Abstract
Transcription of the arginine biosynthetic gene ARG1 is repressed by the ArgR/Mcm1p complex in arginine-replete cells and activated by Gcn4p, a transcription factor induced by starvation for any amino acid. We show that all four subunits of the arginine repressor are recruited to ARG1 by Gcn4p in cells replete with arginine but starved for isoleucine/valine. None of these proteins is recruited to the Gcn4p target genes ARG4 and SNZ1, which are not regulated by ArgR/Mcm1p. Mcm1p and Arg80p were found in a soluble complex lacking Arg81p and Arg82p, and both Mcm1p and Arg80p were efficiently recruited to ARG1 in wild-type cells in the presence or absence of exogenous arginine, and also in arg81Δ cells. By contrast, the recruitment of Arg81p and Arg82p was stimulated by exogenous arginine. These findings suggest that Gcn4p constitutively recruits an Mcm1p/Arg80p heterodimer and that efficient assembly of a functional repressor also containing Arg81p and Arg82p occurs only in arginine excess. By recruiting an arginine-regulated repressor, Gcn4p can precisely modulate its activation function at ARG1 according to the availability of arginine.
Four of the arginine biosynthetic genes in the yeast Saccharomyces cerevisiae (ARG1, ARG3, ARG5,6, and ARG8) are subject to dual regulation by the ArgR/Mcm1p repressor and the transcriptional activator Gcn4p. Transcription of most amino acid biosynthetic genes, including all of the ARG genes, is induced by Gcn4p in cells starved for any amino acid, owing to increased expression of GCN4 at the translational level (reviewed in refs. 1 and 2). The ArgR/Mcm1p complex mediates the repression of specific ARG genes in response to exogenous arginine. It also functions as an inducer of the arginine catabolic genes CAR1 and CAR2, allowing utilization of excess arginine as a nitrogen source. A wealth of genetic and biochemical evidence indicates that the ArgR/Mcm1p repressor binds to the arginine control (ARC) elements located in the promoters of its target genes (reviewed in ref. 3).
A functional ArgR/Mcm1p repressor complex requires Arg80p, Arg81p, and Arg82p, all nonessential for viability, and the essential protein Mcm1. Mcm1p is an MCM1, AGAMOUS, DEFICIENS, and serum response factor (MADS) box protein that cooperates with diverse sequence-specific transcription factors to repress or activate different sets of genes (reviewed in ref. 3). The first 10 bp in the ARC element represents a degenerate P box, the binding site for Mcm1p (4, 5). Arg80p is also a MADS box protein and shows strong similarity to Mcm1p (6). Arg81p belongs to the Zn2C6-cluster family of transcription factors (7), and the last 3 bp of the ARC element corresponds to the CGR motif recognized by other members of this family (4, 5).
None of the subunits of the ArgR/Mcm1p complex can bind individually to an ARC element in vitro. Arginine-dependent binding to an ARC element was reconstituted with the combination of Mcm1p and the N-terminal domain of Arg81p in a manner stimulated by Arg80p. Weaker arginine-stimulated binding also was observed with Arg80p and the Arg81p-N-terminal domain in the absence of Mcm1p (8). Thus, detectable binding to an ARC element in vitro required the combination of Arg81p, either of the two MADS box proteins in the repressor complex (Mcm1p or Arg80p), and arginine. Mcm1p binds to an authentic P box as a dimer (9), and Mcm1p binding to the ARC element is stimulated by Arg80p, even in the presence of Arg81p (8). These findings suggest that Mcm1p and Arg80p bind as a heterodimer of MADS box proteins to the degenerate P box in the ARC element. The fact that Mcm1p and Arg80p can also cooperate individually with Arg81p in DNA binding (8), however, suggests that each protein can bind to the ARC element as a homodimer in concert with Arg81p, albeit with reduced efficiency compared with the Mcm1p/Arg80p heterodimer.
In vitro binding of Mcm1p to an authentic P box is insensitive to arginine, whereas cooperative binding to an ARC element by Mcm1p/Arg81p or Arg80p/Arg81p is arginine-responsive (8). This fact suggests that Arg81p is the arginine sensor in the repressor complex. Consistent with this idea, the Arg81p-N-terminal domain contains a region related to bacterial arginine repressors, and mutating conserved residues in this domain increased the arginine concentration required for DNA binding by ArgR/Mcm1p in cell extracts (8).
Arg82p is capable of phosphorylating various inositol polyphosphates (11), but it was shown that inactivation of Arg82p/Ipk2p kinase activity had little impact on repression of ARG3 or induction of CAR1 by arginine (12). Because Arg80p and Mcm1p are less stable in arg82Δ cells, and the requirement for Arg82p can be partly bypassed by overexpressing Arg80p or Mcm1p, Arg82p may be required primarily to stabilize Arg80p and Mcm1p (10).
In this report, we used chromatin immunoprecipitation (ChIP) assays to analyze binding of the ArgR/Mcm1p repressor in vivo to one of its target genes, ARG1. We found unexpectedly that binding of ArgR/Mcm1p is strongly stimulated when synthesis of Gcn4p, and its attendant binding at ARG1, is induced by amino acid starvation. We show that Mcm1p and Arg80p reside in a stable complex lacking Arg81p and Arg82p, and we present evidence that Mcm1p/Arg80p are recruited by Gcn4p independent of Arg81p and exogenous arginine. Interestingly, recruitment of Arg81p and Arg82p is stimulated by arginine in the medium, implying that Gcn4p efficiently recruits a complete ArgR/Mcm1p repressor only under conditions of arginine excess. These findings suggest a mechanism whereby Gcn4p can dampen its activation function at ARG1 in arginine-replete cells over a continuum of Gcn4p expression levels.
Materials and Methods
Yeast Strains and Plasmids. Yeast strains used in this study are listed in Table 1, which is published as supporting information on the PNAS web site. They were produced by the Saccharomyces Genome Deletion Project from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and purchased from Research Genetics/Invitrogen (Carlsbad, CA), or they were derived from such strains. All deletion alleles were confirmed by PCR amplification (13). We also confirmed that the arg80Δ and arg81Δ strains are defective for arginine-repression of an ARG1-lacZ fusion on plasmid YCp87-ARG1-lacZ (data not shown), and that the arg11Δ strain is an arginine auxotroph. Insertion of the coding sequences for 13 myc epitopes at the C termini of ARG80, ARG81, ARG82, and MCM1 was conducted as described (13), and the myc-tagged alleles were verified by PCR amplification and Western analysis of whole-cell extracts (WCEs), using anti-myc antibodies (data not shown). The MCM1-myc strain grew indistinguishably from the parental untagged strain. The MCM1-myc, ARG80-myc, and ARG81-myc strains were indistinguishable from the untagged strains in the degree of arginine-repression of the ARG1-lacZ fusion (data not shown). The plasmids used in this work are listed in Table 2, which is published as supporting information on the PNAS web site. The details of construction of other plasmids are provided in Table 2.
Biochemical Methods. ChIP assays were conducted as described (14) by using the same primers utilized to amplify the ARG1 upstream activation sequence (UAS) (13), SNZ1 UAS (14), or ARG4 UAS (15). Western analysis of WCEs from cells treated with trichloroacetic acid was conducted as described (14). GST pull-down assays were conducted as described (13) by using goat polyclonal anti-Mcm1p antibodies (Santa Cruz Biotechnology) and rabbit polyclonal anti-Snf5p antibodies (provided by B. Laurent, State University of New York, Brooklyn). Coimmunoprecipitation analysis was performed on the same WCEs by using mouse monoclonal anti-myc antibodies (Roche Applied Science). Briefly, the WCEs were incubated for 1 h at 4°C with 1 μg of anti-myc antibody, 100 μg BSA dissolved in PBS, and MTB buffer [50 mM Hepes/0.2 M potassium acetate/13.5 mM magnesium acetate/1 mM EGTA/20% glycerol/10 mg/ml pepstatin/1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride)/0.01% Nonidet P-40/protease inhibitor cocktail (Roche Molecular Biochemicals)] (16). The immune complexes were washed three times with 1 ml of MTB buffer, dissolved in SDS/PAGE loading buffer (Invitrogen), and subjected to Western analysis.
Results
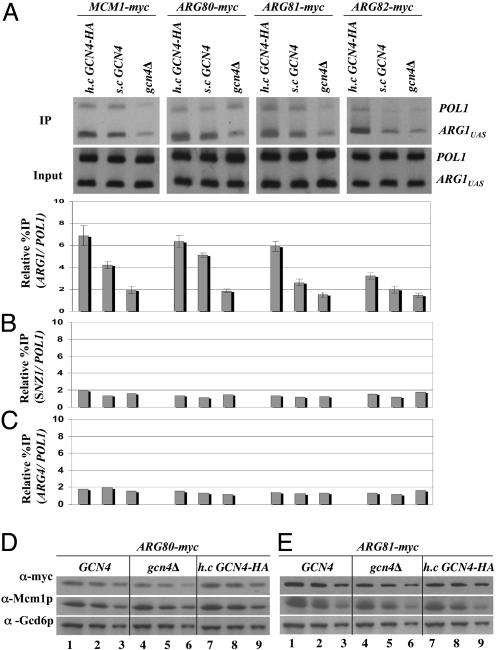
Gcn4p Recruits the Entire ArgR/Mcm1p Complex to ARG1 in Arginine-Replete Cells Through Its Activation Domain. To determine whether Gcn4p influences the binding of the ArgR/Mcm1p complex to the ARG1 promoter in vivo, we conducted ChIP experiments on four sets of isogenic strains containing functional myc-tagged forms of Mcm1p, Arg80p, Arg81p, or Arg82p (see Materials and Methods). For each tagged protein, we compared isogenic strains containing no Gcn4p, native levels of Gcn4p, or overexpressed HA-tagged Gcn4p, grown in synthetic complete medium containing arginine, and treated with sulfometuron methyl (SM) to induce Gcn4p synthesis by starvation for isoleucine and valine. In the gcn4Δ strains, we observed low-level binding of myc-Mcm1p, myc-Arg80p, myc-Arg81p, and myc-Arg82p to the ARG1 promoter only slightly greater than that measured for the coding sequences of the POL1 gene, analyzed as a negative control. A significantly higher level of binding was observed in the strains expressing Gcn4p from the chromosomal allele, and overexpression of Gcn4p from a high-copy (h.c.) plasmid produced even greater binding of these proteins to the ARG1 promoter (Fig. 1A). Overexpressing Gcn4p from this h.c. plasmid produces an ≈2-fold increase in promoter occupancy of Gcn4p at the ARG1 promoter compared with that seen for native levels of Gcn4p (data not shown). These results indicate that Gcn4p stimulates the binding of all four subunits of the ArgR/Mcm1p complex to the ARG1 promoter in cells replete with arginine.
Fig. 1.
Gcn4p recruits the ArgR/Mcm1p complex to the promoter at ARG1 but not at SNZ1 or ARG4. (A) GCN4 strains containing myc-tagged alleles of MCM1 (SY337), ARG80 (SY373), ARG81 (SY375), and ARG82 (SY377) were transformed with empty vector or 2 μm plasmid pHQ1239 harboring GCN4-HA. gcn4Δ strains with myc-tagged MCM1 (SY339), ARG80 (SY374), ARG81 (SY376), or ARG82 (SY378) were transformed with empty vector. Cells were grown to an OD600 of ≈1.0 in synthetic complete-Ura medium (containing 0.5 mM Arg), treated for 2 h with SM (0.6 μg/ml), and cross-linked with HCHO. ChIP assays were conducted by using anti-myc antibodies and PCR primers specific for the ARG1 UAS or POL1 ORF. The ARG1UAS signal in the immunoprecipitate (IP) was normalized for the corresponding POL1 signal and plotted in the histogram. (B and C) The same IP samples described in A were analyzed by using primers to amplify the SNZ1 and ARG4 UAS elements. (D and E) The ARG80-myc or ARG81-myc strains described in A were grown and induced with SM, and WCEs were subjected to Western analysis with antibodies against myc, Mcm1p, or Gcd6p, as indicated on the left. Three different amounts of each extract were loaded in adjacent lanes to ensure that Western signals were dose-dependent.
To determine whether Gcn4p promotes binding of ArgR/Mcm1p at ARG1 by inducing the expression of these proteins, we conducted Western analysis of Mcm1p, Arg80p, and Arg81p in extracts from the strains described above grown in the presence of SM. The amounts of these ArgR/Mcm1p components were quantified and normalized for the levels of Gcd6p, analyzed in parallel as a loading control. The results showed no significant differences in the steady-state levels of Mcm1p and myc-Arg81p between cells harboring single-copy (s.c.) GCN4, gcn4Δ, or h.c.GCN4 (Fig. 1E). The myc-Arg80p level was ≈30% lower in the gcn4Δ strain compared with the GCN4 and h.c.GCN4 strains (Fig. 1D); however, this small difference in expression cannot account for the much greater difference in binding of myc-Arg80p to ARG1 seen in the h.c.GCN4 versus gcn4Δ strain (Fig. 1A). Moreover, we show below that the Gcn4p-stimulated recruitment of Mcm1p can occur at high levels in deletion mutants lacking ARG80 or ARG81. Thus, we conclude that Gcn4p stimulates recruitment of the ArgR/Mcm1p complex to ARG1, rather than inducing the expression of its subunits.
In contrast to the strong Gcn4p-dependent binding of ArgR/Mcm1p at ARG1 shown in Fig. 1A, we observed little or no binding above background levels for all four subunits of the repressor complex at the SNZ1 and ARG4 promoters, irrespective of the Gcn4p expression level (Fig. 1 B and C). Transcription of SNZ1, ARG4, and ARG1 is induced by Gcn4p (2), and Gcn4p binds to the promoters of all three of these genes in vivo (15). However, ARG4 lacks ARC elements and is not repressed by the ArgR/Mcm1p complex (1), and SNZ1 does not encode an arginine biosynthetic enzyme. These last results suggest that the ArgR/Mcm1p complex must interact with the ARC elements in the promoter to be recruited at high levels by Gcn4p to ARG1.
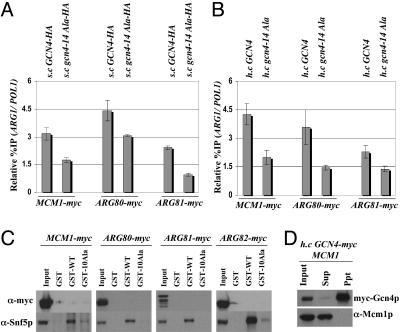
The ability of Gcn4p to interact with coactivators in cell extracts (16, 17) and to recruit coactivators to target promoters in vivo (14, 18) depends on the hydrophobic clusters in its activation domain required for transcriptional activation in vivo (19). To determine whether recruitment of ArgR/Mcm1p by Gcn4p involves its activation domain, we conducted ChIP assays on isogenic strains expressing wild-type Gcn4p or a mutant harboring Ala substitutions in all seven hydrophobic clusters in the activation domain (gcn4-14Ala). We found that binding of myc-Mcm1p, myc-Arg80p, and myc-Arg81p to the ARG1 promoter was greater in cells expressing wild-type Gcn4p versus those expressing the gcn4-14Ala protein (Fig. 2 A and B). In general, the binding observed in the strains containing gcn4-14Ala occurred at the low level seen in gcn4Δ strains (Fig. 1A). We showed previously that gcn4-14Ala binds to the ARG1 promoter in vivo at levels equal to or greater than that seen for wild-type Gcn4p (14). Thus, the hydrophobic residues in the Gcn4p activation domain are required for high-level recruitment of ArgR/Mcm1p to ARG1 by promoter-bound Gcn4p.
Fig. 2.
Hydrophobic clusters in the Gcn4p activation domain are required for recruitment of the ArgR/Mcm1p complex. (A and B) The gcn4Δ myc-tagged strains described in Fig. 1 transformed with s.c. plasmids harboring GCN4-HA (p2382) or gcn4–14Ala-HA (pSY285) (A), or with h.c. plasmids containing GCN4 (pHQ1303) or gcn4–14Ala (pHQ1304) (B), were grown in the presence of arginine and SM and subjected to ChIP analysis as in Fig. 1. (C) GST, GST-Gcn4p (GST-WT), and GST-Gcn4p containing 10 Ala mutations in the activation domain (GST-10Ala) were expressed in Escherichia coli, immobilized on glutathione Sepharose, beads and added to WCEs from the GCN4 myc-tagged strains described in Fig. 1. Proteins bound to the beads were subjected to Western analysis with antibodies against myc or Snf5p. (D) gcn4Δ strain 249 transformed with h.c. plasmids harboring GCN4-myc (pHQ1293) and MCM1 (pED40) were cultured in synthetic complete-Ura-Leu and induced with SM. WCEs were immunoprecipitated with myc antibodies. Ten percent of the input samples (Input), 100% of the immunoprecipitates (Ppt), and 10% of the supernatant (Sup) fractions were subjected to Western analysis with myc antibodies or goat polyclonal antibodies against Mcm1p.
We wished to determine whether Gcn4p can interact directly with the ArgR/Mcm1p complex in cell extracts. We showed (13, 17) that a recombinant GST-Gcn4p fusion can bind specifically to various coactivators that it recruits to ARG1 in vivo, such as SWI/SNF complex. In agreement with previous findings, wild-type GST-Gcn4p fusion, but not the corresponding fusion containing 10 Ala substitutions in the activation domain (10Ala mutant), could bind to the SWI/SNF subunit Snf5p in cell extracts. In contrast, there was no detectable binding of myc-tagged subunits of the ArgR/Mcm1p complex to wild-type or mutant GST-Gcn4p (Fig. 2C). We also failed to observe coimmunoprecipitation of Mcm1p with myc-tagged Gcn4p from cell extracts prepared from a strain overexpressing both proteins (Fig. 2D). (It is shown in Fig. 4A that Mcm1p can be immunoprecipitated with an myc-tagged form of Arg80p.) Thus, we have no evidence that Gcn4p can form a stable complex with ArgR/Mcm1p free of promoter DNA. It is possible that a stable physical interaction between Gcn4p and ArgR/Mcm1p occurs only when Gcn4p and the repressor occupy their respective binding sites in the ARG1 promoter. Alternatively, Gcn4p may facilitate binding of ArgR/Mcm1p by recruiting chromatin remodeling complexes that can increase accessibility of the ARC element.
Fig. 4.
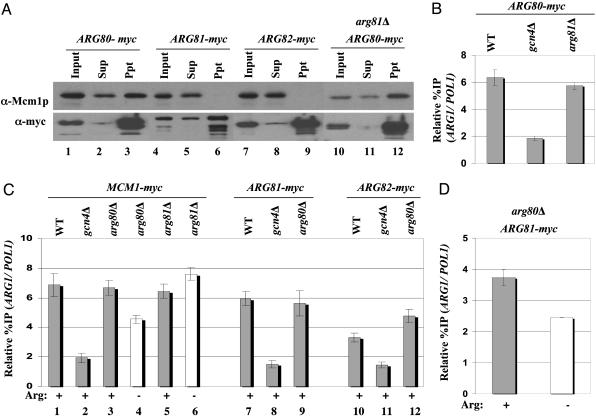
Evidence for Arg81p-independent recruitment of an Arg80p/Mcm1p heterodimer. (A) GCN4 strains with MCM1-myc (SY337), ARG80-myc (SY373), or ARG81-myc (SY375), and the arg81Δ ARG80-myc strain SY384, were grown in YPD, and WCEs were immunoprecipitated with myc antibodies and subjected to Western analysis by using myc antibodies or polyclonal Mcm1p antibodies to probe the blots. (B) Transformants of GCN4 ARG80-myc strain SY373 and GCN4 arg81Δ ARG80-myc strain SY384 harboring episomal GCN4-HA (pHQ1239), and gcn4Δ ARG80-myc strain SY374 containing empty vector, were grown in the presence of SM and arginine and subjected to ChIP analysis, as described in Fig. 1. (C) GCN4 strains harboring episomal GCN4-HA (pHQ1239), or gcn4Δ strains containing empty vector, and harboring the myc-tagged alleles indicated at the top, were cultured in the presence of SM in medium lacking or containing 0.5 mM arginine, as indicated at the bottom, and subjected to ChIP analysis. (D) GCN4 ARG81-myc arg80Δ strain, SY381 grown in the presence (+) or absence (–) of 0.5 mM arginine, was subjected to ChIP analysis.
Gcn4p Recruits Mcm1p and Arg80p to ARG1 Independent of Arginine Concentration. The results presented above suggest that high-level binding of the ArgR/Mcm1p complex at ARG1 should not occur under conditions where Gcn4p is synthesized at low basal levels. In agreement with this prediction, we found that binding of myc-tagged Mcm1p, Arg80p, Arg81p, and Arg82p at ARG1 was strongly induced by SM treatment of cells containing GCN4 and grown in the presence of exogenous arginine (Fig. 3A, compare lanes 1–4for each tagged subunit). The level of binding by these proteins in the absence of SM was similar to that shown above for the gcn4Δ strains treated with SM (Fig. 1A). We have found that treatment of GCN4 cells with SM increases the binding of Gcn4p at ARG1 by at least 10-fold (C.K.G. and A.G.H., unpublished observations).
Fig. 3.
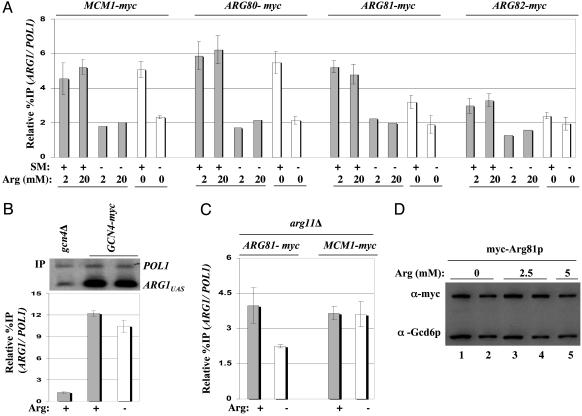
Constitutive recruitment of Mcm1p/Arg80p and arginine-stimulated recruitment of Arg81p/Arg82p by Gcn4p. (A) GCN4 strains harboring the myc-tagged alleles indicated at the top and episomal GCN4-HA (pHQ1239) were subjected to ChIP assays exactly as in Fig. 1, except for the indicated arginine concentrations in the media, and the presence or absence of SM, as indicated in the histograms. (B) Transformants of gcn4Δ strain 249 containing s.c. GCN4-myc plasmid (pSK-1) or empty vector YCp50 were subjected to ChIP analysis after growing in the presence (+) or absence (–) of 0.5 mM arginine. (C) The arg11Δ strains containing myc-tagged MCM1 (SY590) or ARG81 (SY589) and episomal GCN4-HA (pHQ1239) were cultured with 2 mM (+) or 7.5 μM(–) arginine and subjected to ChIP analysis. (D) The ARG81-myc strain SY589 was grown in the presence or absence of arginine as indicated and subjected to Western analysis as described in Fig. 1 D and E. Lanes 2 and 4 were loaded with 50% of the amounts of extract loaded in lanes 1 and 3, respectively.
Binding of ArgR/Mcm1p to the ARC element in vitro depends on arginine (8), and maximum repression of ARG1 by ArgR/Mcm1p in vivo requires an arginine supplement to the medium (5). Accordingly, we investigated whether recruitment of ArgR/Mcm1p by Gcn4p to ARG1 requires exogenous arginine. In the experiments of Figs. 1 and 2, arginine was present in the medium at 0.5 mM, and we found that addition of 4- or 40-fold higher levels of arginine did not provide greater ArgR/Mcm1p binding to ARG1 when Gcn4p is induced with SM (compare Figs. 1A and 3A). Moreover, the higher levels of arginine did not permit detectable binding of ArgR/Mcm1p without induction of Gcn4p by SM. Thus, maximal binding of ArgR/Mcm1p at ARG1 can be achieved with 0.5 mM exogenous arginine. Interestingly, the recruitment of myc-Arg81p and myc-Arg82p in the presence of SM was reproducibly diminished by omitting arginine from the medium (Fig. 3A, compare lanes 1, 2, and 5 for ARG81-myc and ARG82-myc). (Student's t test indicates that the reduction in ARG1 binding of myc-Arg81p and myc-Arg82p produced by exogenous arginine is statistically significant, with P values of 0.001 and 0.02, respectively.) The fact that binding of myc-Arg81p and myc-Arg82p was decreased, but not eliminated, by withdrawal of exogenous arginine is in accordance with previous results (5), indicating that ARG1 transcription is partially derepressed in medium lacking arginine. Importantly, we saw no reduction in binding of myc-Mcm1p or myc-Arg80p at ARG1 when arginine was eliminated from the medium (Fig. 3A), showing that recruitment of these last two subunits of the repressor is insensitive to the cellular arginine concentration. Consistent with the constitutive recruitment of myc-Mcm1p and myc-Arg80p, the binding of myc-tagged Gcn4p at ARG1 was unaffected by the addition of arginine to the medium (Fig. 3B).
It was possible that the recruitment of ArgR/Mcm1p subunits we observed in medium lacking arginine was attributable to the endogenous arginine pool generated by de novo biosynthesis. In an effort to address this possibility, we examined the recruitment of myc-Mcm1p and myc-Arg81p in an arg11Δ mutant grown in the presence of excess arginine (2 mM) or with a minimal amount of arginine (7.5 μM) sufficient to permit growth at a reduced rate. ARG11 encodes a mitochondrial membrane protein required for efficient arginine biosynthesis, most likely functioning in export of ornithine from mitochondria to the cytoplasm (20). Thus, arg11Δ cells have greatly reduced arginine pools (21) and exhibit fully derepressed levels of arginine biosynthetic enzymes regulated by ArgR/Mcm1p in medium lacking arginine (22). Growth of arg11Δ cells on medium containing 7.5 μM arginine was shown previously to produce arginine starvation sufficient to derepress Gcn4p target genes in vivo (23, 24).
In accordance with our findings in Fig. 3A, myc-Arg-81 binding to ARG1 in the presence of SM was lower when the arg11Δ strain was grown on 7.5 μM arginine versus 2 mM arginine (Fig. 3C). However, arginine starvation of the arg11Δ strain did not elicit a greater reduction in myc-Arg81p recruitment compared with that seen in wild-type cells grown in medium lacking arginine (compare Fig. 3 A and C for ARG81-myc). We observed the same high level of myc-Mcm1p binding to ARG1 in the arg11Δ strain grown on 7.5 μM or 2 mM arginine (Fig. 3C), confirming the constitutive recruitment of this protein by Gcn4p, independent of arginine levels (Fig. 3A).
We conducted Western analysis to determine whether the diminished recruitment of Arg81p seen in medium lacking arginine could arise from decreased expression of this protein; however, no significant differences in myc-Arg81p levels were observed between medium containing or lacking arginine (Fig. 3D).
Evidence for Recruitment of an Mcm1p/Arg80p Subcomplex Independent of Arg81p. The above findings raised the possibility that Mcm1p and Arg80p can be recruited by Gcn4p independent of Arg81p and Arg82p. Recalling that Mcm1p and Arg80p contain related MADS box domains and that Mcm1p binds to the P box as a dimer, we considered the possibility that Mcm1p and Arg80p are recruited by Gcn4p as a preformed heterodimer. Supporting this hypothesis, we found that Mcm1p can be coimmunoprecipitated with myc-Arg80p, but not with myc-Arg81p or myc-Arg82p, from extracts of yeast strains expressing these proteins at native levels and grown in medium containing arginine (Fig. 4A, compare lanes 3, 6, and 9). Similar results were obtained in medium lacking arginine (data not shown). Moreover, the stable association of myc-Arg80p and Mcm1p in cell extracts was unaffected by deletion of ARG81 (compare lanes 3 and 12). These findings are consistent with a model in which Gcn4p recruits the preformed Mcm1p/Arg80p complex to the ARG1 promoter independent of the arginine concentration to provide a platform for assembly of the complete ArgR/Mcm1p complex in a manner stimulated by exogenous arginine.
In agreement with this model, we found that recruitment of myc-Arg80p and myc-Mcm1p by Gcn4p was unaffected by deletion of ARG81 (Fig. 4 B and C, lanes 1, 5, and 6). Whereas this finding seems at odds with the previous observation (8) that Arg80p and Mcm1p cannot bind to an ARC element in vitro in the absence of Arg81p, we suggest that the stimulatory effect of Gcn4p on recruitment of Mcm1p/Arg80p can compensate for the absence of Arg81p in vivo. We also observed high-level recruitment of myc-Mcm1p, myc-Arg81p, and myc-Arg82p in arg80Δ cells (Fig. 4C). These last results fit well with the previous in vitro data (8) indicating that Mcm1p and Arg81p can bind to an ARC element in the absence of Arg80p, albeit less efficiently than when Arg80p is present. Here, we presume that Mcm1p binds to the degenerate P box in the ARC element as a homodimer in conjunction with Arg81p, and that Gcn4p compensates for the absence of Arg80p to permit high-level binding of Mcm1p/Arg81p at ARG1 in vivo.
Interestingly, recruitment of myc-Mcm1p was stimulated by arginine in an arg80Δ mutant, but not in arg81Δ cells (Fig. 4C, compare lanes 3–6). Moreover, high-level recruitment of myc-Arg81p remained dependent on arginine in the arg80Δ mutant (Fig. 4D). These findings can be explained by proposing that recruitment of an Mcm1p homodimer in the arg80Δ mutant depends on Arg81p. Hence, when Arg80p is missing, Mcm1p acquires the same dependence on exogenous arginine for optimal recruitment that characterizes the recruitment of Arg81p (Fig. 3A). As expected, recruitment of Mcm1p remained constitutive in the arg81Δ strain (Fig. 4C, lanes 5 and 6), as shown in the previous paragraph in wild-type cells (Fig. 3A).
Discussion
In this report, we showed that binding of the ArgR/Mcm1p repressor at ARG1 is strongly enhanced by Gcn4p, the activator that stimulates transcription of this gene in response to amino acid starvation. Binding of all four components of the ArgR/Mcm1p complex (Mcm1p, Arg80p, Arg81p, and Arg82p) at ARG1 was detected by ChIP assays only when Gcn4p synthesis was induced by starvation for isoleucine and valine (Fig. 3A). The binding of ArgR/Mcm1p at ARG1 was severely reduced under these conditions by deletion of GCN4 (Fig. 1A). Furthermore, the hydrophobic residues in the Gcn4p activation domain required for its recruitment of coactivator proteins (14, 18) are also necessary for recruitment of ArgR/Mcm1p to ARG1 (Fig. 2 A and B).
A role for Gcn4p in recruiting the ArgR/Mcm1p repressor was unexpected because binding to an ARC element has been reconstituted in vitro by using only recombinant Mcm1p, Arg80p, and the Arg-81 N-terminal domain (8). In addition, it was demonstrated (5) that arginine repression of ARG1 can occur under nonstarvation conditions where Gcn4p is produced at low, uninduced levels, and even in gcn4Δ cells. For example, arg81Δ or deletion of ARC elements increased ARG1 transcription 5- to 8-fold on minimal medium containing arginine (5), a condition where Gcn4p synthesis is repressed to low levels by the endogenous amino acids produced biosynthetically (25). Thus, induced levels of Gcn4p are not required for ArgR/Mcm1p repressor function at ARG1 in vivo. How can we reconcile these earlier results with our finding that binding of ArgR/Mcm1p to ARG1 was detected only when Gcn4p was induced by starvation for isoleucine/valine?
To resolve this apparent discrepancy, we note that Gcn4p is the principal activator at ARG1 and it makes a substantial contribution to ARG1 transcription, even in nonstarvation conditions where it is expressed at low levels (5). However, it is difficult to detect Gcn4p binding to the ARG1 promoter by the ChIP assay under nonstarvation conditions. Thus, it is reasonable to propose that a small amount of ArgR/Mcm1p binding, also below the detection limit of the ChIP assay, would be sufficient to repress the low promoter activity of ARG1 that occurs at the uninduced level of Gcn4p. An even smaller amount of ArgR/Mcm1p would be required to repress the basal promoter activity that occurs in gcn4Δ cells.
We presented several pieces evidence leading us to propose that Gcn4p recruits a preformed Mcm1p/Arg80p subcomplex to the ARG1 promoter. First, we showed that Mcm1p and Arg80p, both MADS box proteins, form a stable subcomplex lacking Arg81p and Arg82p that can be coimmunoprecipitated from extracts of wild-type and arg81Δ cells (Fig. 4A). Second, we found that Gcn4p recruits Mcm1p and Arg80p to ARG1 independent of Arg81p (Fig. 4 B and C). Third, we demonstrated that recruitment of Mcm1p and Arg80p occurs constitutively with respect to arginine concentration, whereas recruitment of Arg81p and Arg82p is stimulated by exogenous arginine (Fig. 3 A and C). These results are in accordance with the previous conclusion that Arg81p contains the arginine sensor of the repressor complex (8). Our findings that Gcn4p recruits Mcm1p and Arg80p to ARG1 independent of arginine levels, and in arg81Δ cells, were unexpected in view of previous results indicating that binding of Mcm1p and Arg80p to an ARC element in vitro depended on both Arg81p and arginine (8). We suggest that Gcn4p bound at ARG1 can promote binding of an Mcm1p/Arg80p heterodimer to the ARC elements at ARG1 and bypass the requirement for Arg81p and arginine for high-level promoter binding in vivo.
Whereas recruitment of Mcm1p and Arg80p occurs at high levels in the absence of exogenous arginine, we consistently observed an ≈2-fold lower promoter occupancy of Arg81p and Arg82p in medium lacking arginine compared with medium containing arginine (Fig. 3 A and C). This partial reduction in Arg81p and Arg82p recruitment coincides with the partial derepression of ARG1 transcription that was observed when cells are shifted from medium containing arginine to medium lacking arginine. Indeed, full derepression of ARG1 transcription was observed only in mutants lacking Arg81p or with deletions of the ARC elements in the promoter (5). Thus, we propose that Gcn4p recruits the Mcm1p/Arg80p heterodimer independent of the arginine concentration, and these two proteins provide a platform for arginine-stimulated recruitment of Arg81p and Arg82p. In this way, assembly of a functional ArgR/Mcm1p repressor can be enhanced by Gcn4p and also regulated by arginine levels. We envision that arginine binding to Arg81p produces a conformational change in this protein that stabilizes its association with Arg80p/Mcm1p at high arginine concentrations. However, we cannot rule out an alternative explanation in which recruitment of all four ArgR/Mcm1p subunits occurs constitutively and arginine merely elicits a conformational change that increases the efficiency of formaldehyde-crosslinking of Arg81p/Arg82p to promoter chromatin.
Our model describes an attractive mechanism to modulate the ability of Gcn4p to activate ARG1 transcription according to the demand for arginine. Northern analysis of ARG1 mRNA expression shows that the ability of Gcn4p to activate ARG1 transcription is inhibited by arginine, particularly under nonstarvation conditions where the uninduced level of Gcn4p provides for basal ARG1 transcription (Fig. 5, which is published as supporting information on the PNAS web site). We also found by ChIP analysis that recruitment of TBP by Gcn4p under starvation conditions is partially inhibited by the ArgR/Mcm1p complex (data not shown). Results in accordance with these were obtained (21, 22) from measurements of enzyme activity expressed from ARG1 (5) and ARG3, and from these last studies, it appears that ArgR/Mcm1p represses ARG3 transcription very effectively even when Gcn4p is induced by amino acid starvation (21, 22). These regulatory patterns can be explained by our finding that Gcn4p recruits a fully assembled ArgR/Mcm1p complex to ARG1 in the presence of exogenous arginine, which should dampen its ability to activate transcription. In cells growing in medium lacking arginine, the interaction between Mcm1p/Arg80p and Arg81p/Arg82p is impaired, leading to inadequate recruitment of Arg81p/Arg82p (or a less productive association of Arg81p/Arg82p with Mcm1p/Arg80p). In this situation, Gcn4p recruits a smaller amount of functional repressor and, hence, can achieve a higher degree of transcriptional activation. The ability of Gcn4p to play an active role in recruiting the Mcm1p/Arg80p subcomplex seems to ensure that a constant proportion of Gcn4p and the repressor core subunits are brought to the promoters of ARG genes subject to dual control over a wide range of amino acid concentrations and Gcn4p expression. In this way, Gcn4p can precisely modulate its activation function at these genes according to the availability of arginine.
Acknowledgments
We thank Chris Brandl (University of Western Ontario, London, ON, Canada) for plasmid YCp87-ARG1-lacZ, Francine Messenguy and Evelyn Dubois (Université Libre de Bruxelles, Brussels) for plasmid pED40 and their helpful advice, Brehon Laurent for Snf5p antibodies, and Fan Zhang and other members of the Hinnebusch and Dever Laboratories for helpful suggestions during this work.
Abbreviations: ARC, arginine control; MADS, MCM1, AGAMOUS, DEFICIENS, and serum response factor; ChIP, chromatin immunoprecipitation; WCE, whole-cell extract; UAS, upstream activating sequence; SM, sulfometuron methyl; h.c., high-copy; s.c., single-copy.
References
- 1.Hinnebusch, A. G. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, eds. Broach, J. R., Jones, E. W. & Pringle, J. R. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 319–414.
- 2.Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G. & Marton, M. J. (2001) Mol. Cell. Biol. 21, 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messenguy, F. & Dubois, E. (2003) Gene 316, 1–21. [DOI] [PubMed] [Google Scholar]
- 4.de Rijcke, M., Seneca, S., Punyammalee, B., Glansdorff, N. & Crabeel, M. (1992) Mol. Cell. Biol. 12, 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabeel, M., de Rijcke, M., Seneca, S., Heimberg, H., Pfeiffer, I. & Matisova, A. (1995) Yeast 11, 1367–1380. [DOI] [PubMed] [Google Scholar]
- 6.Jamai, A., Dubois, E., Vershon, A. K. & Messenguy, F. (2002) Mol. Cell. Biol. 22, 5741–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messenguy, F., Dubois, E. & Descamps, F. (1986) Eur. J. Biochem. 157, 77–81. [DOI] [PubMed] [Google Scholar]
- 8.Amar, N., Messenguy, F., El Bakkoury, M. & Dubois, E. (2000) Mol. Cell. Biol. 20, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan, S. & Richmond, T. J. (1998) Nature 391, 660–666. [DOI] [PubMed] [Google Scholar]
- 10.El Bakkoury, M., Dubois, E. & Messenguy, F. (2000) Mol. Microbiol. 35, 15–31. [DOI] [PubMed] [Google Scholar]
- 11.Odom, A. R., Stahlberg, A., Wente, S. R. & York, J. D. (2000) Science 287, 2026–2029. [DOI] [PubMed] [Google Scholar]
- 12.Dubois, E., Dewaste, V., Erneux, C. & Messenguy, F. (2000) FEBS Lett. 486, 300–304. [DOI] [PubMed] [Google Scholar]
- 13.Swanson, M. J., Qiu, H., Sumibcay, L., Krueger, A., Kim, S.-J., Natarajan, K., Yoon, S. & Hinnebusch, A. G. (2003) Mol. Cell. Biol. 23, 2800–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon, S., Qiu, H., Swanson, M. J. & Hinnebusch, A. G. (2003) Mol. Cell. Biol. 23, 8829–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu, H., Hu, C., Yoon, S., Natarajan, K., Swanson, M. & Hinnebusch, A. G. (2004) Mol. Cell. Biol. 24, 4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drysdale, C. M., Jackson, B. M., McVeigh, R., Klebanow, E. R., Bai, Y., Kokubo, T., Swanson, M., Nakatani, Y., Weil, P. A. & Hinnebusch, A. G. (1998) Mol. Cell. Biol. 18, 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan, K., Jackson, B. M., Zhou, H., Winston, F. & Hinnebusch, A. G. (1999) Mol. Cell 4, 657–664. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, M. H., vom Bauer, E., Struhl, K. & Allis, C. D. (2000) Mol. Cell 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, B. M., Drysdale, C. M., Natarajan, K. & Hinnebusch, A. G. (1996) Mol. Cell. Biol. 16, 5557–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crabeel, M., Soetens, O., De Rijcke, M., Pratiwi, R. & Pankiewicz, R. (1996) J. Biol. Chem. 271, 25011–25018. [DOI] [PubMed] [Google Scholar]
- 21.Delforge, J., Messenguy, F. & Wiame, J. (1975) Eur. J. Biochem. 57, 231–239. [DOI] [PubMed] [Google Scholar]
- 22.Messenguy, F. & Dubois, E. (1983) Mol. Gen. Genet. 189, 148–156. [DOI] [PubMed] [Google Scholar]
- 23.Silverman, S. J., Rose, M., Botstein, D. & Fink, G. R. (1982) Mol. Cell. Biol. 2, 1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinnebusch, A. G. & Fink, G. R. (1983) J. Biol. Chem. 258, 5238–5247. [PubMed] [Google Scholar]
- 25.Mueller, P. P. & Hinnebusch, A. G. (1986) Cell 45, 201–207. [DOI] [PubMed] [Google Scholar]