Abstract
Understanding how peptides are selected for presentation by MHC class I is crucial to vaccination strategies based on cytotoxic T lymphocyte priming. We have studied this selection of the MHC class I peptide repertoire in terms of the presentation of a series of individual peptides with a wide range of binding to MHC class I. This series was expressed as minigenes, and the presentation of each peptide variant was determined with the same MHC class I peptide-specific antibody. In wild-type cells, the hierarchy of presentation followed peptide half-life. This hierarchy broke down in cells lacking tapasin but not in cells lacking calreticulin or in cells lacking transporter associated with antigen processing-associated ERp57. We demonstrate a key role for tapasin in shaping the MHC class I peptide repertoire, as enhancement of presentation in the presence of tapasin correlated with peptide half-life.
MHC class I molecules may be the ultimate proteomic microarray (1). On the surface of a single cell, MHC class I molecules provide a readout of the expression level of up to 10,000 proteins (2). This array is interpreted by cytotoxic T lymphocytes and natural killer cells, allowing them to monitor the events inside the cell and detect infection and tumorigenesis.
MHC class I molecules consist of a complex between heavy chain, β2-microglobulin (β2m), and a peptide of usually 8–11 residues (3). Folding of MHC class I heavy chain in the endoplasmic reticulum (ER) is initially assisted by the chaperone calnexin and ERp57, a thiol-dependent oxidoreductase that assists disulfide bond formation (4). When β2m associates with class I heavy chain, calnexin dissociates and class I becomes part of a peptide loading complex comprising transporter associated with antigen processing (TAP), tapasin, calreticulin, and ERp57 (5–9). TAP transports peptides from the cytosol into the ER. Tapasin helps the assembly of class I molecules with peptides (10). Calreticulin is an ER chaperone for the folding of a wide variety of glycoproteins. How the proteins in this complex cooperate in the loading of class I with high-affinity peptide is unclear. High-affinity peptides are of a length that allows both their N and C termini to fit in the class I binding groove and have an allele-specific binding motif (11, 12).
An outstanding problem in the field of antigen presentation is whether there is catalysis of class I peptide loading, by analogy to HLA-DM, which edits the peptides bound to MHC class II according to their binding stability (13). Bulk analysis of class I does indicate impaired loading either in the absence of tapasin (5, 14, 15) or with a mutant class I that fails to associate with TAP (16). This is reflected in lower steady-state levels of class I, faster decay of class I from the cell surface, and increased export of peptide-receptive class I to the cell surface. It is unclear whether this represents class I molecules that are empty or just have a suboptimal peptide cargo. Attempts to compare the repertoire of peptides presented with and without tapasin by peptide elution have failed to detect clear qualitative differences (17–19). Here, we describe a method to sample the repertoire of peptides presented at the cell surface and use this method to evaluate the roles of tapasin, calreticulin, and ERp57 in the loading of class I with peptide.
Materials and Methods
General Reagents. Peptides were synthesized by using F-moc chemistry (Peptide Protein Research, Eastleigh, U.K.) and were >95% pure by HPLC and mass spectrometry. Serum-free media was AIM-V (Sigma).
Antibodies. 25-D1.16 (D1) recognizes H2-Kb-SIINFEKL (20), kindly provided by R. Germain (National Institutes of Health, Bethesda). Y3 recognizes a conformation-sensitive epitope of H2-Kb. 148.3 recognizes human TAP1, kindly provided by R. Tampe (University of Marburg, Marburg, Germany). Antibody to the truncated nerve growth factor receptor (ΔNGFR) was from the hybridoma 8737 (American Type Culture Collection). Monoclonal antibodies were purified from hybridoma supernatant on a protein A-Sepharose or protein G-Sepharose Fast Flow (Amersham Pharmacia) column as appropriate. Calreticulin expression was determined with the rabbit anti-Calregulin antiserum C-17 (Santa Cruz Biotechnology).
Cell Lines. Cells were grown in RPMI medium 1640 with 10% FCS (Globepharm, Surrey, U.K.), 50 units/ml penicillin, 50 μg/ml streptomycin, and 2 mM glutamine (R10) at 37°C with 5% CO2. LBL 721.220 (.220) is a human tapasin-deficient B lymphoblastoid cell line (21). .220 Kb and .220 Kb tapasin were kind gifts of J. McCluskey (University of Melbourne, Melbourne) (17). Fibroblast cell lines, from M. Michalak (University of Alberta, Edmonton, Canada), were generated from wild-type or calreticulin-knockout mice, as described (22). RMA-S is a TAP-deficient mouse T cell line.
Plasmids. The retroviral packaging plasmids CMVbipep-ΔNGFR and CMVbipep-neo have been described (23, 24). Minigene inserts were generated from primers by PCR for insertion at the SpeI and MluI sites of CMVbipep-ΔNGFR and encoded the octameric peptide preceded by methionine, which is cleaved by cytosolic N-aminopeptidase activity. Wild-type Kb or T134K Kb, a kind gift of K. Gould (Imperial College London), was subcloned from pKG into CMVbipep-neo at the XhoI and HincII restriction sites and introduced into cells with retroviral transduction. ΔNGFR-expressing cells were purified by using antibody-coated CEL-Lection Pan Mouse IgG Dynabeads (Dynal, Great Neck, NY). Neo-expressing cells were selected with 0.5 mg/ml G418. pBMN-Zpuro encoding wild-type or C95A human tapasin was a kind gift of P. Cresswell (Yale University, New Haven, CT). KpnI and BamHI sites were added to wild-type and C95A tapasin by PCR with the primers 5′-GTAGGTACCATGAAGTCCCTGTCTCTGCTC-3′ and 5′-GTAGGATCCTCACTCTGCTTTCTTCTTTG-3′ for subcloning into pREP4, a kind gift of F. Momburg (German Cancer Research Center, Heidelberg). pREP4 plasmids encoding wild-type or C95A tapasin were introduced by using Lipofectamine 2000 (Invitrogen). pREP4 transfectants were selected and maintained in 0.3 mg/ml Hygromycin B (Invitrogen). Inserts were confirmed by sequencing in both directions.
Retroviral Transduction. The retroviral packaging cell line Phoenix-Amphotropic (American Type Culture Collection) was transfected on day 1 by using Effectene (Qiagen, Valencia, CA). On day 2, cells were placed in fresh medium at 32°C. On day 3, cell medium was centrifuged at 402 × g for 5 min. Cells to be transduced were resuspended in this supernatant with 5 μg/ml polybrene (Sigma) and 25 mM Hepes and centrifuged at 1,118 × g for 2 h at 32°C. Thereafter, cells were incubated at 37°C and resuspended in fresh medium the next day.
Flow Cytometry. Suspension cells were plated overnight at 3 × 105 cells per ml. Adherent cells were used at 50–70% confluency and trypsinized on ice. Cells were immediately transferred to 4°C to minimize loss of fast-decaying peptides, washed in fluorescence-activated cell sorting (FACS) wash (PBS/2% FCS/0.02% NaN3), and incubated in a saturating concentration of 1° antibody for 40 min. Cells were washed twice and resuspended in fluorescein-labeled goat anti-mouse IgG (Sigma) or phycoerythrin-labeled goat anti-mouse IgG (Abcam) for 30 min. Cells were washed three times and analyzed on a FACScan flow cytometer (Becton Dickinson) by using cellquest software. Data were collected for a minimum of 10,000 cells, and live cells were gated by forward and side scatter.
Peptide Binding Experiments. Cells were incubated overnight at 26°C in a CO2 incubator at 0.5 × 106 cells per ml in R10. After washing twiceat26°C in serum-free RPMI medium 1640, cells were pulsed with peptide for 1 h at the same temperature. For dose–response curves, cells were prepared for flow cytometry by rapidly cooling to 4°C and incubating with 20 μg/ml antibody (25.D1 or Y3) for 45 min followed by phycoerythrin-conjugated goat-anti-mouse Ig for 30 min. To evaluate complex half-life, peptide-pulsed cells were warmed to 37°C, and aliquots were removed at various times, cooled to 4°C, and prepared for flow cytometry as above.
Immunoblotting. Cells were treated with 20 mM N-ethylmaleimide in PBS on ice for 10 min and then lysed in 0.5% Nonidet P-40 in lysis buffer (150 mM NaCl/5 mM EDTA/20 mM Tris-HCl/2mM PMSF/2 mM iodoacetic acid, pH 7.4). Postnuclear supernatants were boiled with 4× loading buffer and run on a nonreducing 8% gel. Proteins were transferred electrophoretically to Hybond-C extra membrane (Amersham Pharmacia), which was then blocked with PBS/2.5% fat-free milk for 1 h. The 1° antibody in PBS with 0.05% Tween 20 (PBST), and 2.5% fat-free milk was added for 1 h. The membrane was washed three times with PBST and incubated with horseradish peroxidase-conjugated 2° antibody for 1 h. The membrane was washed four times with PBST, developed with SuperSignal West Pico chemiluminescent substrate (Pierce), and imaged with a Fluor-S MultiImager (Bio-Rad).
Immunoprecipitation. Cells were starved in Met/Cys-free medium for 30 min and labeled with 35S-Met/Cys (Amersham Pharmacia) for 30 min before lysis with 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) in lysis buffer containing 10 μg of 148.3 (TAP1). Postnuclear supernatants were immunoprecipitated with protein A-Sepharose beads for 1 h. The beads were washed three times with 0.1% CHAPS lysis buffer and then incubated in 1% Triton X-100 lysis buffer for 30 min. The beads were spun out, and the supernatant was added to 10 μg of Y3 (Kb) and fresh beads. After 1 h, beads were washed three times in 0.1% Triton X-100 lysis buffer and boiled in 25 μl of SDS loading buffer, and supernatant was run on a 10% polyacrylamide gel. The gel was fixed, placed in Amplify (Amersham Pharmacia) for 30 min, and dried before exposure on a PhosphorImager screen (Fuji). The screen was imaged with a Fluor-S MultiImager.
Results
Generating a Peptide Hierarchy to Sample the MHC Class I Peptide Repertoire. To sample the class I peptide repertoire, we generated a single detection system with which we could analyze presentation of peptides with a wide range of binding affinities to class I. The use of a single detection system avoided the obfuscation of phenotypic differences between T cells responding to epitopes of different dominance (25). We chose flow cytometry with an MHC-peptide-specific antibody as our readout because of its quantitative signal over a wide range of presentation levels and its independence of costimulation (20). A hierarchy based on the H2-Kb-binding peptide SIINFEKL (chicken ovalbumin residues 257–264) was generated by varying anchor residues to produce peptides with a wide range of binding affinities. Peptides were expressed stably as minigenes in the cytosol, and peptide presentation was detected by flow cytometry with the antibody 25-D1.16 (D1) (20), which could recognize all of the peptide variants.
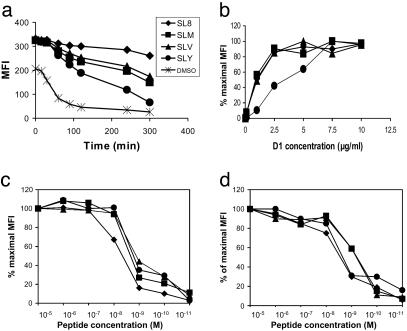
Peptides that bind to and stabilize Kb generally have an aromatic residue at P5 (Tyr or Phe) and a small hydrophobic residue at the C terminus (Leu, Met, Ile, or Val) (11, 26). To generate suboptimal variants of SIINFEKL with a reduced ability to stabilize Kb, peptides with substitutions at P5 and P8 were synthesized. The half-life for dissociation of these peptides from Kb at the cell surface was determined as a relative measure of peptide binding. TAP-deficient cells were loaded with peptide for 1 h, peptide was washed away from the medium, and the loss of Kb from the cell surface was observed as the peptide dissociated over time (Fig. 1a). SIINFEKL (variants at anchor positions 5 or 8 are indicated in boldface) had the slowest half-time for dissociation (360 min), SIINFEKV was faster (294 min), followed by SIINFEKM (270 min), then SIINYEKL, which was significantly faster (120 min). The half-life of Kb, stabilized by low temperature alone (27), was 48 min. This assay for binding therefore established the following hierarchy of peptides: SIINFEKL > SIINFEKV > SIINFEKM > SIINYEKL.
Fig. 1.
Establishing a hierarchy of peptides based on binding quality. (a) Half-life of Kb:SIINFEKL (♦), SIINFEKV (▴), SIINFEKM (▪), and SIINYEKL (•) complexes and empty molecules (×) on peptide-loaded RMA-S cells is expressed as median fluorescence intensity (MFI) after flow cytometry with Y3. The stabilization produced by the peptide at time 0 is 100%, and 0% is the stabilization in the absence of peptide. (b) Affinity of D1 for Kb–peptide complexes. .220 Kb tapasin cells endogenously expressing the SIINFEKL variants were incubated with D1 for 4 h at 4°C. After one wash in FACS wash, they were resuspended in 200 μl of phycoerythrin-labeled 2° antibody for 30 min at 4°C. After one wash in FACS wash, samples were analyzed by flow cytometry. (c) Binding of SIINFEKL derivatives to Kb at the cell surface of .220Kb measured with Mab 25.D1. (d) Binding of SIINFEKL derivatives to Kb at the cell surface of RMA-S measured with Mab Y3.
This hierarchy spanned the range from one of the highest reported affinities (28) to binding that is poorly detectable. Thus, use of this hierarchy of four peptides could allow monitoring of changes in the presentation of peptides that span a broad spectrum of quality.
To test whether the SIINFEKL variants were recognized equally well by 25.D1, we measured the half-maximal antibody concentration for staining cells that endogenously expressed the peptide variants (Fig. 1b). D1 detected all of the peptide variants. The half-maximal concentration was equivalent for SIINFEKL, SIINFEKV, and SIINFEKM (1 μg/ml) but was higher for SIINYEKL (3.5 μg/ml). All subsequent staining with D1 was performed at 20 μg/ml to ensure saturation of complexes of Kb bound to each peptide variant.
Interestingly, when we estimated the relative binding of the peptides to Kb by their ability to stabilize Kb at the cell surface of TAP-deficient cell lines, we observed little difference between them. This was true regardless of whether Kb was assembled with human (Fig. 1c) or murine (Fig. 1d) β2m.
This peptide hierarchy was introduced into cells as minigenes by using transduction with a replication-incompetent retroviral vector, which allowed up to 75% of cells to be stably expressing the minigene within 2 days. A marker gene, ΔNGFR, was driven from the same promoter as the peptide minigene, via an internal ribosome entry site. This marker gene served as a control that cells expressed equivalent amounts of each peptide and allowed transduced cells to be purified to homogeneity. Marker gene expression for the different constructs was equivalent within all hierarchies compared in this study (Table 1). The peptide FILKSINE, which lacks anchor residues for Kb, was included as a negative control for the effect of retroviral transduction.
Table 1.
Mean fluorescence intensity of staining for specific Kb:SIINXEKX complexes with Mab 25.D1, total Kb with Mab Y3, and truncated NGF receptor with anti-NGFR for .220-Tpn (tapasin+/+) and .220 (tapasin–/–) transfected with minigenes encoding SIINFEKL-related peptides
| 25.D1
|
Y3
|
Anti-NGFR
|
||||
|---|---|---|---|---|---|---|
| Peptide | .220-Tpn | .220 | .220-Tpn | .220 | .220-Tpn | .220 |
| SIINFEKL | 1,213 | 31 | 1,081 | 1,023 | 512 | 481 |
| SIINFEKV | 722 | 32 | 1,032 | 1,019 | 527 | 470 |
| SIINFEKM | 604 | 72 | 1,075 | 1,125 | 469 | 482 |
| SIINYEKL | 16 | 5 | 1,031 | 993 | 450 | 420 |
| FILKSINE | 4 | 1.6 | 1,120 | 1,113 | 522 | 438 |
The Effect of Tapasin on the Peptide Repertoire. Tapasin seems to have a key role in the loading of class I with peptide (10). In the absence of tapasin, class I is inefficiently loaded with peptide and decays rapidly from the cell surface. We tested whether our method of analyzing the peptide repertoire would detect differences in the presentation of peptides of different qualities when tapasin was absent.
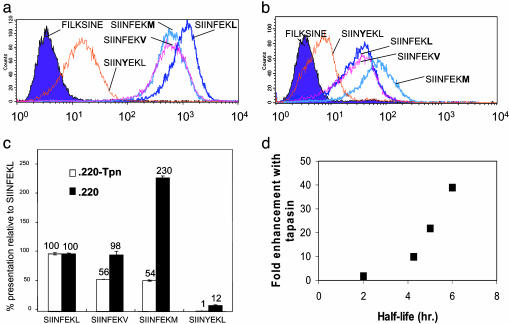
The peptide hierarchy described above was introduced into the tapasin-deficient cell line .220 Kb (Fig. 2b and Table 1) or .220 Kb transfected with tapasin (Fig. 2a and Table 1), and peptide presentation was determined by flow cytometry. All peptides were presented to a greater extent in the presence of tapasin, and their relative presentation followed the hierarchy of peptide binding: SIINFEKL > SIINFEKV > SIINFEKM > SIINYEKL. However, in the absence of tapasin, this hierarchy was lost and a new one emerged: SIINFEKM > SIINFEKL > SIINFEKV > SIINYEKL (Fig. 2c). In the absence of tapasin, presentation of the medium-affinity peptide SIINFEKM was dominant, and presentation of SIINFEKV was similar to the highest affinity variant SIINFEKL.
Fig. 2.
The effect of tapasin on presentation of the hierarchy. Expression level of the SIINFEKL variants in .220Kb-Tpn (a) and .220Kb (b) was determined by flow cytometry using Mab 25.D1. (c) Hierarchy of presentation of the SIINFEKL variants with and without tapasin, expressed as a percentage of SIINFEKL presentation: (peptide MFI – FILKSINE MFI)/(SIINFEKL MFI – FILKSINE MFI) × 100. FILKSINE MFI represents background staining. Means of duplicate measurements are shown ±1 SD. Where error bars are not seen, they are too small to be obvious. (d) Correlation between the enhancement of presentation by tapasin and peptide half-life: enhancement of presentation by tapasin, calculated as (peptide MFI – FILKSINE MFI) with tapasin/(peptide MFI – FILKSINE MFI) without tapasin. Peptide half-life is from Fig. 1a.
Despite the low stability of SIINYEKL, its presentation was clearly detectable, but in tapasin-competent cells it was >100-fold less than that of SIINFEKL. Presentation of SIINYEKL was also the lowest in the series in the absence of tapasin. However, its presentation as a fraction of SIINFEKL presentation was increased >10-fold in the absence of tapasin, supporting a role for tapasin in editing out poor-binding peptides.
Little change was observed in total Kb expression between different transfectants (Table 1), and Y3 staining of .220Kb and .220Kb-Tpn was roughly equivalent. Thus, 25.D1 staining was an indirect measurement of the fraction of Kb molecules at the cell surface occupied by a SIINXEKX-related peptide.
The enhancement of presentation of each peptide in cells expressing tapasin varied greatly depending on the identity of the peptide (Fig. 2c). Importantly, this enhancement correlated specifically with Kb:SIINXEKX half-life, which is probably influenced principally by peptide off-rate (Fig. 2d) and not with peptide affinity measured at equilibrium (Fig. 1 c and d). Tapasin only enhanced to a small extent presentation of the poor stabilizing peptide SIINYEKL. However, as the half-life increased, the enhancement increased dramatically. Thus, a small increase in half-life of a peptide, from SIINFEKV to SIINFEKL, produced a marked increase in the effect of tapasin on its presentation.
The same relationship between peptide half-life and degree of tapasin-mediated enhancement of presentation was seen when the minigenes were transfected into mouse fibroblasts lacking tapasin (a gift from L. Van Kaer, Vanderbilt University, Vanderbilt, TN) compared to equivalent transfections into wild-type C57BL/6 fibroblasts (data not shown).
The Effect of Calreticulin on the Peptide Repertoire. Calreticulin is an ER chaperone that interacts with heavy chain only when it is bound to β2m (9). In calreticulin-deficient cells, class I traffics rapidly from the ER, presents endogenous viral antigens poorly, and has 70% reduced surface expression of class I (29).
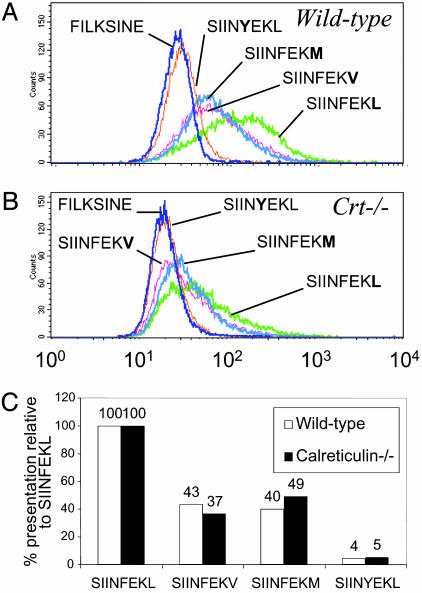
The peptide hierarchy was introduced into wild-type and calreticulin-deficient cells, and the presentation at the cell surface was determined (Fig. 3 A and B). In wild-type cells, the hierarchy followed peptide half-life: SIINFEKL > SIINFEKV > SIINFEKM > SIINYEKL. In calreticulin-knockout cells, the presentation of all of the peptides was reduced. However, unlike with tapasin, in the absence of calreticulin the presentation of each peptide was reduced by a similar amount. Thus, in the absence of calreticulin, the medium-affinity peptides SIINFEKV and SIINFEKM were still presented at approximately half the level of the high-affinity SIINFEKL. The similar peptide hierarchy in the presence and absence of calreticulin (Fig. 3C) is consistent with the similar rate of decay of bulk Kb from the cell surface in the presence or absence of calreticulin, as determined with Brefeldin A (data not shown).
Fig. 3.
The effect of calreticulin on presentation of the hierarchy. (A) Presentation of the SIINFEKL variants on wild-type fibroblasts was determined by flow cytometry with D1. (B) Presentation of the SIINFEKL variants on calreticulin-knockout fibroblasts was determined by flow cytometry with D1. (C) Hierarchy of presentation with and without calreticulin, expressed as a percentage of SIINFEKL presentation.
The expression of mouse or human β2m does not change peptide presentation significantly, as indicated by (i) the similar peptide hierarchy in wild-type mouse fibroblasts (Fig. 3C) and .220 Kb-Tpn (Fig. 2c) and (ii) the same rate of decay of an individual peptide, SIINFEKM, from the surface of these two cell types (data not shown).
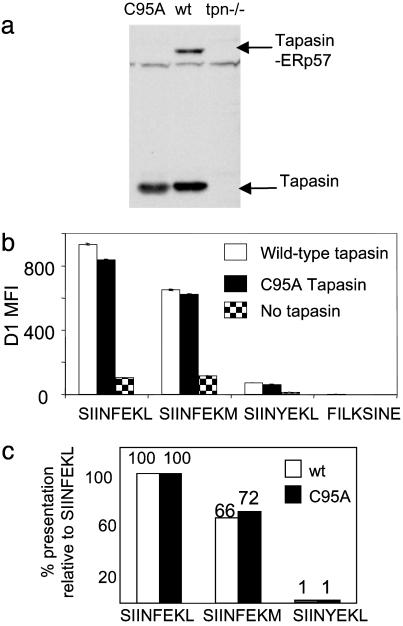
The Effect of ERp57 on the Peptide Repertoire. Class I is believed to assemble with a high-affinity peptide cargo in a complex with TAP, tapasin, calreticulin, and ERp57. Whereas the roles of TAP, tapasin, and calreticulin have been tested by knocking out the respective genes (14, 15, 29, 30), there is currently no system where ERp57 expression has been down-regulated. ERp57 has been recently shown to form a disulfide bond with tapasin, whose formation is blocked by the C95A point mutation in tapasin (31). Dick et al. showed that C95A tapasin allows apparently normal assembly of heavy chain-β2m with TAP, tapasin, and calreticulin, but ERp57 is not recruited to this complex. We analyzed the peptide repertoire of cells expressing the C95A mutant tapasin, which lack ERp57 function in the peptide loading complex.
By immunoblotting for the tapasin-ERp57 covalent intermediate, we first confirmed that the C95A mutation stopped formation of a disulfide to ERp57 in our cells (Fig. 4a). Surprisingly, C95A tapasin restored the hierarchy based on peptide affinity that was lost in the absence of tapasin (Fig. 4 b and c). The degree of enhancement of presentation of each peptide variant by C95A tapasin was comparable to the enhancement by wild-type tapasin. Thus, tapasin in the absence of ERp57 was still able to selectively enhance the presentation of high-affinity peptides. The lack of an effect on the peptide hierarchy was supported by a similar rate of decay of Kb from the cell surface with wild-type or C95A tapasin (data not shown).
Fig. 4.
The effect of recruiting ERp57 to the TAP on presentation of the hierarchy. (a) .220Kb(tpn–/–) was transfected with wild-type (WT) or C95A mutant tapasin, and aliquots of cells were incubated with N-ethylmaleimide. Lysates were fractionated by SDS/PAGE and blotted with antitapasin. A band corresponding to disulfide-bonded heterodimers of ERp57 and tapasin was only seen in cells expressing WT tapasin. (b) Peptide presentation on .220Kb, .220Kb-Tpn, or .220Kb C95A tapasin was determined by flow cytometry with D1, and the MFI is displayed. Data for SIINFEKV were not obtained. (c) Hierarchy of presentation with WT or C95A mutant tapasin, expressed as a percentage of SIINFEKL presentation.
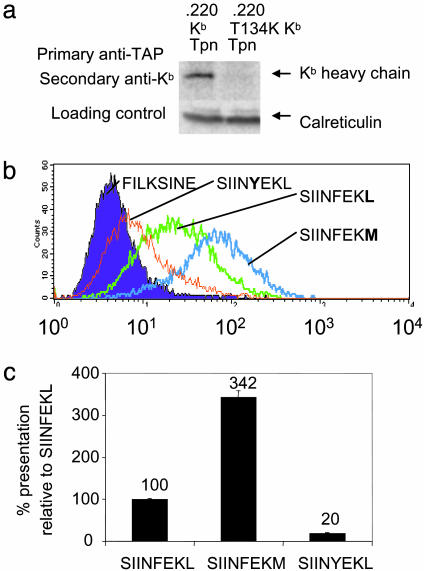
The Peptide Hierarchy for a Mutant Class I That Fails to Associate with TAP. Having established that tapasin modulates the peptide repertoire, we tested whether that was a result of tapasin's effect on TAP or a direct effect on loading of class I with peptide. Thus, we introduced the hierarchy into cells which did express tapasin (.220 tapasin) but expressed a mutant form of class I that was unable to associate with the peptide loading complex. The T134K mutation in HLA-A2 stops class I interacting with the peptide loading complex, and this leads to fast exit of poorly loaded complexes from the ER (16). The interaction between TAP and class I was also disrupted by the T134K mutation in Kb (Fig. 5a). When the peptide series was introduced into .220-Tpn-T134K Kb, a hierarchy based on peptide affinity was not observed (Fig. 5b). In fact, the pattern in these cells was similar to that observed for .220 Kb (Fig. 2b), with highest presentation of the medium-affinity SIINFEKM andahigh percentage of presentation of the low-affinity SIINYEKL relative to SIINFEKL (Fig. 5c). This reinforces the idea that the effect of tapasin on the hierarchy depends on class I directly interacting with the peptide loading complex and is consistent with an editing function for tapasin.
Fig. 5.
The effect of the T134K mutation on presentation of the hierarchy. (a) The effect of the T134K mutation in Kb on TAP association. 35S-Met/Cys-labeled anti-TAP coimmunoprecipitates (148.3) from .220 Kb tapasin or .220 T134K Kb tapasin cells were released by Triton X-100 and reprecipitated with Y3 (Upper). An aliquot of postnuclear supernatant was immunoblotted for calreticulin as a control for equal cell number (Lower). (b) Level of presentation of the SIINFEKL variants with a T134K mutation in Kb. Peptide presentation on .220 T134K Kb tapasin was determined by flow cytometry with D1. SIINFEKV presentation was not determined. (c) Hierarchy of presentation of the SIINFEKL variants with a T134K mutation in Kb, expressed as a % of SIINFEKL presentation. Means of duplicate measurements are shown ±1 SD.
Discussion
We have established a hierarchy of peptides spanning a wide range of half-life that we used as indicators of the repertoire of peptides presented by class I. In tapasin-competent cells, a hierarchy was obtained with greatest presentation of the most stably bound peptide and reduced presentation of peptide variants with shorter half-lives. However, in tapasin-deficient cells, this hierarchy disappeared, and presentation was greatest for a peptide with an intermediate half-life. Because we could detect very little difference in binding of each of the peptides to Kb when measured close to equilibrium conditions, we conclude that the enhancing effect that tapasin has on peptide presentation correlates specifically with complex half-life (which is probably influenced principally by peptide off-rate) and not with peptide affinity per se. These data are relevant to a recent report by Zarling et al. (32), who observed no difference in the average affinity of peptides eluted from HLA-B8 assembled in normal versus tapasin-incompetent cells, despite a significant difference in the cell-surface half-life of B8 in these two cells and argued that their data support a peptide–class I complex-stabilizing function for tapasin. Contrary to this report, we have found the same rate of decay of an endogenously synthesized individual peptide, SIINFEKM, from the surface of these two cell-types (data not shown). The present data show that peptide affinity measurements and cell-surface half-life measurements are not necessarily concordant; therefore, the repertoire of peptides eluted from HLA-B8 assembled in .220 by Zarling et al. may in fact have a shorter half-life than the repertoire eluted from B8 made in .220-Tpn; hence, differences in repertoire may still have arisen as a result of tapasin-mediated peptide editing.
Analyzing tapasin-deficient cells alone is insufficient to resolve the importance of tapasin because, in the absence of tapasin, class I generally fails to interact with ERp57, TAP, and calreticulin (9). However, here we have also explored the peptide hierarchy in calreticulin-deficient cells and cells where ERp57 is not recruited to TAP. The hierarchy of peptides is not greatly changed by the absence of calreticulin, which suggests that it is the interaction between class I and tapasin that is essential for changing the peptide repertoire, and this interaction can still occur in the absence of calreticulin (29). The decrease in surface expression levels in the absence of calreticulin is more likely to reflect increased export in calreticulin-deficient cells of class I free of peptide. Failure to recruit ERp57 to TAP also has little effect on the extent or hierarchy of peptide loading for Kb, pointing to the preeminence of tapasin in the action of the peptide loading complex to optimize peptide loading. Although we do not exclude the possibility that ERp57 molecules not bound to tapasin might have a function in modulating the peptide repertoire, we show that recruitment of ERp57 to TAP is not essential for normal loading of Kb with peptide.
This study provides a clear indication of a reshaping of the peptide repertoire in the presence of tapasin and shows that the presentation of peptides at the cell surface, which relates to the level of T cell priming, depends on the degree of optimization of class I-bound peptides in the ER.
Having established that tapasin is the key chaperone in control of peptide loading quality, we must ask how tapasin carries out its quantitative and qualitative effects. The first critical effect is on TAP function. Tapasin both stabilizes TAP expression and bridges class I to TAP (10). Enhancing the TAP level would explain why all of the peptides are presented better in the presence of tapasin, but not why presentation of high-affinity peptides was preferentially enhanced by tapasin. Tapasin expression does not change the peptide selectivity of TAP (10, 33). Also, when we expressed, in the presence of tapasin, a form of Kb that did not interact with the peptide loading complex, the hierarchy was equivalent to wild-type Kb in the absence of tapasin.
A second suggested role of tapasin is to retain poorly loaded class I molecules in the ER. However, retention is not tapasin's principal mode of action because tapasin has no effect on retention of some alleles but still improves their peptide loading (34). Also, retaining Kb from tapasin-deficient cells in the ER with Brefeldin A for 4 h and then allowing Kb to traffic to the cell surface did not restore the peptide hierarchy found in the presence of tapasin (data not shown), confirming that retention does not explain the effects of tapasin that we observed.
A third role of tapasin is as a catalyst for peptide loading (a loadase function). Key evidence for this comes from work on H2-M3 and HLA-G, where in the absence of tapasin, even when high levels of peptide are supplied, peptide loading is inefficient (35–37). Our results indicate that the loadase activity is greater for optimal peptides, i.e., those that enjoy a long half-life at the cell surface and are therefore most likely to be immunodominant (38). Low-affinity peptides such as SIINYEKL may bind to class I, but tapasin may stabilize a conformation of class I from which the dissociation of suboptimal peptides is enhanced relative to that of suboptimal sequences (39). Overall, the analogy between the loading function of tapasin and HLA-DM is supported by our study and echoes the observation that HLA-DM can reduce presentation by class II of cryptic epitopes and enhance presentation of immunodominant epitopes (40).
The great majority of studies of class I antigen presentation have analyzed high-affinity peptides, so the study here of the presentation of SIINYEKL is of importance. Detection of such low-affinity peptides is vital because of their significance as tumor antigens and in autoimmune disease, as they are less likely to induce negative selection of responding T cells (41). The bearing of our results on immunodominance should also be considered, as the hierarchy of binding strength of the peptides used in this study also represents a hierarchy of immunogenicity (42). Our results indicate that changes in tapasin expression, as occurs with certain viruses (43, 44) and tumors (45), could change dominance hierarchies and thus help determine the success or failure of the immune response.
Acknowledgments
We thank A. Kontouli for excellent technical assistance. This work was supported by the Medical Research Council (M.H.) and the Wellcome Trust (T.E.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β2m, β2-microglobulin; ER, endoplasmic reticulum; TAP, transporter associated with antigen processing; ΔNGFR, truncated nerve growth factor receptor; MFI, median fluorescence intensity.
References
- 1.Shastri, N., Schwab, S. & Serwold, T. (2002) Annu. Rev. Immunol. 20, 463–493. [DOI] [PubMed] [Google Scholar]
- 2.Engelhard, V. H., Brickner, A. G. & Zarling, A. L. (2002) Mol. Immunol. 39, 127–137. [DOI] [PubMed] [Google Scholar]
- 3.Madden, D. R. (1995) Annu. Rev. Immunol. 13, 587–622. [DOI] [PubMed] [Google Scholar]
- 4.Williams, A., Peh, C. A. & Elliott, T. (2002) Tissue Antigens 59, 3–17. [DOI] [PubMed] [Google Scholar]
- 5.Ortmann, B., Copeman, J., Lehner, P. J., Sadasivan, B., Herberg, J. A., Grandea, A. G., Riddell, S. R., Tampe, R., Spies, T., Trowsdale, J. & Cresswell, P. (1997) Science 277, 1306–1309. [DOI] [PubMed] [Google Scholar]
- 6.Hughes, E. A. & Cresswell, P. (1998) Curr. Biol. 8, 709–712. [DOI] [PubMed] [Google Scholar]
- 7.Lindquist, J. A., Jensen, O. N., Mann, M. & Hammerling, G. J. (1998) EMBO J. 17, 2186–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrice, N. A. & Powis, S. J. (1998) Curr. Biol. 8, 713–716. [DOI] [PubMed] [Google Scholar]
- 9.Sadasivan, B., Lehner, P. J., Ortmann, B., Spies, T. & Cresswell, P. (1996) Immunity 5, 103–114. [DOI] [PubMed] [Google Scholar]
- 10.Momburg, F. & Tan, P. (2002) Mol. Immunol. 39, 217–233. [DOI] [PubMed] [Google Scholar]
- 11.Falk, K., Rotzschke, O., Stevanovic, S., Jung, G. & Rammensee, H. G. (1991) Nature 351, 290–296. [DOI] [PubMed] [Google Scholar]
- 12.Cerundolo, V., Elliott, T., Elvin, J., Bastin, J., Rammensee, H. G. & Townsend, A. (1991) Eur. J. Immunol. 21, 2069–2075. [DOI] [PubMed] [Google Scholar]
- 13.Brocke, P., Garbi, N., Momburg, F. & Hammerling, G. J. (2002) Curr. Opin. Immunol. 14, 22–29. [DOI] [PubMed] [Google Scholar]
- 14.Grandea, A. G., III, Golovina, T. N., Hamilton, S. E., Sriram, V., Spies, T., Brutkiewicz, R. R., Harty, J. T., Eisenlohr, L. C. & Van Kaer, L. (2000) Immunity 13, 213–222. [DOI] [PubMed] [Google Scholar]
- 15.Garbi, N., Tan, P., Diehl, A. D., Chambers, B. J., Ljunggren, H. G., Momburg, F. & Hammerling, G. J. (2000) Nat. Immunol. 1, 234–238. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, J. W., Neisig, A., Neefjes, J. & Elliott, T. (1996) Curr. Biol. 6, 873–883. [DOI] [PubMed] [Google Scholar]
- 17.Barnden, M. J., Purcell, A. W., Gorman, J. J. & McCluskey, J. (2000) J. Immunol. 165, 322–330. [DOI] [PubMed] [Google Scholar]
- 18.Tan, P., Kropshofer, H., Mandelboim, O., Bulbuc, N., Hammerling, G. J. & Momburg, F. (2002) J. Immunol. 168, 1950–1960. [DOI] [PubMed] [Google Scholar]
- 19.Barber, L. D., Howarth, M., Bowness, P. & Elliott, T. (2001) Tissue Antigens 58, 363–368. [DOI] [PubMed] [Google Scholar]
- 20.Porgador, A., Yewdell, J. W., Deng, Y., Bennink, J. R. & Germain, R. N. (1997) Immunity 6, 715–726. [DOI] [PubMed] [Google Scholar]
- 21.Greenwood, R., Shimizu, Y., Sekhon, G. S. & DeMars, R. (1994) J. Immunol. 153, 5525–5536. [PubMed] [Google Scholar]
- 22.Mesaeli, N., Nakamura, K., Zvaritch, E., Dickie, P., Dziak, E., Krause, K. H., Opas, M., MacLennan, D. H. & Michalak, M. (1999) J. Cell Biol. 144, 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolstrup, A. B., Duch, M., Dalum, I., Pedersen, F. S. & Mouritsen, S. (2001) Gene 263, 77–84. [DOI] [PubMed] [Google Scholar]
- 24.Duch, M., Tolstrup, A., Dalum, I., Jespersen, T., Mouritsen, S. & Pedersen, F. S. (1999) BioTechniques 26, 1032–1036. [DOI] [PubMed] [Google Scholar]
- 25.Spencer, J. V. & Braciale, T. J. (2000) J. Exp. Med. 191, 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udaka, K., Wiesmuller, K. H., Kienle, S., Jung, G., Tamamura, H., Yamagishi, H., Okumura, K., Walden, P., Suto, T. & Kawasaki, T. (2000) Immunogenetics 51, 816–828. [DOI] [PubMed] [Google Scholar]
- 27.Ljunggren, H. G., Stam, N. J., Ohlen, C., Neefjes, J. J., Hoglund, P., Heemels, M. T., Bastin, J., Schumacher, T. N., Townsend, A., Karre, K., et al. (1990) Nature 346, 476–480. [DOI] [PubMed] [Google Scholar]
- 28.Kageyama, S., Tsomides, T. J., Sykulev, Y. & Eisen, H. N. (1995) J. Immunol. 154, 567–576. [PubMed] [Google Scholar]
- 29.Gao, B., Adhikari, R., Howarth, M., Nakamura, K., Gold, M. C., Hill, A. B., Knee, R., Michalak, M. & Elliott, T. (2002) Immunity 16, 99–109. [DOI] [PubMed] [Google Scholar]
- 30.Van Kaer, L., Ashton-Rickardt, P. G., Ploegh, H. L. & Tonegawa, S. (1992) Cell 71, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 31.Dick, T. P., Bangia, N., Peaper, D. R. & Cresswell, P. (2002) Immunity 16, 87–98. [DOI] [PubMed] [Google Scholar]
- 32.Zarling, A. L., Luckey, C. J., Marto, J. A., White, F. M., Brame, C. J., Evans, A. M., Lehner, P. J., Cresswell, P., Shabanowitz, J., Hunt, D. F. & Engelhard, V. H. (2003) J. Immunol. 171, 5287–5295. [DOI] [PubMed] [Google Scholar]
- 33.Bangia, N., Lehner, P. J., Hughes, E. A., Surman, M. & Cresswell, P. (1999) Eur. J. Immunol. 29, 1858–1870. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, J. W., Sewell, A., Price, D. & Elliott, T. (1998) Eur. J. Immunol. 28, 3214–3220. [DOI] [PubMed] [Google Scholar]
- 35.Park, B. & Ahn, K. (2003) J. Biol. Chem. 278, 14337–14345. [DOI] [PubMed] [Google Scholar]
- 36.Lybarger, L., Yu, Y. Y., Chun, T., Wang, C. R., Grandea, A. G., III, Van Kaer, L. & Hansen, T. H. (2001) J. Immunol. 167, 2097–2105. [DOI] [PubMed] [Google Scholar]
- 37.Chun, T., Grandea, A. G., III, Lybarger, L., Forman, J., Van Kaer, L. & Wang, C. R. (2001) J. Immunol. 167, 1507–1514. [DOI] [PubMed] [Google Scholar]
- 38.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17, 51–88. [DOI] [PubMed] [Google Scholar]
- 39.Springer, S., Doring, K., Skipper, J. C., Townsend, A. R. & Cerundolo, V. (1998) Biochemistry 37, 3001–3012. [DOI] [PubMed] [Google Scholar]
- 40.Nanda, N. K. & Sant, A. J. (2000) J. Exp. Med. 192, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fairchild, P. J. & Wraith, D. C. (1996) Immunol. Today 17, 80–85. [DOI] [PubMed] [Google Scholar]
- 42.Lipford, G. B., Bauer, S., Wagner, H. & Heeg, K. (1995) Vaccine 13, 313–320. [DOI] [PubMed] [Google Scholar]
- 43.Bennett, E. M., Bennink, J. R., Yewdell, J. W. & Brodsky, F. M. (1999) J. Immunol. 162, 5049–5052. [PubMed] [Google Scholar]
- 44.Vertegaal, A. C., Kuiperij, H. B., Houweling, A., Verlaan, M., van der Eb, A. J. & Zantema, A. (2003) J. Biol. Chem. 278, 139–146. [DOI] [PubMed] [Google Scholar]
- 45.Dissemond, J., Kothen, T., Mors, J., Weimann, T. K., Lindeke, A., Goos, M. & Wagner, S. N. (2003) Arch. Dermatol. Res. 295, 43–49. [DOI] [PubMed] [Google Scholar]