Abstract
Despite being critical for energy generation in most forms of life, few if any microbial antibiotics specifically inhibit glycolysis. To develop a specific inhibitor of the glycolytic enzyme Enolase 2 for the treatment of cancers with deletion of Enolase 1, we modeled the synthetic tool compound inhibitor, Phosphonoacetohydroxamate (PhAH) into the active site of human ENO2. A ring-stabilized analogue of PhAH, with the hydroxamic nitrogen linked to the alpha-carbon by an ethylene bridge, was predicted to increase binding affinity by stabilizing the inhibitor in a bound conformation. Unexpectedly, a structure based search revealed that our hypothesized back-bone-stabilized PhAH bears strong similarity to SF2312, a phosphonate antibiotic of unknown mode of action produced by the actinomycete Micromonospora, which is active under anaerobic conditions. Here, we present multiple lines of evidence, including a novel X-ray structure, that SF2312 is a highly potent, low nM inhibitor of Enolase.
Keywords: Glycolysis, Enolase, Enzyme Inhibitor, Phosphonate
INTRODUCTION
Glycolysis is an ancient and conserved metabolic pathway1. Glycolytic enzymes are essential genes in diverse microorganisms2,3. Yet, few antibiotic substances specifically inhibit enzymes of this pathway. Inhibition of glycolysis has gained considerable attention in the field of oncology because cancer cells frequently exhibit a bioenergetics shift towards glycolysis, the so-called Warburg effect4.
We have previously presented proof-of-concept that glioma cells with passenger deletion of the 1p36 locus covering Enolase 1 (ENO1; αEnolase), are highly dependent on Enolase 2 (ENO2; γEnolase5). Enolase is a dimeric enzyme that catalyzes the penultimate step in glycolysis, interconverting 2-phosphoglycerate (2-PGA) and phosphoenolpyruvate (PEP). The most potent enolase inhibitor described in the literature is Phosphoacetohydroxamate (PhAH, Fig. 1a), which has nM IC50 inhibitory activity against Enolase from diverse sources6,7. Although several structures of human ENO2 have been reported8, PhAH-bound structures are only available for yeast and trypanosome Enolase9-11. We modeled PhAH into the active site of human ENO2 and asked what derivatives might improve the binding to the active site.
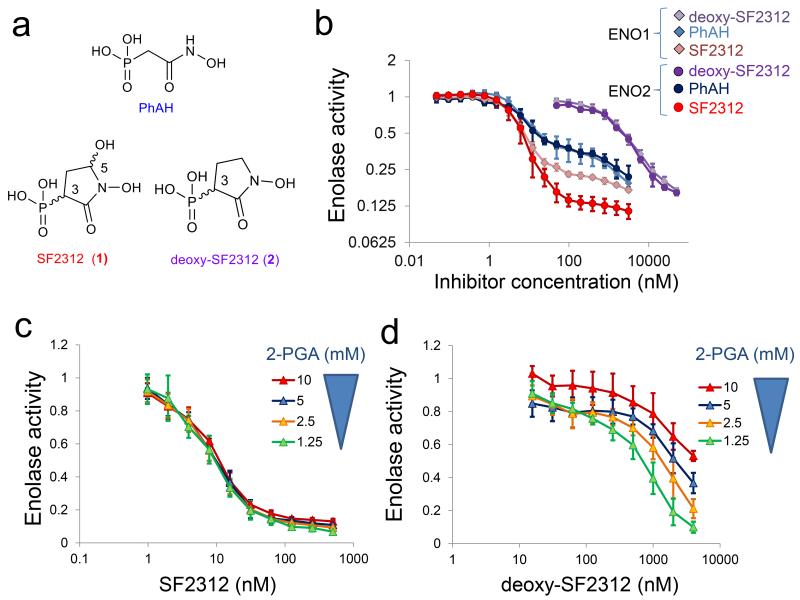
FIGURE 1. SF2312 is a potent inhibitor of Enolase with mixed competitive and non-competitive kinetics.
(a) Chemical structure of PhAH, SF2312 (1), deoxy-SF2312 (2), with chiral centers indicated. (b) Enolase enzymatic activity was measured in lysates of the D423 cell line overexpressing human ENO1 and ENO2. Inhibitors were incubated with enzyme prior to addition of 5 mM substrate. Enzymatic activity was normalized to that in the absence of the inhibitor (y-axis, log2) and plotted as function of inhibitor concentration (nM, x-axis). Traces for ENO2 inhibited with SF2312, PhAH and deoxy-SF2312 are shown as red, blue and purple circles, respectively. Traces for ENO1 are shown crimson, light blue, and light purple diamonds. Each data point shows the mean ± S.D of N = 6 independent measurements. (c, d) Enolase activity (ENO2) was measured as a function of substrate (2-PGA) and inhibitor (c, SF2312; d, deoxy-SF2312) concentration. Data points show mean ± S.D of N = 5 independent measurements.
Molecular docking and modeling studies led us to hypothesize cyclic PhAH derivatives. Structure-based searches revealed an existing molecule bearing strong similarity to such prophetic molecules: SF2312 (1, (1,5-dihydroxy-2-oxopyrrolidin-3-yl)phosphonic acid, Fig. 1a), previously isolated as natural phosphonate antibiotic of unknown mode of action12. Here, we provide multiple lines of evidence that SF2312 is a highly potent inhibitor of Enolase.
RESULTS
Cyclic PhAH derivatives show strong similarity to SF2312
Diverse structures of Enolase have been published, including human ENO2 with the inhibitors tartronate semialdehyde phosphate and lactic acid phosphate8 but no structure of human Enolase with PhAH is currently available. The most current structures of human ENO2 contain its natural substrates: PEP and 2-PGA. In order to probe the binding of PhAH to ENO2, a docking model was created based on the crystal structure of human Enolase 2 (PDB codes 3UJS, 3UJR, 3UJF, 3UJE, 3UCD, 3UCC8) with bound substrates, PEP and 2-PGA. For eleven Enolase inhibitors with known Ki values6,7, we found an r2 = 0.55 for a regression model relating experimental pKi to the Quantum Polarized Linear Dynamic (QPLD) based ΔGbind. (p-value < 0.05). The model for rigid-receptor docking (RRD) plus ΔGbind had a substantially lower value, r2 = 0.42. Using this model, PhAH could be docked at the active site closely resembling the published structures of PhAH bound to trypanosome and yeast enolase (Supplementary Results, Supplementary Fig. 1a,d6,9). The hydroxamate bound closely to the high-affinity Mg atom (Mg(A)), with the carbonyl forming an oxo-bridge between the two Mg atoms. Mg(A) is tight binding whereas Mg(B) comes on and off during the catalytic cycle with a Ka of around 1 mM [7]. Mg(B) was coordinated with the carbonyl and the phosphonate oxygen. We noticed extensive unoccupied space in the active site (Supplementary Fig. 1a, blue dotted circle) in the PhAH bound structure. This space is unoccupied or filled by water in the trypanosome PhAH-bound enolase structure (1ELS, 2PTZ, 2PUO9,10). We theorized that a cyclic derivative of PhAH, which would yield a secondary nitrogen on the hydroxamate moiety, would fill the unoccupied space, increase the rigidity of the molecule and stabilize it in the bound configuration. This 5-membered ring (2, thereafter, deoxy-SF2312, Fig. 1a, Supplementary Fig. 1b,e), was successfully docked and scored very well in the QLP model. Structure based-searches revealed that a very similar molecule, possessing a hydroxyl group on the 5 position, had already been described as SF2312 (1), a natural antibiotic substance active against bacteria under anaerobic conditions (Ref12, Supplementary Fig. 1c,f). Since anaerobic conditions favor dependence on glycolytic fermentation, this suggests that SF2312 might be an inhibitor of glycolysis, likely targeting Enolase. Yet under the RRD+ ΔGbind model, SF2312 had a non-optimal value of −3.76, though the binding configuration appeared reasonable. Changing to QPLD+ ΔGbind model, led to a dramatic shift to −81.6 (pred. pKi = 8.31). SF2312 contains stereocenters at the 3- and 5-positions and both the 3S,5S and 3S,5R but not the 3R,5S 3R,5R isomers could be accommodated in the site. SF2312 showed a number of new interactions with catalytically important residues not seen with PhAH, such as hydrogen bonding with E209 or H370, depending on the S,S/S,R configuration (6Supplementary Fig. 1c,f). These encouraging in silico results prompted us to synthesize and test SF2312 for Enolase inhibitory activity.
SF2312 potently inhibits Enolase enzymatic activity
Both SF2312 and deoxy-SF2312 were synthesized following published procedures (Supplementary Note 113,14). SF2312 was obtained as a racemic mixture of the cis and trans diastereomers that closely mirrors the composition of the natural sample12,13. We attempted to perform chiral separation to generate enantiomerically pure SF2312 (through contract with Phenomenex, Torrence, CA). While SF2312 itself proved impractical to separate due to its high polarity and lack of UV detectable groups, intermediate 3 was successfully separated into its four enantiopure isomers (Supplementary Note 2). However, de-protection reactions (Steps 5 and 6, in Supplementary Note 1) carried out on enantiomerically pure intermediates 3 yielded fully racemic SF2312. Indeed, both stereocenters underwent spontaneous epimerization in aqueous solution (Supplementary Note 2). This was unfortunately expected due to the nature of the two stereocenters, with the C-5 being an anomeric center and the 3-H a highly acidic α-proton. As such, these results suggest that the synthesis of enatiomerically pure SF2312 may not be technically possible.
The effect of SF2312, deoxy-SF2312 and PhAH on the enzymatic activity of Enolase in vitro was determined using an indirect, Pyruvate kinase/Lactate Dehydrogenase linked assay (NADH fluorescence) or directly by measuring the appearance of PEP (absorption at 240 nm). Enolase inhibitory activity was measured in lysates of mouse organs, human cancer cell lines overexpressing ENO1 and ENO2 as well as purified human ENO1 and ENO2 expressed in E. coli. Depending on the source, the IC50 of SF2312 ranged from 10 nM to 50 nM (Supplementary Table I). The shape of the inhibitor titration curves are quite unusual; i.e. an apparent plateau is reached after IC50. SF2312 showed similar IC50 towards ENO1 and ENO2 but at higher concentrations of inhibitor, residual activity was consistently lower for ENO2 than for ENO1 (Fig. 1b). At IC50, SF2312 exhibited non-competitive kinetics with respect to substrate 2-PGA (Fig. 1c) but competitive kinetics at higher concentrations of inhibitor. We speculate that the unusual titration curves and mixed kinetics are related to the anti-cooperative binding behavior of the Enolase dimer8, whereby binding of inhibitor to one active site in the dimer decreases the affinity for inhibitor binding at the other active site8.
The inhibitory potency of SF2312 against Enolase was greatly influenced by whether the inhibitor or the substrate was first added in the assay system. That is, SF2312 acted as a slow-on/slow-off inhibitor. Similar behavior was reported previously for PhAH7, but this was exaggerated for SF2312 (Supplementary Fig. 2). The difference of inhibitor potency against ENO2 and ENO1 was most pronounced for the off-rate, as the differences between the isozymes was evident when the inhibitors were pre-incubated with the enzyme (Fig. 1b and Supplementary Fig. 2a), but were not different when the substrate was added prior to the inhibitors (Supplementary Fig. 2b). Deoxy-SF2312 was much less potent as an enolase inhibitor, with an IC50 of ~2000 nM. Unlike SF2312, deoxy-SF2312 shows clear competitive kinetics with respect to the substrate 2-PGA (Fig. 1d) and minimal difference in inhibitory effectiveness between ENO2 and ENO1 (Fig. 1b). These results suggest that the 5-OH in SF2312 is responsible for the preferential inhibition of ENO2 over ENO1 (above IC50) and mediates tighter binding of the inhibitor to the enzyme.
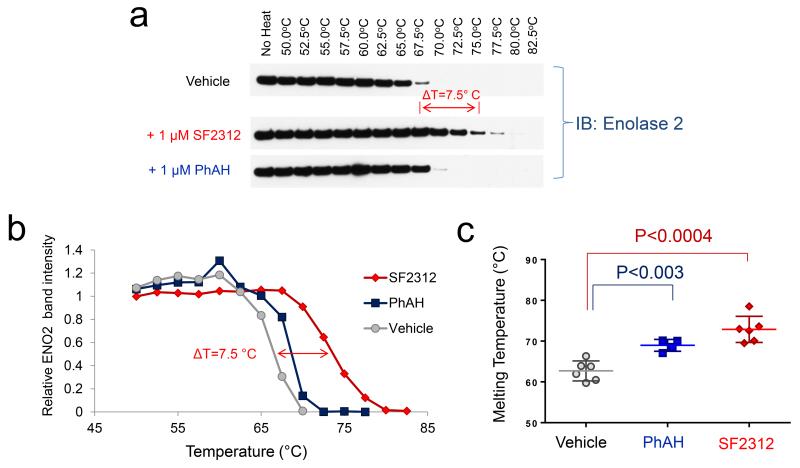
As an additional test of SF2312 binding to the Enolase protein, we performed ligand-induced thermal shift assays15 on cell lysates under the same conditions as we performed the enzymatic assay. Heat-denatured proteins precipitate out of solution when their hydrophobic core is exposed and as a result, disappear from the lysate after centrifugation, whereas native properly folded proteins remain in solution. The levels of specific proteins in the supernatant (i.e. non-denatured) are followed by immunoblotting as a function of increasing temperature. Incubation of cell lysates with 1 μM of SF2312, shifted the melting temperature (Tm) of ENO2 from ~63 °C to ~ 72.5 °C, i.e. a ~ 9.5 °C shift in the thermal denaturation of ENO2 (Fig. 2a, 2b, 2c). The same concentration of PhAH led to a more modest ~ 6 °C stabilization of the protein. Enolase undergoes large conformational changes during catalysis, including opening of the active site8. The stabilization issued by SF2312 could be related to hydrogen bonds formed between the 5-OH and the active site (Fig. 3a, 3b, Supplementary Fig. 3), which likely lock the enzyme in the tighter configuration and minimize instability of the catalytic loop9,16,17. Similar results were obtained for ENO1 (Supplementary Fig. 4), except that the protein is overall less thermally stable than ENO218. Neither PhAH nor SF2312 affected the thermal stability of internal controls, Vinculin or Triosephosphate isomerase (Supplementary Fig. 5). The same experiments were repeated with recombinant human ENO2 protein (the same preparation used for X-ray structures), confirming results obtained in lysates, with SF2312 yielding a ~ 12 °C stabilization as compared to ~ 6 °C by PhAH (Supplementary Fig. 6).
FIGURE 2. SF2312 stabilizes Enolase 2 against thermal denaturation.
(a) Lysates of D423 overexpressing ENO2 cells were incubated with vehicle, 1 μM of PhAH or SF2312 and thermally denatured, with immunoblotting performed in post-centrifugation supernatants (uncropped images Supplementary Fig. 20). (b) Quantification of band intensity (y-axis) versus temperature (x-axis), with the trace in grey being vehicle, blue being PhAH and red being SF2312 treatment groups. (c) Mean melting temperatures (Tm 50) ± S.D for N = 6 control, N = 4 PhAH, N = 6 SF2312 independent measurements. Significant results by unpaired t-test with Bonferroni correction are indicated.
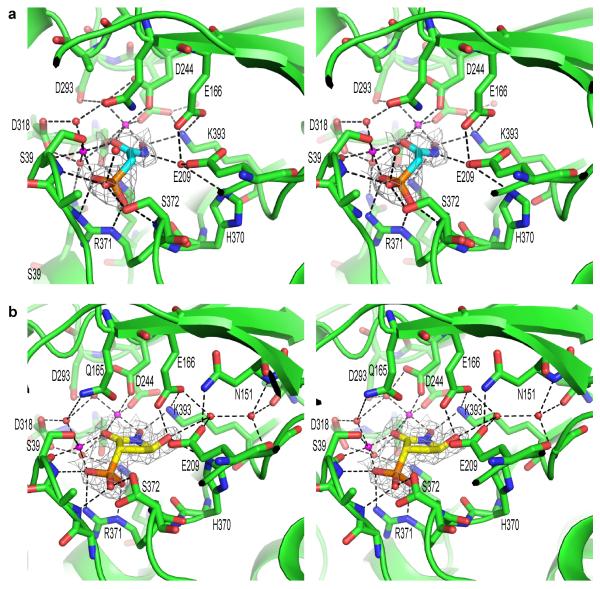
FIGURE 3. PhAH and SF2312 interact with ENO2 through a complex network of electrostatic, metal coordination and hydrogen bond interactions.
(a) Stereo presentation of PhAH and (b) SF2312 binding in the ENO2 active site. The protein backbone is shown using the cartoon depiction with key amino acids for magnesium or ligand binding highlighted using the stick representation. Magnesium ions and water molecules are shown as magenta or red spheres respectively. Hydrogen bonds and metal coordination bonds are both represented by black dashed lines. The 2Fo-Fc unbiased omit electron density map for each ligand, cotoured at 1.5σ, is shown as a grey mesh around the ligand. Coordinates were deposited in PDB (ENO2:PhAH, 4ZA0; ENO2:SF2312, 4ZCW).
Direct structural determination of SF2312 bound to ENO2
Human ENO2 was crystallized with SF2312 and PhAH in order to determine the structure of the enzyme bound to the ligands. Crystals of human ENO2 were generated using recombinant protein expressed in E. coli. They were then co-crystallized with PhAH and SF2312 by soaking for 16 hours in cryoprotectant containing a 2 mM solution of PhAH or a 4 mM solution of SF2312 respectively. The structure of dimeric ENO2:PhAH (4ZA0; Supplementary Table 2) and ENO2:SF2312 (4ZCW; Supplementary Table 2) complexes were analyzed by X-ray crystallography and solved at 2.31 Å and 1.99 Å resolution, with Rfree for the refined structures of 0.195 and 0.202 respectively (Supplementary table 1, 2 for the PhAH and SF2312 structures, respectively). PhAH binds to human ENO2 in a very similar mode to what had been reported previously in yeast and trypanosome Enolase9,11, with the ligand interacting with two magnesium ions in the active site pocket. The SF2312 bound structure revealed the preferential binding of the S,S stereoisomer with the compound adopting a binding mode similar to PhAH. For both PhAH and SF2312, the phosphono group coordinates the catalytic magnesium, Mg(B), and forms salt bridged interactions with R371 (Fig. 3a, 3b and Supplementary Fig. 3). The carbonyl of PhAH and SF2312 make interactions with both Mg(A) and Mg(B) to complete the octahedral coordination of these ions and the hydrogen bonding of the hydroxylamine group is conserved for both inhibitors. In the PhAH bound structure, the hydroxylamine hydrogen binds via a buried water molecule to E209, E166 and H370. E209, E166 and H370 act in concert to remove the hydroxyl from the substrate 2-PGA during the catalytic cycle8 and are critical for Enolase activity. In the SF2312 bound structure, the 5′-hydroxyl group of SF2312 replaces the water molecule and binds directly to E166 and H370. SF2312 binds to E209 via a water mediated hydrogen bond and to the backbone of N151 and G396 through a second water molecule. The position of these two water molecules is conserved in the ENO2:PhAH complex where they play an indirect role in the binding of PhAH by stabilizing the position and orientation of E209, E167 and H370 (Fig. 3a, 3b, Supplementary Fig. 3). The binding mode predicted by the docking molecular modelling is in excellent agreement with the experimentally determined structure. In addition, these water-mediated hydrogen bonds revealed by the crystal structure, provide additional information that was not predicted by molecular modelling, therefore creating a detailed picture of how this natural product is able to inhibit Enolase activity.
SF2312 is selectively toxic to ENO1-deleted glioma cells
We previously demonstrated that ENO1-deleted glioma cells are more sensitive to PhAH than ENO1-intact glioma cells or normal human astrocytes5. We examined the effects of SF2312 on cell proliferation, cell death, as well as a glycolytic flux and bioenergetics parameters in ENO1-deleted and isogenic rescued control cells. To provide a frame of reference, selected experiments were repeated with PhAH.
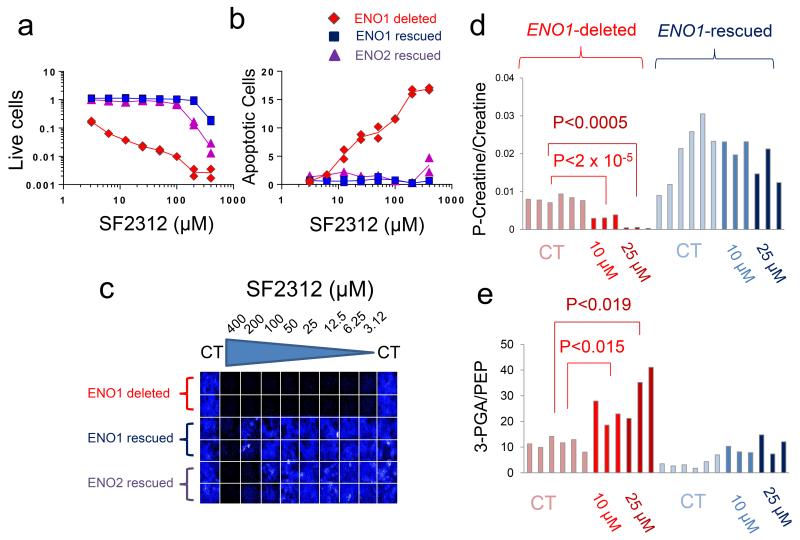
SF2312 displayed strong selective toxicity towards ENO1-deleted glioma cells. SF2312 inhibited the proliferation (2 weeks treatment course) of the ENO1-deleted D423 glioma cell line in the low μM range whist isogenically ENO1-rescued D423 cells, ectopically re-expressing ENO1 (Supplementary Fig. 7) only showed inhibition of proliferation at concentrations of SF2312 above 200 μM (Fig. 4a, 4b, 4c). Isogenically rescued D423 cells overexpressing ENO2 (Supplementary Fig. 7) were somewhat more sensitive than those re-expressing ENO1, but still dramatically more resistant than parental ENO1-deleted D423 cells. Similar selectivity was observed for the induction of cell death. D423 ENO1-deleted glioma cells showed induction of cell death starting at 12.5 μM of SF2312, whilst isogenic ENO1-rescued controls only showed cell death induction at 400 μM inhibitor (Fig. 4b). The same experiments conducted with PhAH recapitulated the same overall patterns (Supplementary Fig. 8). SF2312 and PhAH also exhibited selective toxicity against another ENO1-deleted glioma cell line, Gli56 (Supplementary Fig. 9). We determined the effect of hypoxia on the toxicity of Enolase inhibitors. After 72 hours of treatment with SF2312, ENO1-deleted D423 glioma cells under hypoxia were essentially eradicated at concentrations >6.25 μM (Supplementary Fig. 10). Under normoxia, inhibition of proliferation was evident in D423 ENO1-deleted cells but even at 100 μM inhibitor, no eradication was observed (Supplementary Fig. 10; these effects are less dramatic than those in Fig. 4a, as the later were treated for 2 weeks rather than 72 hours for the former). Toxicity of PhAH was also potentiated by hypoxia, but this was only evident at 200 μM.
FIGURE 4. SF2312 inhibits Enolase activity and is selectively toxic to ENO1-deleted glioma cells.
(a) Effect of SF2312 on cell proliferation (total cell number, Hoechst 33342) and (b) apoptosis (YO-PRO®-1 positive cells). D423 ENO1-deleted (red), D423 isogenic controls expressing ENO1 (dark blue) or overexpressing ENO2 (light blue) were treated for 2-weeks with varying concentrations of inhibitors as indicated (μM). Each condition was conducted in duplicate with each data point represents one biological replicate, expressed as a function of vehicle control. (c) A representative plate image stained with Hoechst 33342. The experiment probing toxicity of PhAH and SF2312 against D423 ENO1-deleted cells was repeated at least four times. (d) D423 ENO1-deleted and isogenic control glioma cells were treated with vehicle, 10 μM and 25 μM SF2312 for 72 hours. Polar metabolites were extracted and quantified by mass spec, showing the ratio of (d) phospho-creatine to creatine and (e) of 3-PGA to PEP. Bars represent individual measurements of independently treated cells. Light blue bars, D423 ENO1-deleted glioma cells CT; blue bars, ENO1-deleted cells treated with 10 μM SF2312; Dark blue bars, ENO1-deleted, 25 μM SF2312; Light crimson bars, D423 ENO1-rescued cells CT; crimson bars, ENO1-rescued, 10 μM SF2312; Brown bars, ENO1-rescued, 25 μM SF2312. Statistically significant comparisons, by unpaired, 2-tailed, t-test with Bonferroni correction are indicated.
Selective toxicity of SF2312 and PhAH to ENO1-deleted versus ENO1-rescued glioma cells was closely paralleled by bioenergetic state and glycolytic flux. Cell death and inhibition of proliferation in ENO1-deleted cells were preceded by decreases in ATP levels (Supplementary Fig. 11). Declines in ATP occurred as early as 8 hours after initiation of treatment with SF2312 (Supplementary Fig. 11), while induction of cell death and inhibition of proliferation only became apparent after 24 hours of treatment (Supplementary Fig. 12). Decreases in ATP levels were mirrored by decreases in other high energy phosphates, such as phosphocreatine (Fig. 4d). These effects were selective for ENO1-deleted glioma cells and were fully reversed by ectopic re-expression of ENO1.
We measured glycolytic flux by determining the conversion of 13C-glucose to 13C-lactate. Treatment with a concentration of SF2312 (10 μM) that is non-toxic to ENO1-intact glioma cells for four days, led to profound inhibition of glucose consumption (isomeric peaks at 97 and 93 ppm) and concomitant inhibition of lactate production (single peak at 20 ppm) in the media of D423 and Gli56 ENO1 deleted but not ENO1-intact glioma cells (Supplementary Fig. 13). This inhibition of glycolysis was strictly dependent on ENO1-deletion as ectopic rescued cells showed no inhibition of 13C-1 glucose to 13C-3 lactate conversion (Fig. 5a, 5b). Similar results were obtained with all atoms 13C glucose. SF2312 dose-dependently reduced the conversion of U-13C glucose to 13C lactate in a manner selective for ENO1-deleted over ENO1-rescued or otherwise ENO1-intact glioma cells (Supplementary Fig. 14). Labeled 13C was observed at all three atoms of lactate and inhibited by SF2312. Similar trends were observed with PhAH but of more modest magnitude.
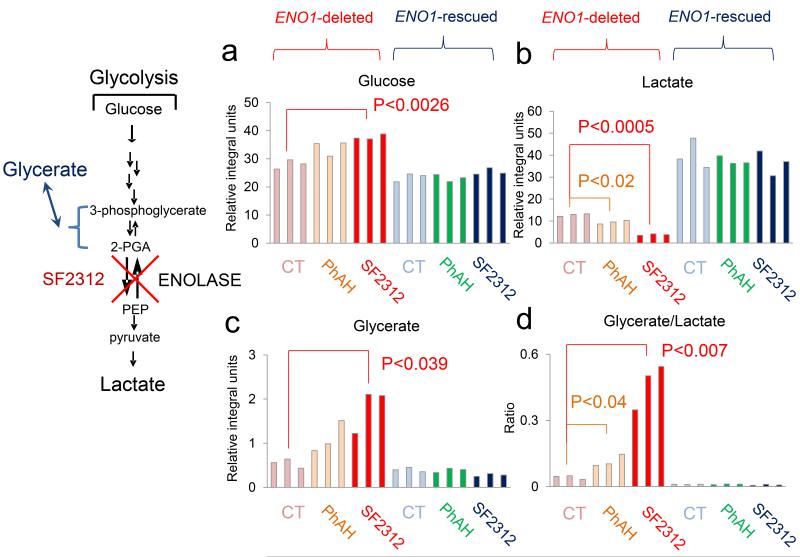
FIGURE 5. SF2312 selectively blocks glycolysis in ENO1-deleted glioma cells.
D423 ENO1-deleted and isogenic rescued cells were supplemented with 13C-1 labeled glucose and treated with 10 μM of SF2312 or PhAH for 72 hours. Integrals (expressed relative to the integral internal standard for NMR in D2O (3-(trimethylsilyl)propionic-2,2,3,3-d4) of peaks at 97 and 93 ppm (glucose, (a)), 20 ppm (C3 of lactate, (b)), 64 ppm (C3 of glycerate, (c)), and the ratio of glycerate to lactate (d) are shown (y-axis). Bars represent individual measurements of biological replicates. Significant differences by unpaired, two-tailed, t-test with Bonferroni correction are indicated. The experiment was conducted only once, but confirmed using 13C-uniformly labeled glucose (Supplementary Figure 9).
SF2312 inhibits Enolase in intact glioma cells
To demonstrate inhibition of Enolase by SF2312 in living cells, we performed two sets of experiments: A) using mass spectroscopy and 13C-NMR isotope tracing, we show accumulation of substrates upstream and depletion of metabolites downstream of the Enolase reaction; B) Using thermal shift assays we provide direct evidence of inhibitor binding to Enolase in intact cells.
We performed mass spectroscopy measurements of the levels of 3-PGA (2-PGA is below detection limit) and PEP, the metabolites immediately upstream and downstream of the Enolase reaction in response to SF2312 in both ENO1-intact and ENO1-deleted glioma cells. Fully consistent with specific inhibition of Enolase, we observed a dramatic increase in the ratio of 3-PGA/PEP in response to SF2312 treatment (Fig. 4e); while both ENO1-deleted and ENO1-rescued cells experienced an elevation in the 3-PGA/PEP ratio, this was much higher in ENO1-deleted cells, as compared to isogenic rescued controls, consistent with the ~90% decreased Enolase activity in deleted versus rescued glioma cells5. Raw data and normalized data for 3-PGA and PEP are shown in Supplementary Fig. 15 and 16.
We demonstrated through mass spec based metabolomics that glycerate accumulates both intracellularly and in media in response to Enolase inhibition by pharmacological (PhAH) as well as genetic (shRNA against ENO2) means in D423 ENO1-deleted cells19. We performed 13C-1 glucose tracing to quantify glycerate production under the same experimental conditions as for the glucose to lactate conversion experiments in ENO1-deleted and ENO1-intact glioma cell lines (Supplementary Fig. 17). A single peak at 64 ppm became visible in the D423 and Gli56 ENO1-deleted cells treated with SF2312 (Supplementary Fig. 17). This is fully consistent with the C-3 atom of glycerate, which has a reported 13C peak of exactly 64 ppm (Spectral Database of Organic Compounds, SDBS No. 18695) and is the atom on which the 13C label derived from the C-1 atom on glucose is expected. Glycerate likely forms in response to enolase inhibition as 2-PGA and 3-PGA accumulate, spontaneously hydrolyze, or are dephosphorylated by the action of glycerate kinase (GLYCTK, Supplementary Fig. 17). We repeated these experiments with D423 ENO1-deleted and isogenic ENO1-rescued control glioma cells (Fig. 5a, 5b, 5c). SF2312 treatment (and to a lesser extent, PhAH) dramatically increases conversion of 13C-1 glucose to 13C-3 glycerate (Fig. 5c). This effect is especially noticeable when expressed as a ratio of 13C-3 glycerate to 13C-3 lactate, metabolites upstream and downstream of the Enolase reaction, respectively (Fig. 5d). Importantly, the production of glycerate was completely abrogated by restoration of ENO1 (D423 ENO1 in Fig. 5c, 5d.), showing strict dependence on deletion of ENO1.
We utilized cellular ligand induced thermal shift assays to demonstrate direct binding of SF2312 and PhAH to Enolase 2, in the setting of intact glioma cells15. To facilitate this assay, we utilized glioma cells overexpressing ENO2 (D423 ENO2); as ENO1-deleted cells are too easily killed by inhibitor treatments. In the absence of inhibitors, ENO2 experienced thermal denaturation in intact glioma cells at ~65 °C (Supplementary Fig. 18). Treatment with 100 μM of SF2312 resulted in a shift of the thermal melting curve of ~15 °C, while the treatment with the same concentration of PhAH resulted in a much more modest shift, of ~ 6 °C. These results are fully consistent with those obtained in vitro (Fig. 2a, 2b, 2c) and provide direct evidence of inhibitor binding to the enzyme in the setting of intact cells.
DISCUSSION
Antibiotic SF2312 was discovered during a screen of natural substances for antibiotic activity under anaerobic conditions12. SF2312 is produced by the actinomycete Micromonospora and is active against a range of bacteria, with strong activity against Salmonella and Staphylococcus, weak activity against E. coli, and no activity against fungi. It likely penetrates bacterial membranes through the Glucose-6-phosphate transporter system in a manner similar to Fosfomycin20, which would explain insensitivity of fungi to SF2312 as this transporter is present only in bacteria. In this manuscript, we present multiple lines of evidence that SF2312 is an enolase inhibitor, both in vitro and in cell based systems. This mechanism of action fully explains increased potency of SF2312 under anaerobic conditions, as ATP generation by the respiratory chain is compromised, forcing the cell to rely exclusively on glycolysis. With an IC50 for Enolase activity inhibition in the low nM (Fig. 1b, Supplementary Table 1), SF2312 is the first reported natural product inhibitor of Enolase. Given that SF2312 was synthesized as a racemic mixture while the X-ray structure is bound by a single enantiomer [((3S,5S)-1,5-dihydroxy-2-oxopyrrolidin-3-yl)phosphonate], it is possible that the actual potency of (S,S)-SF2312 is 4-times stronger than the results presented throughout the manuscript. Despite our efforts, we were unable to isolate enantiomerically pure SF2312, because both stereocenters spontaneously epimerize (Supplementary note 2). SF2312 is similar to the natural products Alahopcin21 and Delalanylalahopcin22, sharing the 1,5 dihydroxypyrrolidin-2-one motif. We attempted to model Alahopcin into the PEP-ENO2 structure (3UCJ) but the alanine-dipeptide portion proved to be too bulky. The Enolase superfamily includes a wide variety of enzymes which display a common chemical step, the abstraction of a proton from a carbon adjacent to a carboxylate with a divalent metallic ion23. We tentatively propose that Alahopcin and Delalanylalahopcin might inhibit members of the Enolase superfamily by chelating the divalent metal with the hydroxamate moiety while hydrogen bonding with the active site catalytic residues (Supplementary Fig. 3).
We have shown that gliomas with passenger deletion of ENO1 are exceptionally sensitive to ablation of ENO25. While an ENO2-specific inhibitor would be ideal, we found that ENO1-deleted glioma cells show selective vulnerability to the pan-enolase inhibitor, PhAH. The reason being that ENO1-deleted glioma cells only retain 10% of normal enzymatic activity (ENO1 is the major isoform), such that even low levels of a pan-enolase inhibitor are sufficient to inhibit the remaining enzymatic activity below toxic threshold (Supplementary Fig. 7, Supplementary Fig. 19c5). Restoration of ENO1 expression or overexpression of ENO2 is thus able to restore resistance to PhAH and SF2312 (5Supplementary Fig. 8). SF2312 proved superior to PhAH for killing ENO1-deleted versus ENO1-rescued glioma cells, especially under anaerobic conditions (Supplementary Fig. 10). This was seconded by biochemical measurements of cellular bioenergetic status, inhibition of glycolysis, and inhibition of enolase. While potentially explained by greater permeability of SF2312, this is not a very satisfying explanation given PhAH and SF2312 show similar toxicity towards ENO1-rescued or ENO1-WT glioma cells. It is also unclear whether inhibitory preference of SF2312 for ENO2 over ENO1 is the explanation. While IC50 of SF2312 does not markedly differ between ENO1 and ENO2 in the in vitro enzymatic assay (Fig. 1b), at higher concentrations, inhibition of ENO2 is higher. This is only apparent when the inhibitor is added before substrate, suggesting that the SF2312:ENO2 inhibitor-enzyme complex is more stable than that of PhAH:ENO2; this is also suggested by ligand-induced thermal stabilization data (Fig. 2a, Supplementary Fig. 6).
Regardless, the main difficulty remains the poor permeability conferred by the phosphonate moiety which will require extensive derivatization in order to generate a credible clinical candidate to treat ENO1-deleted gliomas.
In summary, the main novel finding of our work is the identification of Enolase inhibition as a mechanism of action of the natural phosphonate antibiotic SF2312, making it perhaps the most potent natural product glycolysis enzyme inhibitor reported to date.
ONLINE METHODS
In Silico Docking of PhAH to the ENO2 Crystal Structure
Work was completed on a 2-eight core (16-core) 3.0 Ghz AMD Opteron system using the RHEL 6.x OS and running the Modeling Suite 2013 from Schrödinger LLC (NY). Small-molecule structures and Ki values were taken from multiple sources7,8,24 and adjusted to account for differences at pH = 7.4, when possible. Structures were drawn in ChemBioDraw Ultra 12.0 on a PC and saved as a mol formatted file, transferred to the Linux box, and changed to an sdf formatted file. The ionization state was set to the form expected during binding to two metals, namely (−2) for phosphate, (−1) for carboxylate, and (−1) for hydroxamic acid. A LigPrep (Version 2.8) calculation was performed by using the default criteria, and the top structure for each was saved out as a maegz file. A docking model was created based on the crystal structure of human enolase 2 (PDB code 3UJR). This structure was read into Maestro (Version 9.6) and Chain A was removed. A protein preparation was done with defaults, except that missing side chains were added and waters beyond 5 Å from the ligand were deleted. This involved assigning bond orders, adding hydrogens, and creating zero-order bonds to metals. There were residues missing from C-terminal section, but they were remote from the catalytic site, so no attempt was made to add them. After initial protein preparation, heteroatom states were defined with metal binding states turned. This was followed by hydrogen bond assignment using PROPKA at pH = 7.0. Additional waters with less than three H-bonds to non-waters were then removed. The protein was relaxed by minimization of hydrogens, then minimization of all atoms RMSD = 0.30 using the Impref utility. There were four waters remaining in the binding site, HOH601, HOH602, HOH603, and HOH660. The Grid was created with crystal ligand as reference for the center and default size.
Rigid-Receptor docking (RRD) of the structures was done using the virtual screening workflow,25 which involved docking with Glide XP mode starting with three extra conformations and rescoring with the Prime MM-GBSA ΔGbind.26
A quantum-polarized ligand docking (QPLD) was done with initial Glide XP mode docking and defaults of ligand vdw scaling of 0.8. A maximum of 5 poses per ligand were kept. The charges were calculated with the Jaguar accurate level, which (per the Schrödinger manual) specifies a calculation using the 6-31G*/LACVP* basis sets with the B3LYP density functional and Ultrafine SCF accuracy level. Re-docking was done with Glide XP mode and same scaling factor. A maximum of 10 poses per ligand were kept at this stage and the final selection of compounds was based upon the Emodel score. The resulting receptor and structures were rescored with Prime MM-GBSA using VSGB solvation model and input ligand charges from the QPLD calculation. The default minimization sampling method was used and no protein flexibility was allowed during the calculation. Due to the nature of the calculations and combination of options, there were variations in the scores. Variations were due to subtle differences in the final docked structure. Orientation about the carbon phosphorus bond led to different energies due to the empirical parameters utilized for the oxygen atoms in the phosphate group. The highest scoring configuration and stereoisomer that matched the expected binding mode was used in generating the regression model and evaluating the structure for potential synthesis.
Enolase inhibitors
SF2312 and Deoxy-SF2312 were synthesize with slight modifications of published methods (Supplementary Note 1) while PhAH was synthesized by TCRS (Bristol, PA) as previously described5.
Enolase enzymatic activity
Native lysates of mouse tissues and human cell lines were prepared using 20 mM Tris HCl, 1 mM EDTA, and 1 mM β-mercaptoethanol at pH 7.4 and homogenized using a Polytron homogenizer three times for a period of 10 s followed by sonication, after which the lysates were cleared by centrifugation at 20,000 g for 10 minutes. Enolase activity was measured using two different methods, either by a fluorometric NADH-linked assay or a direct spectrophotometric assay via formation of PEP. In the fluorescent assay, enolase activity was measured via NADH oxidation in a pyruvate kinase–lactate dehydrogenase coupled assay as previously described5. The assay is conducted in 10 mM KCl, 5 mM MgSO4, 100 mM triethanolamine at pH 7.4, with 400 μM NADH and 2 mM ADP. 2-Phosphoglycerate (2-PGA), pyruvate kinase (PK) and lactate dehydrogenase (LDH) are provided in excess, with conversion of 2-PGA to PEP by enolase being rate limiting. PEP (with ADP) is substrate of PK; pyruvate formed by this reaction is linked to NADH oxidation by LDH. Enolase activity is determined by measuring oxidation of NADH fluorescently by excitation at 340 nm and emission at 460 nm. The substrate concentration, if not otherwise indicated, was 5 mM 2-PGA. Fluorescence was measured using Omega Fluorescence Plate Reader (BMG Labtech). Alternatively, Enolase activity was measured directly by the appearance of PEP from 2-PGA via absorption at 240 nm. The assay medium was the same, except that all the auxiliary reagents (PK/LDH, NADH, ADP) are omitted. Both assays were conducted in a 96-well plated format with the direct assay performed in UV-transmissible plates.
Thermal shift assay
Thermal shift assays were conducted as described in ref15, on lysates of D423 cell line overexpressing ENO1 and ENO2, on recombinant human ENO1 and ENO2 purified from E. coli as well as intact D423 ENO2 overexpressing glioma cells. Lysates: The lysates were prepared in 20 mM Tris-HCl, 1 mM EDTA and 1 mM β-mercaptoethanol at pH 7.4 and homogenized using a Polytron homogenizer three times for a period of 10 s followed by sonication, after which the lysates were cleared by centrifugation at 20,000 g for 10 minutes. Lysates were diluted (1:100) in Enolase enzymatic activity buffer as mentioned above. Diluted lysates were treated with SF2312, PhAH or vehicle and heated on a C1000 Thermal cycler (Bio-rad) between 50 to 90 ° C as indicated for 3 minutes. The lysates were then centrifuged at 20,000 g for 10 minutes. Supernatant and pellet were then separated and NuPage LDS Sample buffer (Life Technologies # NP0007) was added. Western Blots were performed as described previously5. The following antibodies were used: ENO1 (1:1000 dilution, Abcam # ab155102), ENO2 (1:1000 dilution, Dako # M087301-2), Vinculin (1:3000 dilution, EMD Millipore # 05-386), pan-ENO (1:1000 dilution, Santa Cruz Biotechnology, sc-7455) and TPI (1:3000 dilution, Proteintech # 10713-1-AP). Recombinant human ENO2: The same procedure was carried out on purified recombinant ENO2 from E. coli¸ utilizing the purified protein generated for the X-ray crystallography studies. Stocks of ENO2 recombinant protein were diluted and treated with PhAH, SF2312, or vehicle in Enolase enzyme activity buffer, followed by thermal denaturation, centrifugation, and immunobloting as detailed for the lysates. Intact glioma cell lines: D423 glioma cell lines over expressing ENO1 and ENO2 were cultured in 10% DMEM. The cell lines were treated with drugs (100 μM PhaH, 100 μM SF2312) or vehicle and after one day, the cells were trypsinized and washed with PBS. These cells were re-suspended in PBS at the concentration of 0.5 × 106 cells/mL. Equal volume of re-suspended live cells were heated on a C1000 Thermal cycler (Bio-rad) with a variable temperature gradient (50 ° C to 90 ° C). Twice the volume of Lysis Buffer (20 mM Tris-HCl, 1 mM EDTA and 1 mM β-mercaptoethanol at pH 7.4) was added to the heat-shocked live cells. These cells were further lysed by a round of freeze-thaw cycle on dry-ice. The lysates were then centrifuged at 20,000g for 10 minutes. Supernatant was collected and NuPage LDS Sample buffer (Life Technologies # NP0007) was added. Western Blots were performed as described previously5.
Glucose 13C Isotope tracing
Glucose labeled with heavy, non-radioactive, 13C at carbon C-1 and 13C at all carbon atoms were purchased from Cambridge Isotopes (CLM-420-1 and CLM-1396, respectively). Glucose free DMEM media was supplemented with 10 mM 13C-1 or 13C-all labeled Glucose. Media was extracted with 80 % methanol at −80 ° C to precipitate proteins and centrifuged with the supernatant recovered. Solvent was subsequently lyophilized and the samples were re-suspended in D2O. Proton-decoupled 13C NMR scans were performed at the M.D. Anderson NMR core using a 500 MHZ Bruker instrument.
Structure Determination of ENO2 and its inhibitors
DNA encoding full length Human Enolase 2 was cloned into pJL-H6 plasmid expression vector using the ligation-independent cloning method 27 and expressed in 1 L of auto-induction media at 20 °C after transformation into E. coli BL21 (DE3). After collecting the E. coli cells by centrifugation, the cells were re-suspended in 50 mM HEPES, 300 mM NaCl, 5 mM MgCl2, 5 mM imidazole, 0.5 mM Tris-(carboxyethyl) phosphine, 10% (v/v) glycerol at pH 7.5. 20 μg mL−1 DNase and 1 mM phenylmethylsulfonyl fluoride were added to the cell suspension prior to sonication at 4 °C. The insoluble cell debris was removed by centrifugation prior to protein purification by His-tag affinity chromatography. The purified Enolase 2 protein was further purified using a Superdex 75 gel filtration column in 20 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 2 mM 2-mercaptoethanol at pH 7.5. Apo crystals of Human Enolase 2 were prepared using the hanging drop vapor diffusion method at 20 ° C, suspending a drop containing 0.5 μL 9.1 mg mL−1 Enolase 2 and 0.5 μL reservoir solution above a 500 μL reservoir containing 200 mM ammonium acetate, 100 mM Bis-Tris and 18-22% (w/v) PEG 3350. Streak seeding in the same solution conditions grew larger crystals. The crystals were soaked overnight in 1 μL drops containing 100 mM Bis-Tis, 200 mM ammonium acetate, 32% (w/v) PEG 3350 at pH 6.5, supplemented with 2-4 mM compound prior to flash freezing in liquid nitrogen. An X-ray diffraction dataset was collected at 100 K using the Advanced Light Source Beamline 8.3.1 equipped with ADSC Q315r detector at a wavelength of 1.11587 Å. For both crystals the MOSFLM strategy algorithm predicted significant overlapping reflections when the detector was positioned sufficiently close to the crystal to collect the highest resolution reflections visible in the diffraction image. The detector was therefore moved further from the crystal to avoid these reflection overlaps.
The diffraction images were indexed and integrated using iMOSFLM28 and scaled using AIMLESS2928. The X-ray structures were solved by molecular replacement using 3029313032. To be consistent with the numbering using in previous ENO2 publications we have used the used residue numbering starting from Serine as in ref31. For ENO2:PhaH, ENO2:SF2312 and ENO2:mSF2312 crystal structures respectively, 97.3%, 97.5% and 97.4% of all amino acids lie within the favored region of the Ramachandran plot. To be consistent with the numbering using in previous ENO2 publications we have used the used residue numbering starting from Serine as residue 1 instead of the N-terminal methionine, a residue that is post-translationally cleaved in human cells.
Cell Culture
The cell line D423-MG (referred to as to D423 throughout the paper) was kindly provided by Dr. Bigner33. The 1p36 homozygous deletion in D423-MG includes the CAMTA1, VAMP3, PER3, UTS2, TNFRSF9, PARK7, ERRFI1, SLC45A1, RERE, ENO1, CA6, SLC2A5, GPR157, MIR34A, H6PD, SPSB1 and SLC25A33 genes. The Gli56 1p36 homozygously deleted cell line was shared by D. Louis, the deletion spans the UTS2, TNFRSF9, PARK7, ERRFI1, SLC45A1, RERE, ENO1, CA6, SLC2A5, GPR157, MIR34A, H6PD, SPSB1, SLC25A33, TMEM201, C1orf200, PIK3CD, CLSTN1, CTNNBIP1, LZIC, NMNAT1, RBP7 and UBE4B genes. The generation of isogenic ENO1 and ENO2 ectopically rescued lines was described previously; (clone pCMV ENO1 5X and clone pCMV ENO2 1X, Supplementary Fig. 7 in ref 5). D423-MG and its sub-clones as well as Gli56 were deposited in the Dept. of Genomic Medicine/IACS Cell Bank, at M.D. Anderson and authenticated by Short Tandem Repeat (STR) testing. The Gli56 cell line does not have a published STR profile so could not be authenticated, but at the same time, its STR profile did not match any known cell line, confirming lack of contamination. Critically for our experiments, absence of ENO1 was confirmed by western blot (Supplementary Fig. 7, 19). A series of ENO1-intact cell lines was used as controls for western blots, enolase activity and sensitivity to enolase inhibitors. These include D502 (Dr. Bigner33), primary human astrocytes (ScienCell), TS543, TS561, TS576 (Cameron Brennan34), HCC1957, COV504, NB1, U343, LN319, and U373 (Dept. Genomic Medicine/IACS Cell Bank, MDACC). Cell lines were authenticated by STR testing. LN319 is a sub-clone of LN-99235, while U373 is a sub-clone of U25136; for the present work, this is acceptable as the these cell lines were used as non-ENO1-homozygously deleted controls. Their exact identity is irrelevant so long as ENO1 is expressed. The expression of ENO1 was verified by western blot (Supplementary Fig. 7, 19). All cell lines were confirmed as mycoplasma negative by ELISA using the MycoAlert PLUS detection kit (Lonza, Basel, Switzerland). Cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS).
Proliferation assays, Apoptosis and ATP determination
Cell proliferation of glioma cell lines was assayed either through crystal violet staining or by live-cell confluence measurements with the IncuCyte (Essen BioScience). Growth curves using the IncuCyte were generated by confluence imaging every 4 hours with duplicate replicates with an initial seeding of 1.5 × 103 cells /well in 96-well plates. At the indicated time point, cells were fixed with 10% formalin and stained with crystal violet. Dye extraction was performed using 10% acetic acid solution, and absorbance was read at 595 nm. To assess cell viability under hypoxic conditions, cells were seeded at a density of 1 × 104 cells cells/well in 96-well plates. The next day, cells were treated with the appropriate drugs and transferred to a controlled hypoxic station (Don Whitley Scientific, Shipley, UK) set at 0.1% O2 concentration and 5% CO2. Following 72 hours incubation plates were stained with crystal violet as described above. Apoptotic cells were counted by staining with YO-PRO®-1 Iodide (491/509, Life Technologies, Y3603). Apoptotic cells become permeable to YO-PRO®-1 while live cells are not stained. Alternatively, Propidium Iodide was employed (Life Technologies) instead of YO-PRO. Total cell number was quantified by Hoechst 33342 (Cat# H3570 Invitrogen). Cells were seeded as 2 × 103 cells/well in 96-well plates and treated in the presence or absence of doxycycline. On the day of apoptosis survey, old media was removed and each well was filled to 100 μl with fresh media. Hoechst and YO-PRO®-1 were mixed in 1:100 PBS and 10 μl of the mixture was added in each well gently without touching the cells for a final dilution of 1:1000 from stock. Plates were then incubated at 37 ° C for 2 hours. Image capture and quantification was done by High Content Screening System- Operetta (Perkin Elmer). ATP content was measured with the luciferase/luciferin CellTiter-Glo® assay. Briefly, 100 μl of CellTiter-Glo® assay media was added to 96-well plates containing 100 μl of media and cells. Lysis was achieved by the perchloric acid in the buffer and vigorous pipetting, and luminescence was determined using an Omega Luminescence Plate Reader (BMG Labtech). Results were expressed relative to vehicle treated control.
Mass spectrometric determination of small molecule metabolites
Small-molecule metabolites from cells in culture were extracted with 80% methanol at −80°C37, following our published methodology38. Methanol extraction recovers polar compounds such as most carboxylic acids, alcohols, sugars but not lipids. After extraction and centrifugation, the samples were dryed by speed-vaccuming. Dryed samples were analyzed using microcapillary liquid chromatography tandem mass spectrometry (LC-MS/MS) using selected reaction monitoring (SRM) with positive/negative polarity switching on a hybrid 5500 QTRAP mass spectrometer (AB/SCIEX). 300 SRM transitions target >260 polar metabolites37 MS-peak quantification was performed using MultiQuant 2.1 software and relative levels of each metabolite in Q3 peak area units across samples.
Statistical Analysis
Statistical significance was tested using the unpaired, 2-tailled, t-test function of Microsoft Excel or Prism Graphpad, using the Welsh correction for unequal variance and the Bonferroni correction for multiple hypothesis testing.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank James Holton and George Meigs for their assistance with X-ray diffraction data collection at the Advanced Light Source, beamline 8.3.1. We thank Dr. Kumar Kalhurachi and Dr. John McMurray for assistance with 13C NMR measurements. We thank Steven Millward, Joe Marszalek and Yuting Sun for critical comments and suggestions. We thank Min Yuan and Susanne Breitkopf for help with the mass spectrometry experiments. We thank Marina Protopopova for assistance with Operetta® cell imaging. We thank Dr. L. Lebioda, Dr. G. Reed and Dr. R. Poyner for useful discussions regarding the catalytic mechanism of Enolase. We thank Rafal Zielinski for assistance with hypoxia experiments. Inhibitors were provided by the Pharmaceutical Chemistry Facility at MD Anderson supported by NIH/NCI under award number P30CA016672. Financial support was provided by NIH-NCI grants 7P01CA095616-10 (R.A.D), CPRIT RP140612 (R.A.D), 5P01CA120964 (J.M.A.), 5P30CA006516 (J.M.A.) and NIH CDP SPORE P50CA127001-07 (F.L.M.). F.L.M. was supported by a Research Scholar Grant, RSG-15-145-01-CDD from the American Cancer Society.
Footnotes
There are NO competing financial interests.
ACCESSION CODES
X-ray structures of ENO2 bound with PhAH (4ZA0) and SF2312 (4ZCW) were deposited in PDB.
AUTHOR CONTRIBUTIONS
D.M. performed structural modeling and docking; F.L.M. and D.M. conceived the cyclized inhibitors, which were synthesized by Z.P., D.S., A.B. and W.B.; P.L., G.L. and T.L performed isolation of recombinant protein and x-ray crystallography; M.E.F, F.P., F.L.M. and B.C. repeated chemical syntheses and wrote synthetic procedures with characterizations. F.P. and F.L.M. performed chiral chromatography. F.L.M. and N.S. performed in vitro enzymatic activity experiments. N.S., N.H., Y-H.L. performed cell culture, western blots, 13C-NMR tracing and biochemical profiling experiments. N.S. performed thermal shift assays. J.A. performed Mass spec small molecule metabolite analysis. R.A.D., Y.A., F.L.M., N.S. and P.L. oversaw overall experimental design, data analysis and wrote the manuscript.
REFERENCES
- 1.Fothergill-Gilmore LA, Michels PA. Evolution of glycolysis. Progress in biophysics and molecular biology. 1993;59:105–235. doi: 10.1016/0079-6107(93)90001-z. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nature genetics. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 3.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller FL, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poyner RR, Reed GH. Structure of the bis divalent cation complex with phosphonoacetohydroxamate at the active site of enolase. Biochemistry. 1992;31:7166–7173. doi: 10.1021/bi00146a020. [DOI] [PubMed] [Google Scholar]
- 7.Anderson VE, Weiss PM, Cleland WW. Reaction intermediate analogues for enolase. Biochemistry. 1984;23:2779–2786. doi: 10.1021/bi00307a038. [DOI] [PubMed] [Google Scholar]
- 8.Qin J, Chai G, Brewer JM, Lovelace LL, Lebioda L. Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity. Journal of inorganic biochemistry. 2012;111:187–194. doi: 10.1016/j.jinorgbio.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de ASNMV, et al. Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics. The FEBS journal. 2007;274:5077–5089. doi: 10.1111/j.1742-4658.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 10.Wedekind JE, Poyner RR, Reed GH, Rayment I. Chelation of serine 39 to Mg2+ latches a gate at the active site of enolase: structure of the bis(Mg2+) complex of yeast enolase and the intermediate analog phosphonoacetohydroxamate at 2.1-A resolution. Biochemistry. 1994;33:9333–9342. doi: 10.1021/bi00197a038. [DOI] [PubMed] [Google Scholar]
- 11.Zhang E, Hatada M, Brewer JM, Lebioda L. Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution. Biochemistry. 1994;33:6295–6300. doi: 10.1021/bi00186a032. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe H, Yoshida J, Tanaka E, Ito M, Miyadoh S, Shomura T. Studies on a new Phosphonic acid antibiotic, SF-2312. Science Reports of Meiji Seika Kaisha. 1986;(No. 25):12–17. [Google Scholar]
- 13.Hanaya, Itoh An effective synthesis of antibiotic SF-2312 (3-dihydroxyphosphoryl-1,5-dihydroxy-2-pyrrolidone. Heterocycles. 2011;82(2):1675–1683. [Google Scholar]
- 14.Liu, et al. “A new approach to cyclic hydroxamic acids: intramolecular cyclization of N-benzyloxy carbamates with carbon nucleophiles,”. Tetrahedron. 2011;67(12):2206–2214. doi: 10.1016/j.tet.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez Molina D, et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 16.Brewer JM, Wampler JE. A differential scanning calorimetric study of the effects of metal ions, substrate/product, substrate analogues and chaotropic anions on the thermal denaturation of yeast enolase 1. International journal of biological macromolecules. 2001;28:213–218. doi: 10.1016/s0141-8130(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhang E, Brewer JM, Minor W, Carreira LA, Lebioda L. Mechanism of enolase: the crystal structure of asymmetric dimer enolase-2-phospho-D-glycerate/enolase-phosphoenolpyruvate at 2.0 A resolution. Biochemistry. 1997;36:12526–12534. doi: 10.1021/bi9712450. [DOI] [PubMed] [Google Scholar]
- 18.Marangos PJ, Parma AM, Goodwin FK. Functional properties of neuronal and glial isoenzymes of brain enolase. Journal of neurochemistry. 1978;31:727–732. doi: 10.1111/j.1471-4159.1978.tb07847.x. [DOI] [PubMed] [Google Scholar]
- 19.Muller FL, Fletcher-Sananikone E, Colla S, Aquilanti E, DePinho R. Collateral gene inactivation biomarkers and targets for cancer therapy. WO2013090732 A2. Patent. Application PCT/US2012/069767, filed 14 Dec. 2012, and issued 20 Jun. 2013.
- 20.Dette GA, Knothe H, Schonenbach B, Plage G. Comparative study of fosfomycin activity in Mueller-Hinton media and in tissues. The Journal of antimicrobial chemotherapy. 1983;11:517–524. doi: 10.1093/jac/11.6.517. [DOI] [PubMed] [Google Scholar]
- 21.Horii S, et al. Structure of alahopcin (nourseimycin), a new dipeptide antibiotic. The Journal of antibiotics. 1985;38:302–311. doi: 10.7164/antibiotics.38.302. [DOI] [PubMed] [Google Scholar]
- 22.Higashide E, Kanamaru T, Fukase H, Horii S. Isolation of dealanylalahopcin, a new amino acid, and its biological activity. The Journal of antibiotics. 1985;38:296–301. doi: 10.7164/antibiotics.38.296. [DOI] [PubMed] [Google Scholar]
- 23.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Archives of biochemistry and biophysics. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 24.Brewer JM. Yeast enolase: mechanism of activation by metal ions. CRC Crit. Rev. Biochem. 1981;11:209–254. doi: 10.3109/10409238109108702. [DOI] [PubMed] [Google Scholar]
- 25.Schrödinger Suite 2013-3 Virtual Screening Workflow. Glide version 5.9, LigPrep version 2.6. Schrödinger, LLC; New York, NY: 2013. (Schrödinger, LLC, New York, NY, 2013.) [Google Scholar]
- 26.Prime. Version 3.4 v. 3.4 Schrödinger, LLC; New York, NY: 2013. [Google Scholar]
- 27.Lee J, Kim SH. High-throughput T7 LIC vector for introducing C-terminal poly-histidine tags with variable lengths without extra sequences. Protein expression and purification. 2009;63:58–61. doi: 10.1016/j.pep.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta crystallographica. Section D, Biological crystallography. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta crystallographica. Section D, Biological crystallography. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan CG, et al. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1:265–277. doi: 10.18632/oncotarget.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stommel JM, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 35.Bady P, et al. DNA fingerprinting of glioma cell lines and considerations on similarity measurements. Neuro-oncology. 2012;14:701–711. doi: 10.1093/neuonc/nos072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torsvik A, et al. U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer medicine. 2014;3:812–824. doi: 10.1002/cam4.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nature protocols. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.