presenting cells, and neutrophil primary granules (1). Defective lysosome-related organelles underlie the pathology of a group of human genetic diseases such as Hermansky–Pudlak syndrome (2). Recent studies have demonstrated the utility of the Drosophila eye pigment granule as a model system for studying biogenesis of lysosome-related organelles (3). In fact, the first clue as to the nature of a vesicular trafficking pathway for the biogenesis of lysosome-related organelles was provided by the discovery that the Drosophila eye-color gene garnet encodes the δ subunit of the adaptor protein 3 (AP3) complex, and garnet mutant flies have abnormal pigment granules (4, 5). Although >80 genes have been identified that affect eye color, garnet is one of the 11 granule group Drosophila eye-color genes whose normal products are not enzymes involved in the biosynthesis of the pigments but, rather, appear to function in the delivery of proteins to pigment granules (3). So far, seven granule group genes have been identified at the molecular level. Among them are genes encoding all four subunits of the tetrameric AP3 adaptor complex, namely garnet for δ3, ruby for β3, carmine for μ3, and orange for σ3 (6–8); the other three genes, light, deep orange, and carnation, encode Drosophila orthologues of yeast vacuolar protein sorting (VPS) gene products VPS41p, VPS18p, and VPS33p, respectively (9–11). It is apparent that identification of additional granule group eye-color gene products may uncover new components of the cellular machinery involved in the biogenesis of lysosome-related organelles.
In the present study, we demonstrate that the granule group gene claret (ca) encodes a protein with regions of homology to regulator of chromosome condensation 1 (RCC1), a guanine nucleotide exchange factor (GEF) for the small GTPase Ran; another granule group gene lightoid (ltd) encodes Rab-RP1, a GTPase of the rab family. We show that Claret, through its RCC1-like domain (RLD), interacts preferentially with the nucleotide-free form of Rab-RP1, and this interaction involves Claret's first three RCC1-like repeats that contain regions critical for Claret's function in pigment granule biogenesis. In addition, the double-mutant analysis suggests that the gene products of claret and lightoid act together in a pathway different from that of garnet and ruby (which encode AP3 subunits). Based on these results, we propose that Claret functions as a GEF for Lightoid/Rab-RP1 in an AP3-independent pathway of pigment granule biogenesis.
Materials and Methods
Fly Strains and EST Clones. The eye-color mutants claret (ca1), lightoid (ltd1), garnet (g1), and ruby (rb1) were obtained from the Bloomington Stock Center (Bloomington, IN). The EST clones RH02355 and SD04942 were obtained from the Berkeley Drosophila Genome Project (Berkeley, CA).
Molecular Biology. The relative amount of the claret mRNA in ca1 flies was determined by semiquantitative RT-PCR with an internal control as follows. The total RNAs were purified from wild-type (Oregon-R) and ca1 flies by using the High Pure RNA isolation kit (Roche Applied Science, Indianapolis) and the RT-PCRs (17 cycles) were performed by using the One-Step RT-PCR system (Roche Applied Science). Two pairs of primers were present in each RT-PCR: one (5′-GGTACAACGAGCTGAACGTC-3′/5′-ACTTGATAGCGTGCACCTGC-3′) for amplifying the claret mRNA and the other (5′-CAACGAAGACACTGATAGTG-3′/5′-GAGGTCAGCTTAAGGATGTG-3′) for amplifying the mRNA of an irrelevant gene (CG7814) as an internal control. The amounts of amplified claret cDNA fragment (582 bp) were normalized to the internal control fragment (819 bp) by staining with ethidium bromide after agarose-gel electrophoresis. Genomic DNA segments were amplified by using the Expand Long Template PCR system (Roche Applied Science). For transgene rescue experiments, we subcloned individually the cDNAs encoding N-terminally hemagglutinin (HA) epitope-tagged full-length and truncated forms of Claret protein and the rab domain of Rab-RP1 into the P element transformation vector pUAST. For expression in mammalian cells, we used pcDNA3 vector (Invitrogen) to express HA-tagged proteins and pEBG vector (a gift of D. Ron, New York University Medical Center, New York) to express GST-fusion proteins.
Transgenic Rescue. By using the standard Drosophila germ-line P element transformation technique, we generated multiple transgenic lines, and those with transgenes mapped to the 2nd or 3rd chromosomes were used in the transgenic rescue experiments. For transgenic rescue of claret and lightoid mutant phenotypes, both upstream activation sequence (UAS) transgene lines and GAL4 driver lines were further crossed into a ca1 or ltd1 background, respectively, by using standard Drosophila genetic techniques and appropriate fly strains.
Quantitative Estimation of Drosophila Eye Pigments. Extraction and measurement of the red (pteridine) and brown (ommachrome) eye pigments of Drosophila were carried out basically as described by Ooi et al. (4) except that 20 fly heads (10 male and 10 female) instead of 10 were used for the extraction of the brown pigments. The absorption spectra were obtained by using a SmartSpec 3000 spectrophotometer.
Cotransfection–Coprecipitation. Human embryonic kidney 293T cells were transfected by using the Lipofectamine reagent (Invitrogen) with appropriate combinations of pcDNA3-based plasmids encoding HA-tagged proteins and pEBG-based plasmids encoding GST-fusion proteins. Forty hours after transfection, the cells were lysed in 1% Triton X-100/50 mM Tris·HCl (pH 7.5)/100 mM NaCl/20 mM MgCl2/10% glycerol and complete protease inhibitor mixture (Roche Applied Science). Supernatants obtained after centrifugation of the cell lysates (12,000 × g for 5 min at 4°C) were incubated (60 min at 4°C) with glutathione-Sepharose beads (Sigma). The precipitates were washed in the lysis buffer (five times at 1 ml each), fractionated by SDS/PAGE, and transferred onto nitrocellulose membranes for Western blotting. The HA-tagged proteins were detected with the HA.11 monoclonal antibodies (Covance, Princeton) and a chemiluminescent detection kit (Amersham Pharmacia). Precipitated GST-fusion proteins present on the nitrocellulose membranes were detected by either staining with Ponceau S or immunoblotting with an affinity-purified rabbit polyclonal antibody against GST (Santa Cruz Biotechnology).
Microscopy. Drosophila heads were bisected and processed for electron microscopy (4). Longitudinal sections (0.5-μm thick) were mounted on glass slides, stained with toluidine blue, and photographed with a Zeiss photomicroscope. Thin sections (10-nm thick) from the same region were collected on carbon-coated grids, contrasted with uranyl acetate and lead citrate, and photographed with a JEOL JEM1200EXII electron microscope.
Results
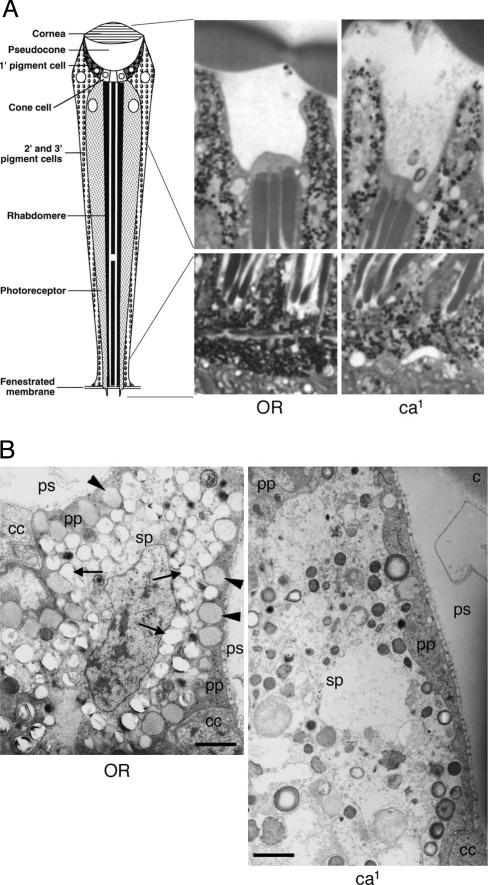
Eye Pigment Granule Phenotypes of claret Mutant Flies. Drosophila claret is a member of the granule group eye color genes (3). To determine the pigment granule defect in the claret mutant, we examined the eyes of wild-type (Oregon-R) and claret (ca1) mutant flies by light and electron microscopy. At the light microscopic level, granules in the claret mutant are fewer in number and less homogenous in size than in the wild type (Fig. 1A). At the electron microscope level, the most salient feature of the claret mutant is that its primary pigment cells lack large electron-lucent xanthommatin granules, which, in the wild type, often appear to be lined up in single file within the rather thin and attenuated cytoplasm. In addition, the granules in the secondary/tertiary pigment cells of the claret mutant were fewer in number and also abnormal in that they varied considerably in shape and the density of their content (Fig. 1B).
Fig. 1.
Eye pigment granule phenotypes of wild-type and claret mutant flies. (A) Light micrographs of toluidine blue-stained longitudinal epon sections of wild-type and claret mutant eyes. The regions shown in the micrographs correspond to those indicated in the schematic diagram of an ommatidium (Left) derived from ref. 32. The overall lower concentration of granules in the pigment cells of the mutant (ca1) is apparent in both the upper and lower portions of the ommatidium. (B) Electron micrographs of wild-type and claret mutant eye sections. (Left) OR. The micrograph shows regions of primary (pp) and secondary (sp) pigment cells of two adjacent ommatidia of a wild-type fly. The primary pigment cells, which face the pseudocone space (ps), contain a layer of large granules with a homogeneous content (arrowheads), which is adjacent to the pseudocone space. The secondary pigment cells are easily distinguished from the primary ones because of their lighter cytoplasm and the presence of numerous granules, which appear empty of content (arrows), probably as a result of a well documented processing artifact (33). (Right) ca1. In the claret mutant the primary pigment cells (pp, adjacent to ps) lack granules, and the secondary pigment cells (sp) are characterized by the presence of abnormal granules that are heterogeneous in size and density. cc, cone cells; c, cornea. (Bars, 1 μm.)
The Gene Defective in the claret Mutant (ca1). The claret locus (99B/C junction) contains two genes, claret and non-claret disjunctional (ncd): The former is required for wild-type eye color and the latter for proper chromosome segregation, and they lie within 1 kb of each other but are transcribed in opposite directions (12). Although ncd has been shown to encode a kinesin-related motor protein, the claret gene sequence and the corresponding product remained unidentified. Aided by this information, we examined the Drosophila genome sequence and EST database and identified the EST clone RH02355 as the candidate cDNA encoding the claret gene product. By DNA sequencing, we found that RH02355 corresponds to a 6.4-kb mRNA whose genomic sequence is only 121 bp away from the start site of ncd transcription in the opposite direction. RH02355 seems to contain a full-length cDNA with a complete open-reading-frame (1,962 codons) preceded by a short (86 bp) 5′ UTR that contains an in-frame stop-codon but no additional ATG, followed by a 3′ UTR that contains a polyadenylation signal near the 3′ end. By semiquantitative RT-PCR with an internal control, we demonstrated that the amount of mRNA corresponding to a region of RH02355 is reduced significantly in the ca1 mutant as compared with the wild type (Oregon-R) (Fig. 2A). By sequencing the chromosomal segment and the RT-PCR products of this putative claret gene in the ca1 mutant, we identified a 14-bp deletion in the second coding exon that causes a frame shift and premature termination of translation (Fig. 2B). The reduced levels of claret mRNA in the ca1 mutant probably result from a nonsense-mediated mRNA decay (13).
Fig. 2.
Molecular identification of the gene defective in the claret mutant (ca1). (A) Semiquantitative RT-PCR analysis of the putative claret mRNA and an internal control (CG7814). The positions of DNA size markers are indicated (Left). (B) Schematic representation of the claret gene and its products in wild-type (WT) and ca1 flies. The top straight line represents the claret locus with the claret and ncd genes indicated as rectangles. The arrows indicate the directions of transcription. The middle series of rectangles represent the 15 exons of the claret gene with the coding regions shaded. The DNA sequence of a portion of the second coding exon is given to indicate the 14 bases (boxed) that are missing in the claret mutant (ca1). Wild-type and mutant Claret proteins are depicted at the bottom as straight lines, with the RCC1-like repeats in the wild type represented as shaded boxes. These repeats (amino acids 1132–1189, 1190–1241, 1242–1293, 1305–1359, 1360–1409, 1519–1570, and 1572–1624) were identified by using the protein repeats identification program (34) with the threshold score set at 2,300. The numbers below the straight lines indicate the first and last Claret amino acid residues present in wild-type and mutant (ca1) proteins. After residue 194, the Claret mutant contains an additional 11 aa due to out-of-frame translation of the mutant mRNA. The numbers above the line for the wild-type protein indicate the sites of truncation used in the transgenic rescue experiments.
The mRNA sequence of the claret gene matches that of a predicted gene CG31037 in the Annotated Drosophila Genome (release 3.1). However, the genome annotation employs the seventh (the fourth in-frame) AUG from the 5′ end of the CG31037 mRNA as initiation codon and predicts an 1,807-codon ORF that corresponds to an incomplete claret ORF missing the first 154 codons. Because the scanning mechanism for initiation of translation predicts that translation should initiate at the AUG codon nearest the 5′ end of the mRNA (14), the predicted translation initiation of the CG31037 mRNA at its seventh AUG from the 5′ end seems unlikely.
The 15 exons of the claret gene encode a 1,961-aa protein that contains seven RCC1-like repeats (Fig. 2B), which are expected to form a seven-bladed propeller structure (15). RCC1 is a GEF for the small GTPase Ran (16), and proteins containing RLDs have been assumed to function as GEFs for particular small GTPases that have yet to be identified (17). Beside the RLD, the rest of the claret protein sequence shows little or no similarity to any other known protein except a predicted mosquito protein (ENSANGP00000019871) whose entire sequence (1,734 aa, incomplete at the N terminus) is 40% identical to that of Claret.
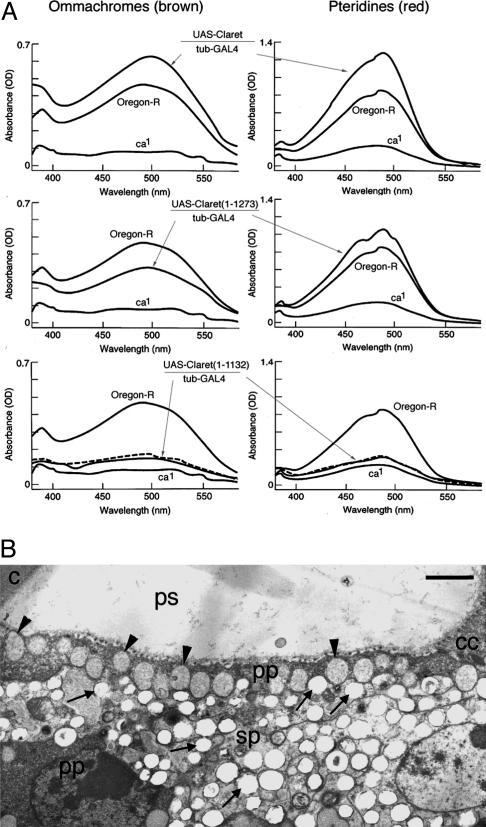
Rescue of the claret Mutant Phenotype by Transformation. By using the GAL4/UAS system (18), we have successfully rescued the claret mutant phenotype by expressing the N-terminally HA epitope-tagged full-length Claret protein in transgenic flies (Fig. 3). We demonstrated that ubiquitous claret transgene expression driven by GAL4 under the tubulin (tub) promoter fully rescued the claret mutant phenotype in transgenic flies. In fact, expression of a truncated Claret protein (amino acids 1–1273) that lacks the C-terminal 688-aa residues with its four RCC1-like repeats and, therefore, cannot form the seven-bladed propeller, also rescued, to a large extent, the claret mutant phenotype (Fig. 3A). This was a fortuitous finding due to the fact that the EST clone SD04942 that we obtained from the Berkeley Drosophila Genome Project and used in the initial assembling of the full-length claret cDNA contains a frame-shifting extra nucleotide (T) not present in the genomic DNA and the mRNA sequences, which was probably due to a reverse transcriptase error during synthesis of the first strand cDNA. This raised the question of whether Claret's function in pigment granule biogenesis requires its RLD at all. We therefore generated transgenic flies expressing a truncated Claret protein (amino acids 1–1132) that lacks all seven RCC1-like repeats and showed that it failed to rescue the claret mutant phenotype (Fig. 3A). These results, thus, demonstrate that the segment (amino acids 1132–1273) encompassing the first three RCC1-like repeats contains regions critical for Claret's function in pigment granule biogenesis.
Fig. 3.
Transgenic rescue of the claret mutant phenotype. (A) Quantitative measurement of eye pigments (ommachromes and pteridines) from wild-type (Oregon-R), claret mutant (ca1), and transgenic flies. The ubiquitous expression of full-length or truncated claret transgenes was driven by GAL4 under control of the tubulin (tub) promoter. The dashed curves represent results from another UAS-Claret (1–1132) transgenic line. (B) Electron micrograph of an eye section of a transgenic fly that expresses in the claret mutant (ca1) background a UAS-claret transgene driven by GAL4 under tubulin promoter control (UAS-claret/tub-GAL4; ca1/ca1). It should be compared with the micrographs from the wild-type (OR) and claret mutant (ca1) flies in Fig. 1B. Both primary and secondary pigment cell granules are restored with their normal appearance. Arrowheads indicate the pigment granules of the primary pigment (pp) cells and arrows those of the secondary pigment (sp) cells. cc, cone cell; ps, pseudocone; c, cornea. (Bars, 1 μm.)
One characteristic of the claret gene, which is also shared by several other granule group eye-color genes, is that its function is required not only within the eye but also outside the eye for the development of normal eye color (19, 20). This would be the case, for example, if the extraocular storage or secretion of a pigment precursor, which also requires functional granule group eye color genes, is necessary for normal eye pigmentation. In their classic Drosophila eye-disk transplantation experiment, Beadle and Ephrussi (19) showed that a wild-type eye disk transplanted into a claret mutant host did not develop into a wild-type color eye, whereas a claret mutant eye disk transplanted into a wild-type host developed autonomously the claret mutant eye color. Consistent with this classic observation, we found that the eye-specific expression of the claret transgene driven by GAL4 under the GMR promoter failed to fully rescue the claret mutant phenotype (data not shown).
Molecular Identification of the Gene Mutated in the lightoid Mutant (ltd1). The fact that the Claret protein contains an RLD suggested that it functions as a GEF for a small GTPase, probably encoded by one of the granule group genes. By searching among the granule group for a mutant that has been mapped to a chromosomal region that contains a gene encoding a small GTPase, we identified the rab GTPase Rab-RP1 as a candidate for the product of the granule group gene lightoid. The lightoid mutation has previously been mapped to the right arm of the second chromosome, between 44E1 and 46E9, a region containing >200 genes. One of the genes in this region encodes Rab-RP1 (21). By sequencing the RT-PCR products and the corresponding chromosomal segment of the Rab-RP1 gene, we found that the Rab-RP1 gene of the lightoid mutant (ltd1) contains a point mutation (C→G) that generates a premature stop codon (Fig. 4). The Drosophila Genome Annotation (release 3.1) predicts that four alternatively initiated and spliced transcripts of Rab-RP1 gene have their last three exons in common, which encode a 216-aa rab GTPase (the rab domain), and these transcripts produce three forms of Rab-RP1 with a common C-terminal rab domain and an N-terminal extension of 6-, 172-, and 463-aa residues, respectively. The point mutation in the lightoid mutant (ltd1) eliminates the C-terminal 203-aa region common to all forms of Rab-RP1 that contains motifs necessary for their GTPase activity (Fig. 4). By electron microscopy, we found that, like the claret mutant (ca1), the lightoid mutant (ltd1) also lacks the large electron-lucent xanthommatin granules in the primary pigment cells, and the granules in the secondary and tertiary pigment cells were fewer in number and abnormal in shape and density of their content (Fig. 5A). In addition, the photoreceptor cells of the lightoid mutant (ltd1) lack the small xanthommatin granules, which appear normal in the claret mutant (ca1) (Fig. 5 C and D). The latter finding suggests that photoreceptor cells have another Rab-RP1 activator.
Fig. 4.
Molecular identification of the gene mutated in the lightoid mutant (ltd1). The top four diagrams are schematic representations of the four alternatively initiated and spliced gene products of Rab-RP1 (CG8024) based on the Drosophila Genome Annotation (release 3.1). The rectangles represent exons with coding regions shaded. The four predicted products of the Rab-RP1 (CG8024) gene have their last three exons in common, which encode a 216-aa rab GTPase. The first 45 nucleotides of the first common exon are depicted at the bottom as a sequence of codons, and the point mutation (C→ G) present in the lightoid mutant (ltd1) is indicated in the box.
Fig. 5.
Transgenic rescue of the lightoid mutant phenotype. (A) Electron micrograph of an eye section of the lightoid mutant (ltd1) fly. Note the complete lack of pigment granules in the primary pigment cells and the presence of abnormal granules in the secondary pigment cells. This micrograph should be compared with that of the claret mutant (ca1) in Fig. 1B.(B) Electron micrograph of an eye section of a transgenic fly that expresses in the lightoid mutant (ltd1) background a UAS-transgene encoding the rab domain of Rab-RP1. This micrograph should be compared with that of the wild-type (OR) fly in Fig. 1B. Arrowheads and arrows point to pigment granules of the primary (pp) and secondary (sp) pigment cells, respectively. cc, cone cell; ps, pseudocone. (C–E) Electron micrographs of eye sections of the wild-type (C), ltd1 mutant (D), and the rescuing transgenic fly (ltd1/ltd1; UAS-RabRP1/da-GAL4) (E). Shown are regions of the photoreceptor cells (ph), close to the rhabdomere (rh), that, in the wild type (C), have small pigment granules (white arrows) that are completely lacking in the lightoid mutant (ltd1) (D) and reappear in the transgenic fly(E) (white arrows). (Bars, 1 μm.) (F) Quantitative measurement of eye pigments (ommachromes and pteridines) from the wild-type (Oregon-R), lightoid mutant (ltd1), and transgenic flies.
Rescue of the lightoid Mutant Phenotype by Transformation. Because the predicted long N-terminal extensions of Rab-RP1 (171 and 470 aa) are at odds with their homologues in other species as well as with rab proteins in general (22), we have used Rab-RP1 without the N-terminal long extensions to rescue the mutant phenotype of lightoid in transgenic flies. We found that the Rab-RP1 transgene expression driven by GAL4 under the daughterless (da) promoter rescued the lightoid mutant phenotype (Fig. 5 B, E, and F). These results also demonstrate that the main functional domain of Rab-RP1 is its rab domain, because it alone can rescue, in transgenic flies, the lightoid mutant phenotype.
Claret Interacts with Rab-RP1 as Well as Its Human Homologues, rab32 and rab38. Our findings that the lightoid gene encodes Rab-RP1 and the claret gene encodes a putative GEF, and the mutant flies with apparently functionally null alleles of claret or lightoid share many phenotypic features, suggest that the two proteins interact and function together in the same vesicular trafficking pathway of pigment granule biogenesis. We, therefore, tested whether the two proteins interact in a cotransfection–coprecipitation assay in which human 293T cells were transfected with plasmids encoding HA-tagged Claret protein and GST-tagged Rab proteins. We found that the Claret protein coprecipitated not only with Rab-RP1 but also with the latter's human homologues, rab38 and rab32 (Fig. 6A). In a similar assay in which GST-tagged Claret and HA-tagged rabs were used, precipitation of a Claret fragment (amino acids 1132–1657) that contains the seven RCC1-like repeats brought down a nucleotide-free mutant form of Rab-RP1 preferentially over its GTPase-deficient mutant form (Fig. 6B), which is consistent with the notion that Claret, through its RLD, may function as a GEF for Rab-RP1. Because the segment of Claret (amino acids 1132–1273) that encompasses most of the first three RCC1-like repeats contains regions critical for Claret's function in biogenesis of the pigment granules, we tested whether this segment contains binding sites for Rab-RP1. Indeed, precipitation of GST-Claret (amino acids 1135–1273) brought down HA-tagged Rab-RP1 and more abundantly its nucleotide-free mutant form (Fig. 6C).
Fig. 6.
Claret interacts with Rab-RP1 and its human homologues, rab32 and rab38, in a cotransfection–coprecipitation assay. (A Top) Immunoblot of HA-tagged Claret coprecipitated with the GST-fusion proteins of human rab38, human rab32, or the rab-domain of Drosophila Rab-RP1, respectively. HA-Claret plus GST cotransfection and HA-Claret single transfection serve as negative controls. (Middle) Immunoblot of HA-Claret present in an aliquot (1/20) of the cell lysates. (Bottom) Ponceau S staining of GST and GST-fusion proteins recovered on glutathione-Sepharose beads. (B and C) Experiments similar to that in A were carried out except that the various rabs were HA-tagged, the Claret protein fragments were GST-tagged, and the latter were detected by immunoblot by using a polyclonal antibody against GST. RabRP1 (GTPase deficient) and RabRP1 (nucleotide free) contain single-point mutations equivalent to the Q61L and N116I mutations in Ras, respectively. Drosophila Rab11 (DmRab11) was included as a negative control.
Analysis of Double Mutants. To determine whether the gene products of claret and lightoid are components of the known AP3-dependent pathway of pigment granule biogenesis, we generated double-mutant flies homozygous for the lightoid mutation (ltd1) together with the claret (ca1), or garnet (g1), or ruby (rb1) (the latter two genes encode AP3 subunits). Quantitative measurement of the eye pigments extracted from the double mutants as well as the parental single mutants shows that, unlike the lightoid (ltd1);claret (ca1) double mutant, whose eye pigment level resembles that of claret single mutant (ca1), the pigment levels of the double mutants garnet (g1);lightoid (ltd1) and ruby (rb1);lightoid (ltd1) are significantly lower than that of their parental homozygous single mutants (Fig. 7), suggesting that claret and lightoid are in the same pathway, which is different from that of garnet and ruby.
Fig. 7.
Quantitative measurement of the eye pigments (pteridines) extracted from double mutants (ltd1;ca1, g1;ltd1, and rb1;ltd1) and their parental single mutants (ltd1, ca1, g1, and rb1). The eye pigment levels of the wild-type (Oregon-R) and a white-eye mutant (w1118) that lacks eye pigments completely (dashed curves) are included for comparison.
Discussion
In the present study, we have identified at the molecular level two granule group genes, lightoid and claret: The former encodes Rab-RP1, and the latter encodes a protein with an RLD. The amino acid sequence of RCC1 consists mainly of seven 51- to 68-residue repeats that form in three-dimensional structure a seven-bladed propeller (15). Proteins containing similar RLDs have been assumed to function as GEFs, but little is known about which GTPases may be regulated by these putative GEFs. The only information available concerning this point is that, in vitro, the N-terminal RLD of HERC1 (a member of the domain homologous to E6 associated protein C terminus and RCC1 protein family) can stimulate guanine nucleotide exchange on Arf1, Rab3A, and Rab5 (23, 24), and the RLD of PRAF1 (an Arabidopsis protein with PH, RCC1, and FYVE domains) can stimulate guanine nucleotide exchange on rab8 (25). Our results that the RLD of Claret interacts preferentially with a nucleotide-free mutant form of Rab-RP1 suggest that Claret functions as a GEF for Rab-RP1.
Recently, Ozaki and coworkers (26) localized Rab-RP1 on the pigment granules of pigment cells and photoreceptor cells of the Drosophila eye and found abnormal accumulation of multivesicular bodies and autophagosome-like structures in the photoreceptor cells that express a dominant-negative mutant Rab-RP1 transgene. These findings are consistent with our discovery that the lightoid gene encodes Rab-RP1.
Of the 60 or so mammalian rab proteins, rab32 and rab38 share the highest sequence similarity with Rab-RP1 (22). By using a cotransfection–coprecipitation assay, we have demonstrated that, like Rab-RP1, both human rab32 and rab38 interact with Claret. The cross-species interaction suggests the existence of Claret-like proteins in mammalian cells and of an evolutionarily conserved biogenetic pathway involving Rab-RP1 and Claret or proteins like them. The recent report (27) that rab38 is a melanosomal protein and a mouse pigmentation mutant, chocolate, contains a missense point mutation in the conserved GTP-binding domain of rab38 supports this notion. Interestingly, rab32 has recently been identified as a mitochondria-associated A-kinase anchoring protein that participates in mitochondrial dynamics, and of all rab proteins, only rab32 and Rab-RP1 contain the sequence motif required for interaction with the R subunits of the PKA holoenzyme (28). Although Rab-RP1 has not been localized to mitochondria (26), the cross-species interaction of rab32 and Claret that we demonstrate in our experiments suggests the involvement of a Claret-like protein in mitochondrial dynamics. Whether Rab-RP1's function in the pigment granule biogenesis depends on its capacity to interact with PKA requires further investigation.
Abundant evidence indicates that, in addition to AP3-mediated protein sorting and trafficking, an AP3-independent pathway(s) exists that plays a major role in transport to lysosome-related organelles (29). Which trafficking pathway of the pigment granule biogenesis, AP3-dependent or AP3-independent. do the gene products of claret and lightoid regulate? We suggest that it is the AP3-independent pathway for the following reasons. First, we found that the eye pigment levels of double-mutants garnet (g1);lightoid (ltd1) and ruby (rb1);lightoid (ltd1) are much lower than that of their single-mutant parents, whereas the eye pigment levels of the lightoid (ltd1);claret (ca1) double mutant resemble that of lightoid and claret single mutants (Fig. 7). By using a different method of quantifying eye pigments, Silva and Mensua (30) made similar observations. Interpreting these data in the current context would place claret and lightoid in the same pathway different from that of garnet and ruby (which encode AP3 subunits). Second, the mutation of rab38 in the mouse pigmentation mutant, chocolate, causes inefficient targeting of tyrosinase-related protein-1 to melanosomes (27). Because targeting of tyrosinase-related protein-1 to melanosomes has been shown to be independent of AP3 (31), it thus appears that rab38 regulates an AP3-independent trafficking pathway to the melanosome. Further studies to elucidate other components of the Claret/Rab-RP1 pathway may link pigment granule biogenesis with the Drosophila orthologues of the Hermansky–Pudlak syndrome genes, because most of them are involved in the biogenesis of lysosome-related organelles by mechanisms independent of AP3 (29).
Acknowledgments
We thank the Bloomington Stock Center for the fly strains and Berkeley Drosophila Genome Project for the EST clones.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VPS, vacuolar protein sorting; HA, hemagglutinin; UAS, upstream activation sequence; GEF, guanine nucleotide exchange factor; RCC1, regulator of chromosome condensation 1; RLD, RCC1-like domain; AP3, adaptor protein 3.
References
- 1.Dell'Angelica, E. C., Mullins, C., Caplan, S. & Bonifacino, J. S. (2000) FASEB J. 14, 1265–1278. [DOI] [PubMed] [Google Scholar]
- 2.Huizing, M., Anikster, Y. & Gahl, W. A. (2000) Traffic 1, 823–835. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd, V., Ramaswami, M. & Kramer, H. (1998) Trends Cell Biol. 8, 257–259. [DOI] [PubMed] [Google Scholar]
- 4.Ooi, C. E., Moreira, J. E., Dell'Angelica, E. C., Poy, G., Wassarman, D. A. & Bonifacino, J. S. (1997) EMBO J. 16, 4508–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson, F., Peden, A. A., Christopoulou, L. & Robinson, M. S. (1997) J. Cell Biol. 137, 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzschmar, D., Poeck, B., Roth, H., Ernst, R., Keller, A., Porsch, M., Strauss, R. & Pflugfelder, G. O. (2000) Genetics 155, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullins, C., Hartnell, L. M., Wassarman, D. A. & Bonifacino, J. S. (1999) Mol. Gen. Genet. 262, 401–412. [DOI] [PubMed] [Google Scholar]
- 8.Mullins, C., Hartnell, L. M. & Bonifacino, J. S. (2000) Mol. Gen. Genet. 263, 1003–1014. [DOI] [PubMed] [Google Scholar]
- 9.Warner, T. S., Sinclair, D. A., Fitzpatrick, K. A., Singh, M., Devlin, R. H. & Honda, B. M. (1998) Genome 41, 236–243. [PubMed] [Google Scholar]
- 10.Shestopal, S. A., Makunin, I. V., Belyaeva, E. S., Ashburner, M. & Zhimulev, I. F. (1997) Mol. Gen. Genet. 253, 642–648. [DOI] [PubMed] [Google Scholar]
- 11.Sevrioukov, E. A., He, J. P., Moghrabi, N., Sunio, A. & Kramer, H. (1999) Mol. Cell 4, 479–486. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto, A. H., Komma, D. J., Shaffer, C. D., Pirrotta, V. & Endow, S. A. (1989) EMBO J. 8, 3543–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maquat, L. E. (2002) Curr. Biol. 12, 196–197. [DOI] [PubMed] [Google Scholar]
- 14.Kozak, M. (2002) Gene 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renault, L., Nassar, N., Vetter, I., Becker, J., Klebe, C., Roth, M. & Wittinghofer, A. (1998) Nature 392, 97–101. [DOI] [PubMed] [Google Scholar]
- 16.Bischoff, F. R. & Ponstingl, H. (1991) Nature 354, 80–82. [DOI] [PubMed] [Google Scholar]
- 17.Hadano, S., Hand, C. K., Osuga, H., Yanagisawa, Y., Otomo, A., Devon, R. S., Miyamoto, N., Showguchi-Miyata, J., Okada, Y., Singaraja, R., et al. (2001) Nat. Genet. 29, 166–173. [DOI] [PubMed] [Google Scholar]
- 18.Brand, A. H. & Perrimon, N. (1993) Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 19.Beadle, G. W. & Ephrussi, B. (1936) Genetics 21, 225–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clancy, C. W. (1942) Genetics 27, 417–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh, A. K., Tokunaga, F. & Ozaki, K. (1997) FEBS Lett. 404, 65–69. [DOI] [PubMed] [Google Scholar]
- 22.Pereira-Leal, J. B. & Seabra, M. C. (2001) J. Mol. Biol. 313, 889–901. [DOI] [PubMed] [Google Scholar]
- 23.Rosa, J. L., Casaroli-Marano, R. P., Buckler, A. J., Vilaro, S. & Barbacid, M. (1996) EMBO J. 15, 4262–4273. [PMC free article] [PubMed] [Google Scholar]
- 24.Rosa, J. L. & Barbacid, M. (1997) Oncogene 15, 1–6. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, R. B., La Cour, T., Albrethsen, J., Nielsen, M. & Skriver, K. (2001) Biochem. J. 359, 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujikawa, K., Satoh, A. K., Kawamura, S. & Ozaki, K. (2002) Zool. Sci. 19, 981–993. [DOI] [PubMed] [Google Scholar]
- 27.Loftus, S. K., Larson, D. M., Baxter, L. L., Antonellis, A., Chen, Y., Wu, X., Jiang, Y., Bittner, M., Hammer, J. A., III, & Pavan, W. J. (2002) Proc. Natl. Acad. Sci. USA 99, 4471–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alto, N. M., Soderling, J. & Scott, J. D. (2002) J. Cell Biol. 158, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dell'Angelica, E. C., Aguilar, R. C., Wolins, N., Hazelwood, S., Gahl, W. A. & Bonifacino, J. S. (2000) J. Biol. Chem. 275, 1300–1306. [DOI] [PubMed] [Google Scholar]
- 30.Silva, F. J. & Mensua, J. L. (1985) Drosophila Information Service 61, 156. [Google Scholar]
- 31.Huizing, M., Sarangarajan, R., Strovel, E., Zhao, Y., Gahl, W. A. & Boissy, R. E. (2001) Mol. Biol. Cell 12, 2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cagan, R. L. & Ready, D. F. (1989) Dev. Biol. 136, 346–362. [DOI] [PubMed] [Google Scholar]
- 33.Stark, W. S. & Sapp, R. (1988) Can. J. Zool. 66, 1301–1308. [Google Scholar]
- 34.Andrade, M. A., Ponting, C. P., Gibson, T. J. & Bork, P. (2000) J. Mol. Biol. 298, 521–537. [DOI] [PubMed] [Google Scholar]