Abstract
Diphthamide and the tRNA wobble uridine modifications both require Dph3 (DiPHthamide biosynthesis 3) protein as an electron donor for the iron-sulfur clusters in their biosynthetic enzymes. Here, using a proteomic approach, we identified Saccharomyces cerevisiae cytochrome B5 reductase (Cbr1) as a NADH-dependent reductase for Dph3. The NADH- and Cbr1-dependent reduction of Dph3 may provide a regulatory linkage between cellular metabolic state and protein translation.
Dph3 (DiPHthamide biosynthesis 3), also known as Kti11 (Kluveromyces lactis Toxin Insensitive 11), is required for two highly conserved modifications in eukaryotes: diphthamide, a unique protein post-translational modification on eukaryotic elongation factor 2 (eEF2), and the tRNA wobble uridine modification, 5-carboxymethyl-2-thiouridine (mcm5s2U). These two distinct modifications were both suggested to be important for translation fidelity.1,2
Studies of the diphthamide biosynthesis pathway have elucidated the molecular function of Dph3. Formation of diphthamide in eukaryotes takes four steps (Supplementary Results, Supplementary Fig. 1a), requiring seven proteins (Dph1-Dph7).3-5 The first step requires an unconventional radical SAM enzyme, the Dph1-Dph2 heterodimer. Dph1-Dph2 contains [4Fe-4S] clusters and relies on Dph3 as an electron donor to keep the [4Fe-4S] clusters in the active and reduced state.6,7 The functions of Dph5, Dph6, and Dph7 in the subsequent steps of the biosynthesis pathway are well characterized4,5,8 while the role of Dph4 is still unknown.
In eukaryotes, approximately 25% of cytoplasmic tRNAs have the wobble uridine modified to 5-methoxycarbonylmethyluridine (mcm5U), 5-carbamoylmethyl-2’-O-methyluridine (ncm5U) or mcm5s2U. Synthesis of a common intermediate, 5-carboxymethyluridine (cm5U), requires the eukaryotic elongator complex consisting of six subunits (Elp1-Elp6) and seven other associated proteins (Kti11-Kti14, Sit4, Sap185 and Sap190) in eukaryotes (Supplementary Fig. 1b).9,10 Elp3, a radical SAM enzyme, is the catalytic subunit for the first step. Recombinant archaeal Elp3 from Methanocaldococcus infernus catalyzes the formation of cm5U in the presence of SAM and sodium dithionite in vitro via a radical mechanism.11 Given that Dph3 (also known as Kti11) is an electron donor for the radical SAM enzyme Dph1-Dph2 in diphthamide biosynthesis, it is believed that Dph3 also acts as an electron carrier for Elp3 in the tRNA modification reaction.12 Thus, Dph3 connects the two modifications that are important for translation elongation. However, the physiological reductase(s) that ultimately provides electrons to Dph3 is not known.
To identify candidate reductase(s) for Dph3, we performed a protein interactome study on Dph3 in Saccharomyces cerevisiae (Supplementary Fig. 2a) using stable isotope labeling by amino acids in cell culture (SILAC).13 We generated a yeast BY4741 strain expressing endogenous level of FLAG-tagged Dph3 by inserting a C-terminal triple flag tag on the endogenous dph3 gene and cultured this strain in heavy media. A BY4741 strain expressing untagged Dph3 was cultured in light media. The cell lysates were immunoprecipitated with anti-FLAG resins. The eluted fractions from both immunopreicipitation were mixed, precipitated, digested with trypsin, and analyzed by mass spectrometry to identify proteins with high heavy to light (H/L) ratios, which would be potential interacting partners of Dph3. Among the list of proteins with high H/L ratios (Supplementary Fig. 2b), we found known interactors of Dph3, such as Dph1-Dph2 and the elongator complex subunits Elp1-Elp3.14 Our SILAC also identified Kti13. This is consistent with the recent findings reporting that Kti13 forms a heterodimer with Dph3.15,16 The identification of known Dph3 interacting partners suggested that our SILAC results were reliable. We suspected that the reductase is likely a flavin-containing protein because electron transfer in cells from the common two-electron donors (NADH or NADPH) to Fe-containing one-electron acceptors typically require flavin cofactors, which are capable of both two-electron and one-electron transfers. To our delight, we found an NADH dependent flavoprotein, cytochrome b5 reductase (Cbr1), among the proteins with high H/L ratio (Supplementary Fig. 2b). Cbr1 is a transmembrane protein embedded in the endoplasmic reticulum (ER) membrane and mitochondrion outer membrane with the catalytic domain residing on the cytosolic side (Fig. 1a).17 Therefore, it is plausible that Cbr1 reduces the cytosolic Dph3.
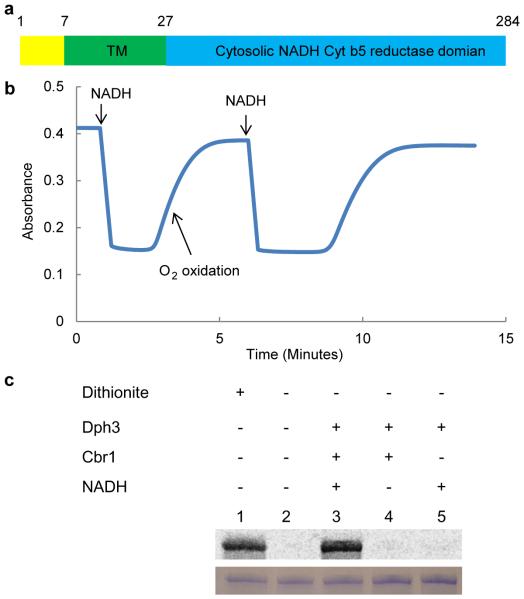
Figure 1.
Cbr1 reduces Dph3 in vitro. (a) Schematic view of the domain structure of yeast Cbr1. (b) Reduction of Dph3 monitored using the 488 nm absorption of oxidized Dph3. (c) In vitro reconstitution of first step of diphthamide biosynthesis on eEF2 using Dph1-2, Dph3, Cbr1, NADH and carboxy-14C-SAM. Autoradiography shows labeled eEF2. Bottom panel shows eEF2 stained with Coomassie blue. Figures are representative images from three experimental repeats.
To test if Cbr1 could reduce Dph3 in vitro, we cloned, expressed and purified the recombinant Cbr1. We first monitored the reduction of Dph3 by detecting the 488 nm absorption of oxidized Dph3 as previously described.7 Upon addition of NADH to initiate the reaction, Dph3 was rapidly reduced by Cbr1 (Fig. 1b). In contrast, addition of NADPH did not lead to reduction of Dph3, suggesting that Cbr1 is an NADH-specific enzyme (Supplementary Fig. 3). To confirm Cbr1’s role as a Dph3 reductase, we tested if this reduction system can be used to reduce Dph1-Dph2 and reconstitute the first step of diphthamide biosynthesis in vitro. Using 14C-SAM, the substrate eEF2 was radioactively labeled in the presence of Dph1-Dph2, Dph3, Cbr1 and NADH (Fig. 1c, and Supplementary Fig. 7, lane3). The Cbr1/NADH/Dph3 reduction system was able to reduce Dph1-Dph2 similarly to dithionite, a chemical reductant for Fe-S clusters (Fig. 1c, and Supplementary Fig. 7, lane 1). These results demonstrated that Cbr1 can reduce Dph3 using NADH in vitro.
To confirm that Cbr1 is the reductase of Dph3 in vivo, we examined the formation of diphthamide in CBR1 knockout (cbr1Δ) yeast. Diphthamide is targeted by the diphtheria toxin (DT) which inactivates eEF2 and kills the cells.18 Thus, we used the established DT sensitivity assay19 to test if cbr1Δ strain abolished diphthamide formation. Interestingly, the cbr1Δ strain conferred no resistance to DT (Supplementary Fig. 4), suggesting that formation of diphthamide is not affected. One possibility is that multiple proteins could serve as reductases for Dph3. Thus, we used the Basic Local Alignment Search Tool (BLAST) to search for other potential reductases in Saccharomyces cerevisiae using the protein sequence of Cbr1. Three proteins with highly similar sequence to Cbr1 were found: Mitochondrial cytochrome b5 reductase (Mcr1), Plasma membrane-associated coenzyme Q6 reductase (Pga3) and Altered inheritance of mitochondria protein 33 (Aim33). Since both Cbr1 and the NADPH dependent Cytochrome P450 reductase (Ncp1) were reported to provide electrons for Cytochromes P450 involved in ergosterol biosynthesis,20 we also tested Ncp1 for Dph3 reduction. We cloned, overexpressed and purified the recombinant Mcr1, Pga3 and Ncp1 (over-expression of the putative protein Aim33 in E.coli or yeast did not yield any protein) and tested their Dph3 reduction activity by monitoring the 488 nm absorption. Interestingly, we found that both Mcr1 and Ncp1 reduced Dph3 at a slower rate compared to that of Cbr1 under similar reaction conditions (Supplementary Fig. 5a and 5c). Pga3 displayed no Dph3 reduction activity (Supplementary Fig. 5b). We found that both the Cbr1/NADH/Dph3 or Ncp1/NADPH/Dph3 reduction systems were able to reduce Dph1-Dph2 and support the first step of diphthamide in vitro (Supplementary Fig. 5d and Supplementary Fig. 8). These results suggested that Mcr1 and Ncp1 could also be reductases for Dph3.
We next examined diphthamide formation in multiple reductase deletion strains by DT sensitivity assay. We failed to generate a cbr1Δmcr1Δncp1Δ strain as NCP1 was found to be an essential gene by the Saccharomyces Genome Deletion Project (http://wwwsequence.stanford.edu/group/yeast_deletion_project/Essential_ORFs.txt). Surprisingly, the cbr1Δmcr1Δ strain still did not confer any resistance to DT (Supplementary Fig. 4). The lack of obvious phenotype in diphthamide biosynthesis for the double deletion strain prompted us to investigate the tRNA modifications in these reductase deletion strains. We reasoned that the synthesis of the much more abundant wobble uridine modifications (about 25% of the tRNA population)12 is likely to depend more heavily on the efficient reduction of Dph3 compared to the formation of the irreversible diphthamide modification on the eEF2 with a slow protein turnover rate.21 The mcm5s2U modified tRNAs are targeted by the Kluyveromyces lactis killer toxin which cleaves the modified tRNAs, leading to cell-cycle arrest. Therefore, we tested if the reductase(s) deletion strains conferred resistance to inducible expression of γ-toxin, the killer toxin catalytic subunit, using a reported assay.22,23 As expected, BY4741 wild type and dph2Δ strain which contained the mcm5s2U tRNA modification were unable to grow when the expression of γ-toxin was induced (Fig. 2a). The dph3Δ or elp3Δ strain lacking the mcm5s2U tRNA modification survived under such conditions. Remarkably, we found that CBR1 deletion alone conferred some resistance to γ-toxin, suggesting that formation of tRNA modifications was partially impaired. Furthermore, while mcr1Δ strain did not confer any resistance, cbr1Δmcr1Δ strain exhibited greater resistance to γ-toxin. To confirm that the mcm5s2U formation is impaired in cbr1Δ strain, we isolated total tRNAs and treated the tRNAs with γ-toxin to probe for mcm5s2U. Using northern blot, we found that the glu-tRNA from cbr1Δ strain had significantly lower cleaved glu-tRNA fragment compared to that from BY4741 WT strain (Fig. 2b, and Supplementary Fig. 6). Taken together, these results support a role for Cbr1 as the major reductase of Dph3 under normal physiological conditions.
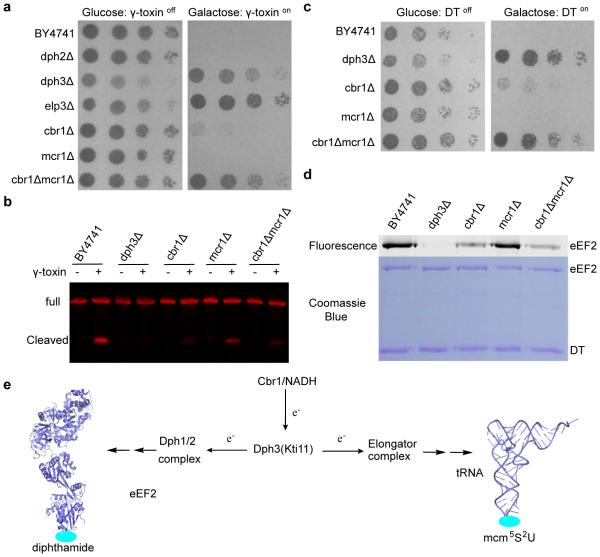
Figure 2.
Cbr1 is the major Dph3 reductase in vivo. (a) In vivo γ-toxin sensitivity assay. The strains used are specified on the left. Each row represents a serial dilution from left to right. (b) In vitro γ-toxin treatment of isolated tRNA. Samples were analyzed by northern blot probing for 5’ glu-tRNA. (c)In vivo DT sensitivity assay for cells over-expressing eEF2. The strains used are specified on the left. Each row represents a serial dilution from left to right. (d) In vitro ADP-ribosylation to detect diphthamide formation on purified eEF2 from cells over-expressing eEF2. Fluorescence label indicates formation of diphthamide. (e) Reduction of Dph3 by Cbr1 is important for the two modifications. Figure 2a and 2c are representative of three biological triplicates. Figure 2b and 2d show representative images from two experimental repeats.
Interestingly, a small fraction of mcm5s2U is still formed in the cbr1Δmcr1Δ strain (Fig. 2b, and Supplementary Fig. 6), indicating some residual reduction of Dph3 by other reductase, possibly by Ncp1. We hypothesized that this residual reduction of Dph3 is sufficient to support the diphthamide modification, which needs much fewer electrons compared to the more abundant tRNA wobble uridine modifications. To test this, we checked if diphthamide formation in the cbr1Δmcr1Δ cells would be affected when eEF2 was over-expressed. While the BY4741 WT cells with over-expressed eEF2 do not confer any resistance to DT, cbr1Δ cells with over-expressed eEF2 are partially resistant to DT, and CBR1 and MCR1 double deletion render the cells almost full resistance to DT (Fig. 2c). Furthermore, purified eEF2 from cbr1Δ strain over-expressing eEF2 showed a significant decrease in diphthamide modification level (Fig. 2d, and Supplementary Fig. 9). These results suggested that Cbr1 is required for efficient reduction of Dph3 to support the increased electron demand in diphthamide biosynthesis when eEF2 is over-expressed.
In summary, we identified Cbr1 as the major physiological reductase of Dph3, the electron carrier for the radical SAM enzymes required for two distinct modifications that are important for translation fidelity (Fig. 2e). Because Cbr1 is a NADH dependent reductase, it is plausible that the overall amount of reduced Dph3 is regulated by the redox state of the cell via the NAD+/NADH ratio. Moreover, the abundant tRNA wobble uridine modifications are dependent on the availability of reduced Dph3. Thus, the Cbr1 and NADH-dependent reduction of Dph3 may provide a regulatory linkage between the metabolic state of the cells and protein translation. In bacteria, it is known that flavodoxin serves as the reductase for radical SAM enzymes.24 However, in eukaryotes, the identity of the physiological reduction system for radical SAM enzymes is largely unknown. The Cbr1/Dph3 system is the first physiological reduction system identified for radical SAM enzymes in eukaryotes. This finding may be useful for the identification of other eukaryotic reduction systems for radical SAM enzymes.
Methods
Yeast Strains
All strains used in this study are listed in Supplementary Table 1. The HL1352Y strain expressing endogenous FLAG-tagged Dph3 was generated using PCR-based tagging method as previously described.25 Briefly, the PCR fragment amplified from the plasmid pFA6a-6xGLY-3xFLAG-HIS3MX6 (Addgene plasmid 20753) with primers ZL210 and ZL211 (Supplementary Table 2) was transformed into BY4741 strain and plated on synthetic complete agar plates with histidine dropout for selection. The DPH2, DPH3, CBR1 and MCR1 single deletion strains were obtained from Open Biosystems (GE Dharmacon). The cbr1Δmcr1Δ strain was generated from cbr1Δ strain by Longtine PCR-based method as previously described.26 The PCR fragment amplified from the plasmid pFA6a-NATMX6 with primers ZL244 and ZL245 was transformed into the cbr1Δ strain. Transformed cells were grown in YPD media for three hours to recover and plated on YPD plates with 100 μg/mL nourseothricin for selection. The cbr1Δmcr1Δaim33Δ strain was generated by transforming cbr1Δmcr1Δ strain with PCR fragment amplified from pFA6a-His3MX6 (Addgene plasmid 41596) using primers ZL242 and Zl243 and selecting with synthetic complete histidine drop out plates. The HL1352Y strain with endogenous FLAG-tagged Dph3 was verified by anti-Flag western blot. Single deletion strains obtained from Open Biosystems were verified by PCR method using strain associated barcode primers. Deletion of MCR1 or AIM33 by Longtine fragments were confirmed by PCR method using 5’ UTR and 3’ UTR primers (Supplementary Table 2).
Sample preparation of Dph3 SILAC study
Saccharomyces cerevisiae BY4741 strain was cultured in synthetic complete media (200 mL) with light lysine and arginine with an initial A600 of 0.02 untill the A600 reached approximately 0.5. The HL1352Y strain expressing Flag-tagged Dph3 was first cultured in synthetic complete media with heavy lysine (Sigma 608041) and arginine (Sigma 608033) for about eight generation. The overnight culture was then used to inoculate 200 mL heavy synthetic complete media with an initial A600 of 0.02 untill the A600 reached approximately 0.5. Cells were harvested by centrifugation at 4,000 g and lysed with 600 μL of glass beads (OPS Diagnostics BAWG400-200-04) and 1 mL lysis buffer containing 50 mM Tris pH 8.0, 0.2% NP-40, 150 mM sodium chloride, 5 mM EDTA and 1 mM phenylmethanesulfonyl fluoride. Cells were lysed by 5 intervals of vortexing for 2 minutes with 2 minutes cooling on ice between intervals. Total cell lysates were cleared by centrifuging for 10 min at 13,000 g and 4 °C. The supernatants containing 2.5 mg of proteins were incubated with 25 μL anti-flag resins (Sigma A2220) for 4 hours at 4°C. The resins were washed with 1 mL of lysis buffer five times and eluted with 90 μL of elution buffer (50 mM Tris pH 8.0 and 1% SDS) and heated at 95 °C for 5 minutes. The eluted fractions were reduced by DTT (10 mM) for 30 minutes at room temperature and then alkylated by iodoacetamide (40 mM) for 30 minutes at room temperature. The heavy (HL1352Y) and light (BY4741) eluates from equal amounts of beads loaded with equivalent amounts of total lysate (2.5 mg) from the two cultures were mixed. Proteins were precipitated by adding 600 μL of precipitation buffer containing 50% Acetone, 49.9% ethanol and 0.1% acetic acid and cooling on ice for 30 minutes. The protein pellet was washed with 400 μL of ice-cold precipitation buffer and air-dried. The resultant pellet was resuspended in 50 μL of resolubilization buffer containing 8 M urea and 50 mM Tris pH 8.0, and then diluted with 400 μL 50 mM Tris pH 8.0 and digested by 1 μg trypsin overnight at 37 °C.
Nano LC/MS/MS and data Analysis of Dph3 SILAC interactome sample
The SILAC tryptic digests were reconstituted in 50 μL of 0.5% formic acid (FA) estimated at 0.1 μg/μL for nanoLC-ESI-MS/MS analysis, which was carried out on an Orbitrap Elite mass spectrometer (Thermo-Fisher Scientific, San Jose, CA) equipped with a “CorConneX” nano ion source device (CorSolutions LLC, Ithaca, NY). The Orbitrap was interfaced with a Dionex UltiMate3000RSLCnano system (Thermo, Sunnyvale, CA). Each SILAC peptide sample (5 μL) was injected under “User Defined Program” onto a PepMap C18 trap column-nano Viper (5 μm, 100 μm × 2 cm, Thermo) at 20 μL/min flow rate and then separated on a PepMap C18 RP nano column (3 μm, 75 μm × 25 cm, Thermo) which was installed in the nano device with a 10-μm spray emitter (NewObjective, Woburn, MA). The peptides were eluted with a 120 minutes gradient of 5% to 38% acetonitrile (ACN) in 0.1% formic acid at a flow rate of 300 nL/min, followed by a 7-min ramping to 95% ACN-0.1% FA and a 8-min hold at 95% ACN-0.1% FA. The column was re-equilibrated with 2% ACN-0.1% FA for 25 minutes prior to the next run. The Orbitrap Elite was operated in positive ion mode with nano spray voltage set at 1.6 kV and source temperature at 250 °C with external calibration for FT mass analyzer being performed. The instrument was operated in parallel data-dependent acquisition (DDA) under FT-IT mode using FT mass analyzer for one MS survey scan from m/z 375 to 1800 with a resolving power of 120,000 (FWHM at m/z 400) followed by MS/MS scans on top 20 most intensive peaks with multiple charged ions above a threshold ion count of 10,000 in FT mass analyzer. Dynamic exclusion parameters were set at repeat count 1 with a 30 s repeat duration, an exclusion list size of 500, 60 s exclusion duration with ±10 ppm exclusion mass width. Collision induced dissociation (CID) parameters were set at the following values: isolation width 2.0 m/z, normalized collision energy at 35 %, activation Q at 0.25, and activation time 10 ms. All data were acquired under Xcalibur 2.2 operation software (Thermo). All MS and MS/MS raw spectra were processed and searched using Sequest HT software within the Proteome Discoverer 1.4.1.14 (PD 1.4, Thermo Scientific). The Saccharomyces cerevisiae RefSeq sequence database (5,847 entries, downloaded on 5/17/2015 from NCBInr) was used for database searches. The database search was performed under a search workflow with the “Precursor Ions Quantifier” node for SILAC 2plex (Arg10, Lys8) quantitation. The default settings for protein identification in Sequest node were: two mis-cleavages for full trypsin with fixed carbamidomethyl modification of cysteine, variable modifications of 10.008 Da on Arginine and 8.014 Da on lysine, N-terminal acetylation, methionine oxidation and deamidation of asparagine and glutamine residues. The peptide mass tolerance and fragment mass tolerance values were 15 ppm and 0.8 Da, respectively. Only high confidence peptides defined by Sequest HT with a 1% FDR by Percolator were considered for the peptide identification. The mass precision for expected standard deviation of the detected mass used to create extracted ion chromatograms was set to 3 ppm. The SILAC 2-plex quantification method within PD 1.4 was used to calculate the heavy/light ratios of all identified proteins. Only unique peptides were used for quantification. The final protein list was filtered by proteins with at least two peptides identified. 500 was set as the maximum H/L ratio to make it mathematically meaningful for peptides essentially not present in the light sample. Table in Supplementary Figure 2b lists protein with H/L ratio greater than 10.
Cloning, expression and purification of Cbr1, Mcr1, Pga3, and Ncp1
Genomic DNA was extracted from Saccharomyces cerevisiae BY4741 strain using Pierce Yeast DNA Extraction Kit. The DNA sequence encoding for Cbr1 with an N-terminal truncation of 27 amino acids was amplified by PCR with primers ZL224 and ZL222 (Supplementary Table 2) from the genomic DNA. The amplified gene fragment was inserted into the pET28a vector and transformed into the Escherichia coli expression strain BL21 pRARE2. A single colony was used to inoculate an overnight starter culture, which was used to inoculate 2 liters of lysogeny broth (LB) containing 50 μg/mL kanamycin and 20 μg/mL chloramphenicol. Cells were grown at 37°C to A600 of approximately 0.6 and cooled to 16°C. Expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 50 μM riboflavin, and grown overnight at 16°C. Cells were harvested by centrifugation and lysed using the EmulsiFlex-C3 cell disruptor (Avestin, Inc., Canada). The protein was purified on BioLogic DuoFlow 10 System (Bio-Rad, Hercules, CA). The purification was performed on a HisTrap HP column (GE Healthcare, Piscataway, NJ) with a linear gradient from 30 mM imidazole to 500 mM imidazole in 30 minutes. The yellow protein fractions were collected and dialyzed against 25 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl. Protein concentration was determined by standard Bradford assay.
The DNA sequence encoding for Pga3 with an N-terminal truncation of 61 amino acids was amplified by PCR with primers ZL234 and ZL235 (Supplementary Table 2) from the genomic DNA. The DNA sequence encoding Ncp1 with an N-terminal truncation of 33 amino acids was amplified by PCR with primers ZL316 and ZL318 from the genomic DNA. Yeast complementary DNA was used as template for amplification of the mcr1 gene. Yeast RNA was purified using the TRIzol® reagent (Thermo Fisher Scientific). DNA was removed from the purified RNA using DNase I (New England Biolabs). The RNA was extracted with phenol/chloroform to inactivate DNase I. Total cDNA was synthesized from the purified RNA using SuperScript® III Reverse Transcriptase (Thermo Fisher Scientific). The DNA sequence encoding for Mcr1 with an N-terminal truncation of 31 amino acids was amplified by PCR with primers ZL240 and ZL241 (Supplementary Table 2) from the synthesized cDNA. Subsequent cloning and expression of recombinant Pga3, Ncp1, and Mcr1 were similar to that of Cbr1.
Ultraviolet–visible spectroscopy of Dph3 reduction reactions
Recombinant yeast Dph3 was purified as previously described.7 The reaction was monitored on a Cary 50 Bio UV-Vis spectrophotometer (Varian) at 488 nm. Cbr1 (0.5 μM) was mixed with 100 μM of Dph3 in a cuvette. The reaction was initiated by addition of NADH or NADPH at a final concentration of 0.2 mM or as indicated. Reduction of Dph3 by Mcr1 or Pga3 was monitored under the same conditions as that of Cbr1. Figure 1b and Supplementary Figure 5a-5c show a representative image from three experimental repeats.
Anaerobic reconstitution of the first step of yeast diphthamide biosynthesis
The Dph3 protein, Dph1-Dph2 complex and eEF2 proteins from BY4741 dph2 deletion strain were expressed and purified as previously described.7 The reaction mixture was assembled in the anaerobic chamber under strictly anaerobic conditions. Aerobically purified Dph3, Cbr1 and eEF2 were degassed by a schlenk line. The reconstitution reactions were set up in an anaerobic chamber. Dph1-Dph2 (5 μM), Dph3 (10 μM), eEF2 (2 μM), Cbr1 or Mcr1 or Ncp1 (5 μM), and NADH or NADPH (200 μM) were added to a buffer containing 150 mM NaCl and 200 mM Tris-HCl at pH 7.4. Reactions without NADH or without reductases were also carried out as negative controls. The reaction vials were sealed before being taken out of the anaerobic chamber. 14C-SAM (ARC 0343-50, 18 μM) was injected into each reaction vial to initiate the reaction. The reaction mixtures were mixed by brief vortexing and incubated at 30 °C for 60 minutes. The reactions were stopped by adding protein loading dye and subsequently heating at 95 °C for 5 minutes, and then resolved by 12% SDS–polyacrylamide gel electrophoresis. The dried gel was exposed to a Phosphor Imaging screen and scanned using a Typhoon FLA 7000 (GE Healthcare Life Sciences). Figure 1c and Supplementary Figure 7 show a representative image from three experimental repeats. Supplementary Figure 5d and Supplementary Figure 9 show a representative image from two experimental repeats.
Purification of eEF2 and its in vitro ADP-ribosylation by DT
Cells transformed with p423Met25-eEF2 (allowing over-expression of eEF2 with a 8 His C-terminal tag) were cultured in synthetic complete media with histidine dropout at 30 °C with an initial A600 of 0.02 until the A600 reached approximately 1.0. Cells harvested were lysed and eEF2 was purified as previously described.5 Purified eEF2 (1 μM) and Rh-NAD (25 μM) were incubated with DT (1μM) at 30 °C in 50 mM NaCl, 30 mM dithiothreitol (DTT), 2 mM ethylenediaminetetraacetic acid (EDTA), and 25 mM Tris-HCl at pH 8.0 for 15 minutes as previously described.4 Figure 2d shows a representative image from two experimental repeats.
DT and γ-toxin sensitivity assays
For DT sensitivity assays, cells were transformed with plasmid pHL1015, which allows for galactose-inducible, glucose-repressible expression of diphtheria toxin as previously described.4 Transformed cells were cultured in synthetic complete media with uracil dropout at 30 °C overnight, adjusted to A600 of 0.2 with autoclaved water, and then diluted serially in 4-fold increments. Aliquots of each dilution were spotted on glucose-containing or galactose-containing agar plates using a replica plater. Plates were incubated at 30 °C. Cell growth was recorded 2-3 days after plating. DT sensitivity assay for cells co-transformed with p423Met25-eEF2 and pHL1015 were performed similarly and plated onto galactose-containing or glucose-containing synthetic complete with histidine and uracil dropout agar plates. For γ-toxin sensitivity assays, cells were transformed with plasmid pLF16 which allows for galactose-inducible, glucose-repressible expression of γ-toxin as previously described.23 Cultures were grown in synthetic complete media with leucine dropout at 30°C. Plating of cells on agar plates was performed similar to that of DT assays. Figure 2a, 2c and Supplementary Figure 4 are representative of three biological triplicates.
Cloning, expression and purification of γ-toxin
The DNA sequence encoding for γ-toxin with an N-terminal truncation of 19 amino acids was amplified by PCR with primers ZL436 and ZL437 (Supplementary Table 2) from pLF16. The amplified gene fragment was inserted into the pET28a vector and transformed into the Escherichia coli expression strain BL21 pRARE2. A single colony was used to inoculate an overnight starter culture, which was used to inoculate 2 liters of LB containing 50 μg/mL kanamycin and 20 μg/mL chloramphenicol. Cells were grown at 37°C to A600 of approximately 0.6 and cooled to 16°C. Expression was induced with 0.5 mM IPTG and grown overnight at 16°C. Subsequent protein purification was performed similar to that of Cbr1.
Bulk tRNA isolation and in vitro γ-toxin treatment
Yeast cells were cultured in 2 liters YPD from initial A600 of 0.02 till A600 reached approximately 0.5. Cells were harvested and bulk tRNA was purified as previously described.27 The total tRNAs (5μg) were incubated withγ-toxin (5 μM) in 10 mM NaCl, 10 mM MgCl2, 1 mM DTT and 10 mM Tris-HCl at pH 7.4 for 2 hours at 30°C. The time course of γ-toxin treatment in Figure S5 was performed under the same reaction conditions with specified incubation time. The samples were separated on 12% polyacrylamide, 8M urea gels, and transferred to nylon membrane (GE Health, rpn119b) for northern blot. The oligonucleotide used to detect 5’ of glu-tRNA was ordered from IDT (/5AmMC6/GTGATAGCCGTTACACTATATCGGA) and conjugated to Alexa Fluor® 680 (ThermoFisher, A37567). Northern blots were visualized by Odyssey® CLx imaging system (LI-COR). Figure 2b shows a representative image from two experimental repeats.
Supplementary Material
Acknowledgment
We thank Dr. Sheng Zhang (Cornell Proteomics & Mass Spectrometry Facility) for the help with MS experiment. We are grateful to Dr. Stewart Shuman (Memorial Sloan Kettering Cancer Center) for providing us with the plasmid pLF16. pFA6a-NATMX6 was a gift from Dr. Scott Emr (Cornell University). This work is supported by NIH (R01GM088276). Z.L. is supported by a Singapore Agency for Science, Technology and Research (A∗STAR) scholarship.
Footnotes
Author contributions
Z.L. and H.L. designed the experiments and wrote the paper. M.D., Y.Z. and E.A.L. purified Dph1-Dph2, Dph2 and eEF2 proteins. Z.L. performed the SILAC interactome study of Dph3, in vitro Dph3 reduction experiments and in vivo toxin sensitivity assays.
Competing financial interests
The authors declare no competing financial interests.
Reference
- 1.Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S, et al. Proc Natl Acad Sci USA. 2012;109:13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Mol Cell Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su X, et al. Proc Natl Acad Sci USA. 2012;109:19983–19987. doi: 10.1073/pnas.1214346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, et al. J Am Chem Soc. 2014;136:6179–6182. doi: 10.1021/ja5009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong M, et al. J Am Chem Soc. 2014;136:1754–1757. doi: 10.1021/ja4118957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattheakis LC, Shen WH, Collier RJ. Mol Cell Biol. 1992;12:4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Johansson MJ, Bystrom AS. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jablonowski D, et al. Genetics. 2001;159:1479–1489. doi: 10.1093/genetics/159.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selvadurai K, Wang P, Seimetz J, Huang RH. Nat Chem Biol. 2014;10:810–812. doi: 10.1038/nchembio.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlsborn T, et al. RNA Biol. 2014;11:1519–1528. doi: 10.4161/15476286.2014.992276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann M. Methods Mol Biol. 2014;1188:1–7. doi: 10.1007/978-1-4939-1142-4_1. [DOI] [PubMed] [Google Scholar]
- 14.Bar C, Zabel R, Liu S, Stark MJ, Schaffrath R. Mol Microbiol. 2008;69:1221–1233. doi: 10.1111/j.1365-2958.2008.06350.x. [DOI] [PubMed] [Google Scholar]
- 15.Kolaj-Robin O, McEwen AG, Cavarelli J, Seraphin B. FEBS J. 2015;282:819–833. doi: 10.1111/febs.13199. [DOI] [PubMed] [Google Scholar]
- 16.Glatt S, et al. Structure. 2015;23:149–160. doi: 10.1016/j.str.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Du M, Shirabe K, Takeshita M. Biochem Biophys Res Commun. 1997;235:779–783. doi: 10.1006/bbrc.1997.6873. [DOI] [PubMed] [Google Scholar]
- 18.Honjo T, Nishizuka Y, Hayaishi O. J Biol Chem. 1968;243:3553–3555. [PubMed] [Google Scholar]
- 19.Chen JY, Bodley JW, Livingston DM. Mol Cell Biol. 1985;5:3357–3360. doi: 10.1128/mcb.5.12.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb DC, Kelly DE, Manning NJ, Kaderbhai MA, Kelly SL. FEBS Lett. 1999;462:283–288. doi: 10.1016/s0014-5793(99)01548-3. [DOI] [PubMed] [Google Scholar]
- 21.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Proc Natl Acad Sci USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keppetipola N, Jain R, Meineke B, Diver M, Shuman S. RNA. 2009;15:1036–1044. doi: 10.1261/rna.1637809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bianchi V, et al. Biochem Biophys Res Commun. 1993;197:792–797. doi: 10.1006/bbrc.1993.2548. [DOI] [PubMed] [Google Scholar]
- 25.Funakoshi M, Hochstrasser M. Yeast. 2009;26:185–192. doi: 10.1002/yea.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine MS, et al. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.de Crecy-Lagard V, et al. Mol Biol Evol. 2010;27:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.