Abstract
Complete genome annotation relies on precise identification of transcription units bounded by a transcription initiation site (TIS) and a polyadenylation site (PAS). To facilitate this process, we developed a set of two complementary methods, 5′ Long serial analysis of gene expression (LS) and 3′LS. These analyses are based on the original SAGE and LS methods coupled with full-length cDNA cloning, and enable the high-throughput extraction of the first and the last 20 bp of each transcript. We demonstrate that the mapping of 5′LS and 3′LS tags to the genome allows the localization of TIS and PAS. By using 537 tag pairs mapping to the region of known genes, we confirmed that >90% of the tag pairs appropriately assigned to the first and last exons. Moreover, by using tag sequences as primers for RT-PCRs, we were able to recover putative full-length transcripts in 81% of the attempts. This large-scale generation of transcript terminal tags is at least 20–40 times more efficient than full-length cDNA cloning and sequencing in the identification of complete transcription units. The apparent precision and deep coverage makes 5′LS and 3′LS an advanced approach for genome annotation through whole-transcriptome characterization.
Keywords: genome annotation, full-length cDNA, transcription analysis
The complete genome sequences of human and other model organisms (1–5) have raised questions as to the accuracy of previously used gene annotation algorithms. Current genome annotations are mostly based on the growing but still limited cDNA sequence databases. Computational gene prediction algorithms, even when trained based on current cDNA data sets, may not provide reliable ab initio predictions of new genes, and all predictions need to be validated by further experimental evidence (6, 7).
Efforts in large scale full-length cDNA sequencing (8–11; reviewed in ref. 12) provide not only the complete sequences of expressed genes but also the ability to locate transcription initiation sites (TISs), polyadenylation sites (PASs), as well as splicing junctions. The localization of these functional sites related to transcripts in the context of the genome can greatly help to define the surrounding regulatory elements such as promoters. However, sequencing all possible full-length transcript clones is an expensive and cumbersome process. Despite current progress, complete disclosure of the transcriptome has yet to be achieved. It is clear, therefore, that the precise identification of all genes and their regulatory elements will require more comprehensive and facile technologies with greater throughput to provide the necessary empirical evidence.
Serial analysis of gene expression (SAGE) (13, 14) and massively parallel signature sequencing (MPSS) (15) are high-throughput methods that use short tags (14–21 bp of internal transcript signatures) to count transcripts in transcriptomes. When mapped to assembled genome sequences, these short tags help to locate transcription units in the context of the genome. Because concatenation of these short tags and their subsequent cloning greatly increases sequencing efficiency, large volumes of transcript data can be generated, and all transcripts, including rare ones, can theoretically be detected. However, tags generated by SAGE and MPSS, although closer to the 3′ side of the cDNA, often reside several hundred bp upstream of the 3′ ends. When mapped to the genome, such “internal” tags are often ambiguous in defining transcription units because they do not specify where the putative transcripts start and end on the genome landscape.
To retain the efficiency of the short-tag strategy, and at the same time increase the specificity and information content of short transcript tags, we have developed two protocols, 5′ and 3′ Long-SAGE (LS). These protocols are based on the original SAGE and LS methods, but enable the extraction of the first and last 20 bp of each transcript. This transcript terminal tag data can then be assembled to map the TIS and PAS of each transcript in the genome. Here, we describe this approach with data generated from mouse embryonic stem cells.
Materials and Methods
Detailed protocols are available in Supporting Text, which is published as supporting information on the PNAS web site. All oligonucleotide sequences are listed in Table 2, which is published as supporting information on the PNAS web site.
RNA Sample. E14 mouse embryonic stem cells (16) were first expanded on fibroblast feeder subconfluent cultures and were then trypsinized and replated in the presence of leukemia inhibitory factor in DMEM. The cells were subsequently harvested and used for RNA purification by using TRIzol reagent (Invitrogen).
Construction of a 5′LS Library. First-strand cDNA synthesis and full-length cDNA selection. This procedure was carried out essentially as described for the “cap-trapper” procedure (17). A NotI-dT20 oligonucleotide was used to prime first-strand cDNA synthesis from 20 μg of mRNA template.
Synthesis of double-stranded cDNA and addition of MmeI/BamHI adapter. In this step, a unique adapter containing BamHI and MmeI recognition sites was ligated to the 5′ terminus of cDNA. As in the standard SAGE procedure, to avoid PCR inhibition, the cDNA pool was first divided into two aliquots, each of which was then ligated to an adapter differing in the 5′ PCR primer annealing region.
Formation of 5′LS ditags. NotI digestion was used to create a cohesive site at the 3′ end for the addition of a biotinylated linker. After size fractionation, the selected cDNA was immobilized and digested with MmeI (New England Biolabs, Beverly, MA) to release the 5′-terminal tags. The released tags were then pooled and ligated to form 5′LS ditags that were amplified by PCR. The size of the PCR-derived ditags was ≈120 bp.
Concatenation and cloning of 5′LS ditags. After large-scale PCR amplification, the 5′LS ditags were PAGE purified and digested with BamHI to generate short ditags (≈50 bp) with 4-bp cohesive 5′ overhangs. The eluted ditags were ligated to form concatemers and cloned into BamHI-digested pZErO-1 vector (Invitrogen) for sequencing analysis.
Construction of a 3′LS Library. Synthesis of cDNA and addition of MmeI/BamHI adapter. A GsuI-dT16 oligonucleotide was used to prime the cDNA synthesis from 5 μg of mRNA template. The Superscript RT kit (Invitrogen) was used for cDNA synthesis. Double-stranded cDNA was ligated at the 5′ end to biotinylated SalI adapter, followed by a GsuI digestion to remove the poly(A) tail, leaving an AA dinucleotide overhang. Similar to the 5′LS method, the digested and purified cDNA was divided into two equal aliquots. Mme/BamHI adapters A and B were each ligated to an aliquot. Common to both adapters A and B were MmeI and BamHI recognition sites at the 3′ end, but their sequences differed at the 5′ end. The rest of the 3′LS library protocol was the same as in the 5′LS protocol.
Tag Sequence Analysis and Mapping to the Genome. Clones of 5′LS and 3′LS libraries were plated out on low-salt LB agar media (Lennox L) containing Zeocin (25 μg/ml) and incubated overnight at 37°C. Individual colonies were picked in 384-well plates and grown overnight in LB media. The SprintPrep plasmid purification system (Agencourt, Beverly, MA) was used to prepare DNA templates for sequencing. The raw sequences were processed by vector trimming and quality base calling with standard phred/phrap software. Ditag sequences were extracted with a modified version of the usage program (18). Unique 5′LS and 3′LS tag sequences were mapped to the assembled mouse genome (mm3) to obtain genome coordinates for each tag sequence.
Preliminary mapping indicated that the terminal nucleotides tended to be nonspecific. Accordingly, a shorter length (17 bp) of perfect tag-to-genome match was allowed. Based on the genome coordinates of known transcript information compiled in the University of California, Santa Cruz (UCSC), mouse genome (19), tag sequences were assigned to known reference transcripts whether they overlapped, or were in close proximity (<1,000 bp) in the genome. Different tag sequences, according to their genome coordinates, could be clearly clustered if their nucleotide sequences overlapped. Furthermore, nonoverlapping tag sequences that were close to each other on the genome landscape (<200 bp), or were related to the same reference transcript, were grouped together as well. Such grouped tags were given a distinctive cluster ID to reflect their relation to a particular transcript unit. Paired 5′LS and 3′LS tags along the genome sequences were recognized whether they fit the following parameters: on the same chromosome, in the same direction, in correct order (5′ → 3′), and within an arbitrary range of <1 million base pairs.
Results
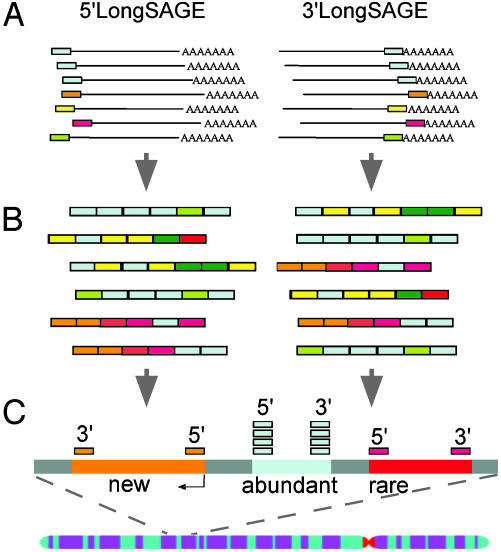
Experimental Strategy. The outline of the 5′LS and 3′LS is to extract terminal transcript tags, concatenate them for efficient sequencing, and map the tag sequences to the genome to define the transcript boundaries and expression levels (Fig. 1). The principal strategy in capturing the first and last 20-bp nucleotide sequences of transcripts in the 5′LS and 3′LS methods is the introduction of an MmeI(atype IIS restriction endonuclease) recognition site immediately flanking the intact 5′ or 3′ ends of cDNA fragments. In the 5′LS method, a linker/primer containing an MmeI site was introduced at the 5′ most end of each cDNA derived by the biotinylated cap-trapper method (17) during second-strand synthesis (Fig. 4A, which is published as supporting information on the PNAS web site). In the 3′LS method, a GsuI (another type IIS restriction endonuclease) site was included in the oligo(dT) primer used for first-strand cDNA synthesis. The poly(A) tails of cDNA were excised by GsuI digestion, but a 3′ AA dinucleotide overhang was retained to facilitate subsequent adapter ligation. An adapter containing MmeI and BamHI sites was then added to the cDNA flanking the polyadenylation site (Fig. 4B). After MmeI digestion, the extracted 5′ or 3′ tags of cDNA were concatenated and cloned to construct the 5′LS and 3′LS libraries, respectively, for sequencing analysis. The tag sequences can then be mapped to the genome sequences to define TIS and PAS. Based on the genome coordinates, the tags can be efficiently assigned to known transcripts and partial cDNA sequences that are located in the same genomic regions. More importantly, the juxtaposition of a 5′LS tag with a 3′LS tag would provide evidence for a likely transcription unit bracketed by these tag markers. To demonstrate the validity of this approach, we constructed a 5′LS library and a 3′LS library from the mRNA of the mouse E14 cell line (16). We analyzed 10,465 tags from the 5′LS library and 10,528 tags from the 3′LS library, representing 7,329 unique 5′LS and 7,825 unique 3′LS tags, respectively.
Fig. 1.
Schematic overview of the 5′LS and 3′LS methods for mapping TISs and PASs. (A) The first and last 20-bp nucleotides of full-length transcripts were extracted as 5′LS and 3′LS tags, respectively (see the detailed protocols in Supporting Text). (B) The 5′LS and 3′LS tags were concatenated and cloned as separate 5′LS and 3′LS libraries for sequencing analysis. (C) The 5′ and 3′ tags were concurrently mapped to the assembled genome sequences to define the TIS and PAS of transcripts and determine expression levels.
Mapping Specificity of Tags to Genome. Unique tag sequences were mapped to the mouse genome sequences (mm3) by using blast (20). A perfect 5′LS tag should contain the first 20 bp of a transcript at the 5′ cap site. By requiring a perfect 20-bp match of the tag sequence to the genome, we found only 1,039 tags that matched to the genome: 687 tags matched a single locus, 92 to two loci, 38 to three loci, and the rest to more than three loci. This number of tags accounted for only 14% of the 7,329 unique 5′LS tags. For better mapping efficiency, we allowed mismatches in the first 3 bp and in the last 3 bp only. However, a minimal 17-bp continuous match was still required when aligning the tags to the genome. Based on this revised criterion, we found that 5,622 (76.7%) of the 5′LS tags mapped to the genome. Of these tags, 4,087 (72.7% of 5,622) mapped to single loci, 709 had two matches, and 270 had three locations in the genome. This increase in the number of alignments indicated that nontemplated nucleotides were incorporated into the ends of these tag sequences, presumably during the tag cloning process.
To understand the nature of mismatches at the ends of the tag sequences, we analyzed in detail the tags that mapped to single loci in the genome (Table 3, which is published as supporting information on the PNAS web site). Of the 4,087 5′LS tags that had a single location in the genome, we found that the majority of the tags (2,059 >50%) had a perfect match in the genome from nucleotides 2–20 of their sequences [hereinafter referred to as tag position 2–20 (tp 2–20)] suggesting that the first nucleotide of these tags was a mismatch to the genome sequences. In effect, these tags contained only 19 bp of transcript-specific signatures. In addition, 2,002, or 97% of these 2,059 tags (tp 2–20), were mapped to known genes or supported by EST data. If we summated all tags that had possible mismatches in the first to third positions (tp 2–20, 2–19, 2–18, 3–20, 3–19, and 4–20; see Table 3), these tags accounted for 75% of all of the single-locus 5′LS tags. Even considering just the tags with mismatches in the first two and the first three nucleotide positions (tp 3–20, 18 bp; tp 4–20, 17bp), we found that >80% of them aligned to known transcripts and EST sequences. This finding is in agreement with observations that extra nucleotides, mostly one or more dC, are added to the 3′ end of the first-strand cDNA by Moloney murine leukemia virus and other reverse transcriptases (21, 22). Indeed, we observed that of the 2,059 tags that contained a nontemplated first nucleotide, 97% (1,998 of 2,059) of these were dG (sense strand).
In contrast to the higher mismatch rate at the 5′ end (which resulted from nontemplated nucleotide addition during reverse transcription, and so is referred to as the RT end) of the 5′LS tags, the 3′ ends of the 5′LS tags created by MmeI digestion (hereinafter referred to as the MmeI digestion end) were relatively stable. Only 17.2% of the tags had mismatches (tp 1–19, 1–18, 1–17, 2–19, 2–18, and 3–19 in Table 3) at the MmeI digestion end. These mismatches can be attributed to the difficulty of determining the precise boundary between each tag in a ditag during data extraction. This finding is due to imprecise cleavage (slippage) by MmeI, a phenomenon that has been observed in other Type IIS restriction endonucleases (13, 23, 24). Therefore, allowing mismatches in the first 3 and the last 3 nucleotide positions of each tag sequence could significantly increase the 5′LS mapping efficiency and specificity to the genome.
The 3′LS tags contained the last 18 bp of transcripts before the polyadenylation site, plus an AA dinucleotide residual as an orientation indicator. A blast search of the 7,826 unique 3′LS tags to the mouse genome found perfect matches (tp 1–18) for 3,873 (50% of 7,826) tags, within which 2,627 tags mapped to single loci in the genome, 876 matched to two loci, and 370 to three loci. When we allowed one mismatch either at the first or the last nucleotide position, we found 1,607 more tags that mapped one or more times to the genome. In the case of tags that mapped only once to the genome, we found that 430 tags matched in tp 1–17, and 632 tags mapped in tp 2–18. This finding suggested that only 17.1% of the tags had one nonspecific nucleotide in the first position (the MmeI digestion end), and 11.7% had a mismatched nucleotide at the last position [the oligo(dT) priming end] in 3′LS tags.
In comparison with 5′LS tags, the 3′LS tags exhibited far greater specificity. This finding could be attributed to the fact that the 3′LS tags did not contain the RT ends. It also appeared that the MmeI digestion effect at the MmeI-cleaved ends of both the 5′LS and 3′LS tags was very consistent: the mismatch rates at the MmeI digestion end for 5′LS and 3′LS tags were almost identical; 17.2% and 17.1%, respectively.
Although allowing a shorter length of tag sequence when matching to the genome increased the total number of tags that could be assigned, the possibility of nonspecific mapping might also increase. Therefore, care should be taken when interpreting the mapping data. Further efforts are required to determine the specificity and false-positive mapping rate for shorter tag sequences with outskirt nucleotide mismatching, because we are not entirely certain as to the exact cause of the mismatches in every case. A number of other factors such as sequencing errors in the tag or genome sequences, or transcript polymorphisms, would also contribute to the mismatches.
Mapping TISs and mRNA PASs. A major application of 5′LS and 3′LS analysis is to map and identify TIS and the PAS of genes in genome sequences. After mapping the tags to the mouse genome sequence, we correlated the genome coordinates of the tags obtained by blast to those of known gene exons, EST sequences, and predicted genes compiled in the mouse genome database of the UCSC genome browser, so as to relate the tags to transcript information. By using this annotation method, we assigned 2,954 5′LS tags to known transcripts (from databases of RefSeq, Mammalian Gene Collection full-length cDNA sequences, National Center for Biotechnology Information mRNA, and Swiss-Prot), 621 tags to EST records, and 287 to predicted genes. Similarly, we also assigned 2,078 of the 3′LS tags to known genes, 603 to ESTs, and 847 to predicted genes (Table 1). Unlike the traditional SAGE tag mapping procedure, this tag → genome → gene mapping strategy is independent of existing cDNA sequences in databases, and therefore, allows for the identification of new transcripts, as well as new TISs and PASs of known genes.
Table 1. Tag positions relative to exons of known genes, ESTs, and predicted genes.
| Tags to exon/intron | Known transcripts | Percent | ESTs | Percent | Predicted genes | Percent | Total | Percent |
|---|---|---|---|---|---|---|---|---|
| 5′LS | ||||||||
| First exon | 2,730 | 92.4 | 466 | 75.0 | 60 | 20.9 | 3,256 | 79.7 |
| Other exon | 58 | 2.0 | 57 | 9.2 | 14 | 4.9 | 129 | 3.2 |
| Intronic | 166 | 5.6 | 98 | 15.8 | 213 | 74.2 | 477 | 11.7 |
| Total | 2,954 | 100.0 | 621 | 100.0 | 287 | 100.0 | 3,862 | 100.0 |
| 3′LS | ||||||||
| Last exon | 1,112 | 53.5 | 300 | 49.8 | 41 | 4.8 | 1,453 | 41.2 |
| Other exon | 559 | 26.9 | 119 | 19.7 | 63 | 7.4 | 741 | 21.0 |
| Intronic | 407 | 19.6 | 184 | 30.5 | 743 | 87.7 | 1,334 | 37.8 |
| Total | 2,078 | 100.0 | 603 | 100.0 | 847 | 100.0 | 3,528 | 100.0 |
Known transcripts include sequences of known genes, RefSeq genes, MGC FL cDNA, and GenBank mRNA. ESTs include mouse EST and ENSEMBL EST. Predicted transcripts include ENSEMBL predictions, Twinscan, SGP, Geneid, Fgenesh++, and GENSCAN predictions, as compiled in the UCSC genome browser databases. The percentages are based on the total numbers in each category.
Of the 2,954 5′LS tags that hit known genes, 2,730 tags either overlapped with, or were in close proximity (<1,000 bp) to, the first exons of known genes. These data suggested that over 90% of the 5′LS tags would probably represent the true 5′ end of transcripts. We noticed that the proportion of tags that matched to the first exons defined in the mouse EST database was lower at 75%, reflecting the well known fact that many ESTs were generated from partial cDNA clones. This tag-to-first-exon match rate further dropped to 20.9% when using gene prediction to assign first exons (Table 1). Conversely, we observed that >74% (213 of 287) of the tags mapped to predicted genes were “intronic,” whereas 15.8% of the tags matched to EST sequences, and only 5.6% of the tags assigned to known transcripts were intronic. This observation confirms the observations of others that standard gene prediction algorithms perform poorly when compared with empirical information (25), and that the EST sequences available in the current databases remain incompletely characterized.
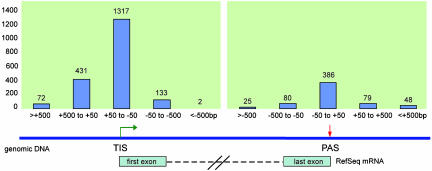
We then measured the distances in base pairs from tags to exons of known genes. If a tag is located within the exon sequence, the distance of the tag to exon is presented as a negative (–) number, indicating that the transcript represented by the tag is shorter than the reference transcript sequence. Conversely, whether a 5′LS tag is further 5′ upstream of the first base pair of the first exon, or whether a 3′LS tag is further downstream of the last base pair of the last exon, then the distance is presented as a positive (+) number, to denote a larger-than-reference transcript suggested by the tags. As shown in Fig. 2, the majority (1,317 or 67%) of the 5′LS tags that mapped to the first exons of RefSeq genes fall in the region of plus or minus 50 bp around the transcription start sites. However, there were 135 tags located significantly inside the first exons, which might represent either downstream alternative transcription initiation sites or truncated transcript 5′ ends due to incomplete reverse transcription or partial mRNA templates. Nonetheless, >3-fold more tags were found further 5′ upstream of the known TIS, including 72 tags that mapped >500 bp upstream of known TIS. For example (Fig. 5A, which is published as supporting information on the PNAS web site), a 5′LS tag (5′LS1484, GAGGGCGGCTGAGACGAGAG) was mapped to mouse chromosome 2 (chr2:18826912–18826893) and located 210 bp upstream of a Ref-Seq gene Bup (accession no. NM_147778). This tag mapping suggests that a longer novel transcript related to this RefSeq sequence might be expressed in mouse E14 cells.
Fig. 2.
Mapping positions of 5′LS tags relative to TISs and 3′LS tags relative to PASs of RefSeq mRNA on genome sequences. The position of each 5′LS and 3′LS tag is indicated by the number of base pairs relative to the corresponding known RefSeq sequence. Negative numbers on the horizontal axis indicate that tags are either downstream of known TISs (for 5′LS tags), or upstream of known PASs (for 3′LS tags). Positive numbers indicate that tags are either upstream of known TISs (for 5′LS tags), or downstream of known PASs (for 3′LS tags). Values above each bar represent the number of tags within that particular range (in bp) in relation to known TISs and PASs.
Furthermore, we identified many tags that mapped to predicted genes and desert regions in the genome. For instance, a set of multiple overlapping tags matched to a locus in chromosome 14 that was 30 bp from an ENSEMBL-predicted gene (Fig. 5B). The multiple tag matching strongly suggested that this was not merely a random match, and provided validation for this gene prediction.
In addition, like SAGE and MPSS, 5′LS should also have the same ability to quantify gene expression by counting tag numbers. Although the total number of tags sampled in this study may not be large enough for a strong statistical argument, we indeed observed, at first glance, that the most abundant transcripts in this 5′LS library were from well known housekeeping genes, such as ornithine decarboxylase antizyme 1 (Oaz1), ribosomal protein S11 (RPS11), and ATP synthase (Atp5a1). Furthermore, we were able to detect several genes known to be specifically expressed in mouse embryonic stem cells within the 10,467 tags of the 5′LS library, and could validate these in a parallel MPSS experiment. For example, we detected Oct4 (26), zfp42 (27), and Utf1 (28) transcripts at 1, 3, and 4 tag counts, respectively, in the 5′LS library, and registered proportionately at 501, 407, and 2,009 tags per million by MPSS (C.-L.W., unpublished observations). These numbers demonstrated a reasonable correlation, considering the difference in sampling size between these two tag-based experimental systems. However, a more in-depth comparison with data sets generated by other techniques, such as microarray and real-time PCR, is needed to fully validate the quantitative aspects of this method.
Finally, the combination of mapping the genome location and counting the copy number of tags could represent an advantage over other short tag approaches by providing a quantitative measure of differential use of alternative TIS. As observed (Fig. 5C), there were 15 unique tags (total 21 tag counts) that were mapped to chromosome 1 in a 279-bp region (chr1:9194671–9194950) and, therefore, clustered together. Based on sequence overlapping, these tags could be viewed as five distinctive groups possibly representing five alternative transcription initiation sites, designated TIS1-TIS5 (the tag counts were 12, 2, 2, 1, and 4, respectively), for the transcripts initiated in this region of the genome. TIS1 and TIS2 aligned with an exon suggested by a number of EST sequences (accession nos. CA493594, BY079805, and BG078406). Because the TIS1 site appeared to be the most abundant, it could be considered the main transcription initiation site of the putative gene encoded in this region.
The 3′LS tags showed a similar trend. The majority of tags that matched to known genes matched terminal exons (Table 1). In contrast, the tags related to predicted genes were mostly located in the putative introns, again highlighting the inadequacies of the current gene prediction methods. Similarly, of the 618 3′LS tags that hit to the last exons of RefSeq genes, we found 386 (62%) tags that mapped close to known mRNA polyadenylation sites (Fig. 2). Many tags were found aligned further downstream of known polyadenylation sites. As an example shown in Fig. 6A, which is published as supporting information on the PNAS web site, two tags were aligned to the last exon of the RefSeq gene Mtpn (myotrophin) on mouse chromosome 6 (chr6:35483262–35483244 and 35484252–35484234). Chromosome 3LS5376 was aligned at the tip of the exon sequence, whereas 3LS1791 was located 990 bp downstream, representing two alternative polyadenylation sites of the gene.
It is apparent that more 3′LS tags matched to internal exons than did the 5′LS tags (21% for 3′ tags vs. 3.2% for 5′ tags, Table 1). Indeed, we had observed many instances in which multiple 3′LS tags were aligned within the same genes. We suspect that this occurrence was mostly due to alternative use of different polyadenylation signals. These tag mappings may suggest multiple alternative PAS. For example, as shown in Fig. 6B, the Hspa4 gene on mouse chromosome 11 was matched by four tags (the genome mapping coordinates are chr11:53876575–53876557, 53860182–53860164, 53858884–53858866, and 53855753–53855735). Chromosome 3LS1963 matched to exon 7, 3LS7706 to exon 17, while 3LS5071 was located in the intron between exons 16 and 17, and 3LS1564 was found 1,736 bp downstream of Hspa4. We found that all these tag-identified PAS had supporting EST sequence data in the databases. We further analyzed the genomic DNA sequences around the tag sites and confirmed that all had recognizable poly(A) signals, such as AATAAA, within 100 bp upstream of the tag sites, and no poly(A) stretches immediately downstream. This finding ruled out the possibility of internal mispriming by oligo(dT) during cDNA synthesis. The status of 3LS5071 is interesting. Although there was a clear poly(A) signal (AATAAA) 16 bp upstream of this tag site, and two cDNA sequences (Riken cDNA records: AK054211 and BB551834) that terminated at this tag site, we did find a 17-bp poly(A) stretch immediately after the tag site, which made it uncertain whether this tag site represented a true PAS or was merely an artifact caused by internal mispriming of oligo(dT) during cDNA synthesis. Nevertheless, our preliminary observations emphasize the view that alternative polyadenylation is far more complex than we understand currently. It is certain that different alternative polyadenylation can result in significant differences in transcript structure, and therefore may have important functional implications.
Pairing 5′LS and 3′LS Tags to Identify Transcription Units. Once the 5′LS and 3′LS tags were mapped to common genome sequences, the two data sets could be merged into a single set, based on the premise that the 5′ and 3′ tag originating from the same transcript would be colocated in close proximity along the chromosome. Hence, independently generated 5′LS and 3′LS tags could be paired according to their genome coordinates.
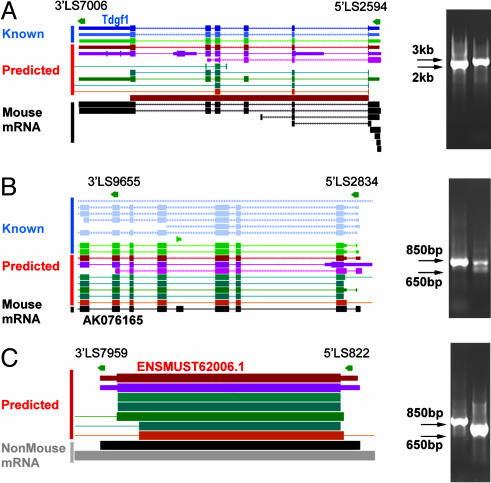
From the current data set of 5′LS and 3′LS tags, we identified 701 pairs of 5′ and 3′ tags based on these criteria: they were on the same chromosome, in the same direction, in the correct order (5′ → 3′), and within an arbitrary range of <1 million base pairs. Of these, 537 pairs were aligned to known transcript sequences, 164 pairs were associated with EST sequence data, 75 pairs supported predicted genes, and 25 pairs were located in desert regions where no reference sequences were recorded. Of the 537 tag pairs aligned to known transcripts, >94% or 508 pairs mapped to the first exons, and 77% (413 pairs) matched the last exons of the corresponding known genes. As mentioned earlier, we believe that the relatively lower rate of tag to last exon for 3′LS tags reflected more the complex issue of alternative polyadenylation sites, than the quality of the tag data. Nevertheless, these match rates essentially validated our strategy of using the 5′LS and 3′LS tags to define the boundaries of transcript units by means of mapping the TIS and PAS. Fig. 3 illustrates three applications of the paired 5′LS and 3′LS tags for validating known transcripts, identification of putative splicing variants, and validation of predicted genes. An obvious implication is that mapping paired tags provides a convenient means to identify novel transcription units.
Fig. 3.
Transcription units identified by paired-tag analysis. (A) Tag pair 5′LS822/3′LS7959 mapped closely to a predicted gene (ENSMUST62006.1) on chromosome 11. (B) Tag pair 5′LS2834/3′LS9655 identified a possible splice variant of eukaryotic translation initiation factor 3 subunit (AK076165) on chromosome 7. (C) Tag pair 5′LS2594 and 3′LS7006 identified a transcript of Tdgf1 teratocarcinoma-derived growth factor on chromosome 9. (Insets) RT-PCR validations of these putative transcript units; primary PCR products are to the left of secondary PCR products.
To validate the pairing accuracy to transcripts, and to test the efficacy of direct PCR by using primers designed from the paired tag sequences to amplify putative transcripts, we selected 90 pairs of 5′LS and 3′LS tags. These 90 pairs included four pairs aligned only to predicted genes, 21 pairs that matched but did not directly overlap known transcripts, and 64 pairs whose tag sequences overlapped with known transcript sequences. We first amplified the putative transcripts by RT-PCR using the paired tag sequences as primers, and then confirmed the primary PCR products by a secondary PCR using the nested primers derived from the genomic DNA sequences encompassed by the paired tags (Fig. 3). From the 90 PCRs, we were able to obtain an 81% (73 of 90) overall retrieval success rate (see Table 4, which is published as supporting information on the PNAS web site). These PCR tests also validated three ENSEMBL-predicted genes of four analyzed.
Discussion
Complete genome annotation relies on comprehensive transcriptome characterization. Apart from the tens of thousands of genes that might be expressed in a cell, the additional complexity of a transcriptome is mostly contributed to by three major mechanisms, namely alternative transcription initiation, alternative splicing, and alternative polyadenylation. Our data presented in this study demonstrated that the 5′LS and 3′ LS approach can effectively and efficiently identify the alternative TIS and PAS, and quantify the differential use of these sites. The effectiveness was reflected by the observation that the majority of the tags faithfully mapped either close to, or further upstream of, the reference TIS (for 5′LS tags), or downstream of the reference PAS (for 3′LS tags) of known transcripts in the genome. Compared with cDNA EST sequencing, the 5′LS and 3′LS approach is 20- to 40-fold more efficient in determining the TIS and PAS on a genomic scale. Hence, large-scale production of 5′LS and 3′LS tags would greatly complement the current full-length cDNA sequencing effort by providing further information on UTR regions that might be missed in full-length cDNA cloning and sequencing, as well as identify new transcripts.
A direct outcome of high-throughput TIS and PAS mapping would be the identification of many alternative TISs and PASs, and therefore, the identification of new 5′ and 3′ UTR regions of known transcripts, as illustrated in this study. Increasing evidence suggests that 5′ and 3′ UTR regions of transcripts are important for translational regulation, transcript stability, and subcellular targeting (review in refs. 29–31). Dense mapping of TISs on chromosomes would also help to provide quantitative measurements of differential TIS use and aid in the identification of putative promoter regions.
A major advantage of 5′LS and 3′LS is the ability to identify new transcription units, which is particularly useful if these units are expressed either transiently or at low levels. Although they were generated independently, simultaneous mapping of 5′LS and 3′LS tags to the same genome sequences allows one to discern the relationship of 5′ tags to 3′ tags that were derived from the same transcripts. Furthermore, as we demonstrated in this study, the paired 5′LS and 3′LS tag sequences can be used directly as PCR primers to amplify the full-length transcript clone of interest for further study, avoiding tedious and inefficient molecular cloning steps such as RACE and library hybridization screening.
As is inherent in any short-tag approach, the length of these tags could still cause some ambiguity when mapped to the genome. Due to nontemplated nucleotide incorporation at the 5′ end of transcripts and other variations, including MmeI slippage, the actual useable length of 5′LS and 3′LS tag sequences could be as short as 17 bp. Further effort is needed to develop better methodology and enzymatic reagents to increase the tag length and therefore tag specificity. However, a balance should be maintained, because tag length is inversely proportional to the efficiency of the short-tag strategy. We are also optimizing the “tags → genome → genes” mapping approach for the 5′LS and 3′LS tags. This mapping approach, as we demonstrated, is clearly advantageous over the traditional “tag → unigene” mapping approach in its ability to identify tags that represent new TISs and PASs of transcripts, and to distinguish between alternative transcripts of the same gene with different TISs and/or PASs. As the assembled genome sequences become more accurate and more transcript sequence data are compiled along the genome landscape, this tags → genome → genes annotation approach will become more streamlined. We envisage that a systematic and large scale production of 5′LS and 3′LS tags will provide an invaluable dataset for the thorough characterization of transcriptomes and the annotation of complex genomes.
Acknowledgments
We thank Mr. H. Thoreau, Mr. L. Lim, the Sequencing Group [Genome Institute of Singapore (GIS)], Mr. C. S. Chan (GIS), Mr. H. L. Hor and Mr. M. Hirwan (Bioinformatics Institute) for technical support; and Dr. S. K. Lim (GIS) for providing E14 cells. We also thank Dr. K. Boon and Ms. J. Shoemaker in Dr. Gregory Riggins' laboratory (Duke University, Durham, NC) for providing hands-on experience with SAGE protocols. This work was supported by the Agency for Science, Technology, and Research of Singapore.
Abbreviations: SAGE, serial analysis of gene expression; LS, LongSage; MPSS, massively parallel signature sequencing; TIS, transcription initiation site; PAS, polyadenylation site; UCSC, University of California, Santa Cruz; tp, tag position.
Note. While this manuscript was in preparation, Shiraki et al. (32) published their work on cap analysis of gene expression (CAGE), that is very similar to the 5′LS method described here. It should be noted that whereas both CAGE and 5′LS are useful for identifying TISs and possibly promoter regions, the combined 5′LS and 3′LS mapping strategy as outlined here possesses an obvious advantage in identifying new transcript units with greater confidence than the single-tag mapping.
References
- 1.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 3.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 4.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 5.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 6.Cruveiller, S., Jabbari, K., Clay, O. & Bemardi, G. (2003) Brief. Bioinform. 4, 43–52. [DOI] [PubMed] [Google Scholar]
- 7.Rust, A. G., Mongin, E. & Birney, E. (2002) Drug Discov. Today 7, S70–S76. [DOI] [PubMed] [Google Scholar]
- 8.Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. (2001) Genome Res. 11, 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S., Nikaido, I., Osato, N., Saito, R., Suzuki, H., et al. (2002) Nature 420, 563–573.12466851 [Google Scholar]
- 11.Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. (2001) Nature 409, 685–690. [DOI] [PubMed] [Google Scholar]
- 12.Das, M., Harvey, I., Chu, L. L., Sinha, M. & Pelletier, J. (2001) Physiol. Genomics 6, 57–80. [DOI] [PubMed] [Google Scholar]
- 13.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 14.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508–512. [DOI] [PubMed] [Google Scholar]
- 15.Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D. H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. (2000) Nat. Biotechnol. 18, 630–634. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, M., Hardy, K., Handyside, A., Hunter, S. & Monk, M. (1987) Nature 326, 292–295. [DOI] [PubMed] [Google Scholar]
- 17.Carninci, P. & Hayashizaki, Y. (1999) Methods Enzymol. 303, 19–44. [DOI] [PubMed] [Google Scholar]
- 18.van Kampen, A. H., van Schaik, B. D., Pauws, E., Michiels, E. M., Ruijter, J. M., Caron, H. N., Versteeg, R., Heisterkamp, S. H., Leunissen, J. A., Baas, F., et al. (2000) Bioinformatics 16, 899–905. [DOI] [PubMed] [Google Scholar]
- 19.Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., Roskin, K. M., Schwartz, M., Sugnet, C. W., Thomas, D. J., et al. (2003) Nucleic Acids Res. 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt, W. M. & Mueller, M. W. (1999) Nucleic Acids Res. 27, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, P. H. & Preston, B. D. (1994) Proc. Natl. Acad. Sci. USA 91, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata, Y., Carninci, P., Sato, K., Hayatsu, N., Shiraki, T., Ishii, Y., Arakawa, T., Hara, A., Ohsato, N., Izawa, M., et al. (2001) BioTechniques 31, 1042, 1044, 1048–1049. [DOI] [PubMed] [Google Scholar]
- 24.Cho, S.-H. & Kang, C. (1990) Mol. Cells 1, 81–86. [Google Scholar]
- 25.Reymond, A., Friedli, M., Henrichsen, C. N., Chapot, F., Deutsch, S., Ucla, C., Rossier, C., Lyle, R., Guipponi, M. & Antonarakis, S. E. (2001) Genomics 78, 46–54. [DOI] [PubMed] [Google Scholar]
- 26.Rosner, M. H., Vigano, M. A., Ozato, K., Timmons, P. M., Poirier, F., Rigby, P. W. & Staudt, L. M. (1990) Nature 345, 686–692. [DOI] [PubMed] [Google Scholar]
- 27.Rogers, M. B., Hosler, B. A. & Gudas, L. J. (1991) Development (Cambridge, U.K.) 113, 815–824. [DOI] [PubMed] [Google Scholar]
- 28.Okuda, A., Fukushima, A., Nishimoto, M., Orimo, A., Yamagishi, T., Nabeshima, Y., Kuro-o, M., Boon, K., Keaveney, M., Stunnenberg, H. G., et al. (1998) EMBO J. 17, 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignone, F., Gissi, C., Liuni, S. & Pesole, G. (2002) Genome Biol. 3, REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuersten, S. & Goodwin, E. B. (2003) Nat. Rev. Genet. 4, 626–637. [DOI] [PubMed] [Google Scholar]
- 31.Pesole, G., Mignone, F., Gissi, C., Grillo, G., Licciulli, F. & Liuni, S. (2001) Gene 276, 73–81. [DOI] [PubMed] [Google Scholar]
- 32.Shiraki, T., Kondo, S., Katayama, S., Waki, K., Kasukawa, T., Kawaji, H., Kodzius, R., Watahiki, A., Nakamura, M., Arakawa, T., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]