Abstract
FtsZ, the ancestral homolog of eukaryotic tubulins, is a GTPase that assembles into a cytokinetic ring structure essential for cell division in prokaryotic cells. Similar to tubulin, purified FtsZ polymerizes into dynamic protofilaments in the presence of GTP; polymer assembly is accompanied by GTP hydrolysis. We used a high-throughput protein-based chemical screen to identify small molecules that target assembly-dependent GTPase activity of FtsZ. Here, we report the identification of five structurally diverse compounds, named Zantrins, which inhibit FtsZ GTPase either by destabilizing the FtsZ protofilaments or by inducing filament hyperstability through increased lateral association. These two classes of FtsZ inhibitors are reminiscent of the antitubulin drugs colchicine and Taxol, respectively. We also show that Zantrins perturb FtsZ ring assembly in Escherichia coli cells and cause lethality to a variety of bacteria in broth cultures, indicating that FtsZ antagonists may serve as chemical leads for the development of new broad-spectrum antibacterial agents. Our results illustrate the utility of small-molecule chemical probes to study FtsZ polymerization dynamics and the feasibility of FtsZ as a novel therapeutic target.
Bacterial cells divide by binary fission, a process that involves invagination of the cell envelope at the division site to form a septum. The earliest known step in bacterial cytokinesis is the assembly of the essential division protein FtsZ into the contractile Z ring on the inner face of the cytoplasmic membrane at the site of division (1–3). Another 12 proteins in Escherichia coli then congregate at the Z ring in a sequentially dependent pathway, causing assembly of the septal ring that guides circumferential septum synthesis (2–5). The septal ring constricts in concert with septal progression, culminating in complete disassembly once the septum matures. FtsZ is a structural homolog of eukaryotic tubulin (6) and, similar to tubulin, it is a GTPase (7–9) that polymerizes in a GTP-regulated manner into dynamic structures that resemble microtubule protofilaments (PFs) (10–16). GTP-dependent FtsZ assembly is coupled to GTP hydrolysis (12, 14, 16, 17). Hydrolysis destabilizes the polymer, promoting disassembly into individual subunits. Once all of the GTP is converted to GDP, the polymer system reverts back to monomers (11, 12, 16).
In eukaryotic pathogens as well as in higher eukaryotic cells, cell division has been a productive area for finding drugs that combat infection or uncontrolled cell proliferation (18, 19). The majority of such drugs have been found to target microtubules (18). However, to date, the essential and conserved components of the bacterial cell division machinery have remained largely unexploited for therapeutic purposes. Because of the essential role of FtsZ in prokaryotic cell division, its widespread conservation in the Bacterial kingdom, its absence in the mitochondria of higher eukaryotes, its evolutionary distance from tubulin, and its known biochemical activity and atomic structure, we and others (20, 21) consider FtsZ an attractive target to develop agents that may cause selective toxicity to bacterial pathogens. The goal to develop novel antibacterial countermeasures gains immediacy in the context of the global menace of multiple antibiotic resistance, emerging novel pathogens, and the potential threat of virulent or engineered organisms in the environment. Chemical inhibitors against FtsZ from Mycobacterium tuberculosis (20) and E. coli (21) have been reported recently. However, these molecules did not exhibit high affinity, their mechanisms of action were not described, and it was not proven whether they perturbed FtsZ assembly in vivo. Thus, much remains to be done to exploit FtsZ as a new target.
To this end, we screened chemical libraries against FtsZ GTPase in vitro and identified five small molecules, structurally dissimilar to GTP and to each other, that inhibit the polymerization-GTP hydrolysis-depolymerization cycle of FtsZ through distinct mechanisms. One class destabilizes FtsZ polymers, whereas the other class induces pairing or bundling of the PFs. Furthermore, the FtsZ inhibitors perturb Z ring assembly in E. coli cells and kill a variety of bacteria in broth cultures, indicating that they may be useful as chemical probes to study FtsZ dynamics and as chemical scaffolds to develop new therapeutic agents.
Materials and Methods
FtsZ Expression and Purification. Untagged E. coli FtsZ (FtsZEc) was purified by using previously published methods (7, 15, 22). M. tuberculosis ftsZ was PCR-amplified from the cosmid clone Y270 (a kind gift of K. Eiglmeier and S. T. Cole, both from Institut Pasteur, Paris) and cloned into pET-16b (Novagen) to obtain pRM4, which yielded soluble His6-FtsZ. Further details of FtsZ expression and purification are described in Supporting Text, which is published as supporting information on the PNAS web site.
High-Throughput Screen. A real-time assembly-dependent FtsZ GTPase assay was developed by using coupled enzymes pyruvate kinase and lactate dehydrogenase in the presence of phosphoenolpyruvate (PEP) and NADH (ref. 23; see Results). The reaction mixture contained 1 mM PEP, 200 μM NADH, 68 units/ml pyruvate kinase, 68 units/ml l-lactate dehydrogenase, 0.1% Tween 20, and 2 μM FtsZEc in assembly buffer E (50 mM 4-morpholinepropanesulfonic acid, pH 6.5/50 mM KCl/5 mM MgCl2). A 25-μl reaction mixture was dispensed per well of 384-well plates (Labsystems, Chicago), with one column designated for controls. One-hundred-nanoliter library compounds (in DMSO) were robotically pin-transferred from 384-well library plates to the assay plates such that the final concentrations ranged between 20 and 50 μM, depending on the molecular weights of the compounds. Control wells received either 0.3% DMSO or AlF–4, which was previously shown to inhibit FtsZ GTPase (24). The reactions were initiated at ≈28°C by adding 1 mM GTP to the wells and monitored over 25–40 min by the decrease in fluorescence emission as NADH underwent oxidation (355-nm excitation, 460-nm emission). Two small-molecule libraries (16,320-molecule DiverSet E, ChemBridge, San Diego; 2,000-molecule Diversity Set, National Cancer Institute, Bethesda) were screened against FtsZEc by using this assay.
GTPase Assays. FtsZ GTPase activity was measured by using either a radioactive charcoal-based assay (25) or the nonradioactive malachite green-phosphomolybdate assay (26). The assays are described in Supporting Text.
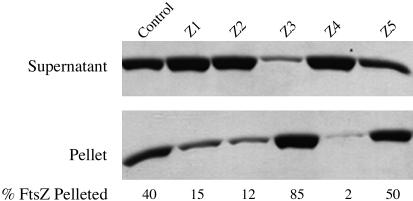
Sedimentation Assay. Sedimentation analysis of FtsZ polymers was performed as described (22), and the conditions are in the legend to Fig. 4.
Fig. 4.
Effects of Zantrins on steady-state FtsZ polymer mass and structure. Ten micrograms of FtsZEc was preincubated with Zantrins or with 1% DMSO in 50-μl buffer E for 3 min, before assembly was initiated with 1 mM GTP. After a 5-min incubation at 25°C to establish the steady-state, the polymers were pelleted by using a TLA 100.2 rotor (Beckman Coulter) (80,000 rpm, 12 min, 25°C). The pellets were dissolved in 50 μl of buffer E containing 0.01% SDS. Twenty microliters each of the supernatant and pellet fractions were analyzed by 15% SDS/PAGE, the FtsZ bands were visualized by Coomassie staining, and the relative amounts of FtsZ in the bands were quantified by using theimagequant Ver. 1.2 software (Amersham Biosciences). Zantrins were used at 2× IC50:10 μM Z1, 20 μM Z2, 30 μM Z3, 50 μM Z4, and 10 μM Z5.
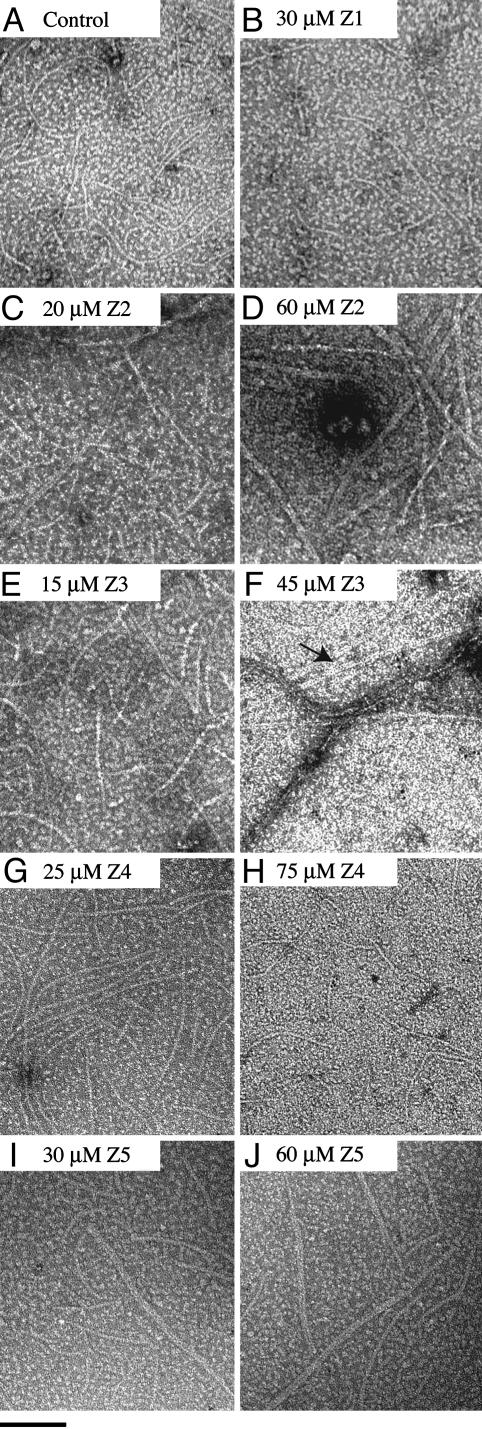
Electron Microscopy. FtsZEc protein stocks were precleared by centrifugation (350,000 × g, 15 min, 4°C) and diluted in buffer E. Alternatively, FtsZ was put through a cycle of calcium-aided assembly and disassembly to select for active nonaggregated protein (15). FtsZ assembly reactions and electron microscopy of the FtsZ polymers are described in the legend to Fig. 3.
Fig. 3.
Effects of Zantrins on the ultrastructure of FtsZ polymers. Assembly reactions containing 2 μM FtsZEc in buffer E were preincubated with varying concentrations of Zantrins or with 5% DMSO for 3 min, before adding 1 mM GTP (pH ≈7.0). After 2 min at 25°C, 10-μl aliquots were applied to glow-discharged carbon-coated copper grids (400 mesh size), which were stained with 0.75% uranyl formate. The filaments were photographed by using a Philips 401 (Eindhoven, The Netherlands) electron microscope at ×50,000. The arrow in F indicates clustered doublets of FtsZ PFs. An adjacent FtsZ bundle is also visible. (Bar = 100 nm.)
Immunofluorescence Microscopy. Immunofluorescence microscopy of DRC39 cells to visualize Z rings (22) is described in the legend to Fig. 5. DRC39 was obtained by P1vir transduction of ΔacrAB::Tn903kanr null mutation from JZM120 (27) into E. coli MC1000.
Fig. 5.
Effects of Zantrins on Z ring assembly in E. coli. Log-phase cultures of DRC39 (≈5–7 × 107 cells per ml) were treated with Zantrins (0.5× MIC or MIC) or with 0.66% DMSO. Culture aliquots were withdrawn at 30- or 60-min intervals, fixed with 80% methanol, and immunostained with polyclonal anti-FtsZ rabbit antibody, followed by FITC-conjugated goat anti-rabbit secondary antibody (The Jackson Laboratory). Nucleoids were visualized by treating the immunostained cells with 200 ng/ml 4′, 6-diamidino-2-phenylindole (DAPI). Z rings are shown in green, and the DAPI-stained nucleoids have been pseudocolored red. Shown are control cells treated with DMSO for 30 (a and a′) and 60 (b and b′) min, with 10 μM Z1 for 30 min (c and c′), with 2.5 μM Z2 for 60 min (d and d′), with 5 μM Z2 for 30 min (e), with 10 μM Z3 for 30 min (f), and with 40 μM Z5 for 60 min (g). The white arrowhead in c indicates a cell with polar FtsZ foci, whereas the gray arrowheads in g indicate cells in which Z rings are visible over background. (Bar = 4.5 μm.)
Determination of Minimum Inhibitory Concentration (MIC). MICs were determined by using a standard 2-fold serial dilution format. Bacterial growth was assessed visually and the lowest inhibitor concentration that caused >99% growth inhibition was designated the MIC. Bacterial culture conditions for MIC determination have been described in Supporting Text.
Results
Screening for FtsZ Antagonists. To develop a nonradioactive real-time high-throughput assay, we took advantage of the classic enzyme-coupled reactions involving pyruvate kinase and lactate dehydrogenase (23). Pyruvate kinase converts GDP/ADP (products of GTPase/ATPase) to GTP/ATP in the presence of phosphoenolpyruvate (reaction 1). Pyruvate formed in reaction 1 is then reduced to lactate by l-lactate dehydrogenase in the presence of NADH (reaction 2). Thus, for each mol of GDP converted to GTP, 1 mol of NADH is oxidized to NAD+. Because FtsZ undergoes facile GDP exchange with free GTP in solution (9, 28), we could rapidly capture GDP associated with FtsZ using the enzyme-coupled reactions. This allowed a real-time FtsZ GTPase assay to be developed by monitoring the decrease in fluorescence emission upon NADH oxidation as the read-out for GTP hydrolysis.
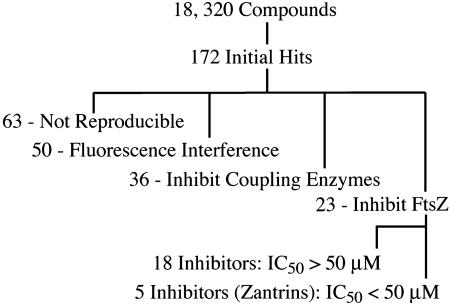
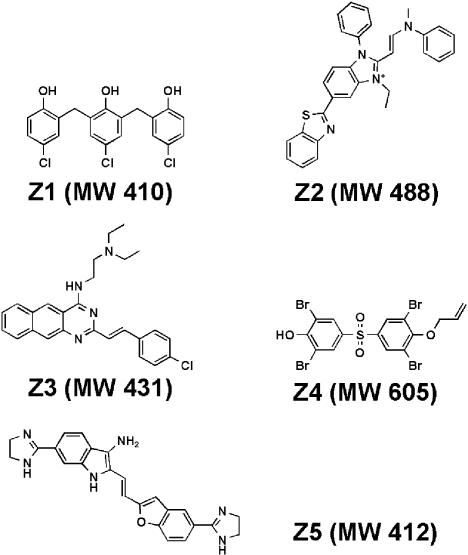
Of 18,320 compounds screened, 172 molecules were identified that apparently caused ≈50% inhibition or 2-fold activation of NADH oxidation (Fig. 1). To determine whether the observed inhibition or activation was due to an effect on FtsZ or the coupling enzymes, the small molecules were retested in a coupled assay where the reactions were driven either with FtsZ and GTP or with GDP alone. Of 172 initial hits identified in the primary screen, 63 compounds failed to reproduce the initial effects, 36 compounds inhibited the coupling enzymes (≥40% inhibition), and 50 compounds interfered with the fluorescent read-out by causing the fluorescence signal to increase in the absence of FtsZ (Fig. 1). The remaining 23 compounds that inhibited NADH oxidation, at least 30% upon retesting, were assayed directly against FtsZ GTPase using a charcoal-based radioactive assay. We identified five compounds that inhibited E. coli FtsZ GTPase activity ≈50% at concentrations <50 μM, whereas the remaining 18 compounds showed a similar level of inhibition at concentrations between 50 and 100 μM (Fig. 1). We chose five FtsZ inhibitors showing higher potency for further study and named them Zantrins (FtsZ guanosine triphosphatase inhibitors). The chemical structures and the molecular weights of Zantrins are shown in Fig. 2.
Fig. 1.
High-throughput screen summary. A real-time, enzyme-coupled, fluorescent assay in the 384-well plate format was used for screening chemical libraries against FtsZEc. A total of 18,320 compounds were screened at 20–50 μM against 2 μM FtsZ.
Fig. 2.
Chemical structures of Zantrins. Molecular weights (MW) are shown within parentheses.
Potency of Zantrins in Vitro. The IC50 (50% inhibitory concentration) of each Zantrin against FtsZ GTPase activity was determined by using the malachite green-phosphomolybdate colorimetric assay that measures inorganic phosphate released upon GTP hydrolysis. For dose–response experiments, we used 2 μM FtsZEc, a concentration that allows robust FtsZ assembly (Fig. 3 and ref. 15) and found that the IC50 values of Zantrins ranged between 4 and 25 μM (Table 1). We also tested Zantrins against FtsZ from M. tuberculosis (FtsZMt) to examine whether the small molecule inhibitors identified against FtsZEc could inhibit a diverged FtsZ ortholog from a clinically important bacterial pathogen. FtsZMt shares ≈46% amino acid identity with FtsZEc and has been shown to be a markedly slower GTPase in vitro (more than three orders of magnitude lower, Vmax) (14). We chose 12.5 μM FtsZMt to monitor the effects of Zantrins on GTP hydrolysis over a 2-h period. Remarkably, the majority of Zantrins inhibited FtsZMt GTPase with IC50 values up to one order of magnitude higher than the corresponding values against FtsZEc (Table 1). Given the dissimilarities between the two FtsZ orthologs, inhibition of FtsZMt by Zantrins in the IC50 range 30–70 μM is significant and suggests that future optimized FtsZ inhibitors might display a broad spectrum of antibacterial activity.
Table 1. IC50 values of Zantrins against FtsZEc and FtsZMt GTPases.
| IC50, μM
|
||
|---|---|---|
| Zantrins | FtsZEc | FtsZMt |
| Z1 | 5 | 55 |
| Z2 | 10 | 70 |
| Z3 | 15 | 50 |
| Z4 | 25 | >100 |
| Z5 | 4 | 30 |
FtsZ was preincubated with varying concentrations of Zantrins or with 3% DMSO in buffer E for 5 min at 30°C, before adding 1 mM GTP to the reactions. Reaction kinetics was followed by measuring inorganic phosphate released upon GTP hydrolysis. The IC50 values were extrapolated from log dose—response curves and are averages from three independent determinations.
Zantrins Perturb FtsZ Assembly Through Distinct Mechanisms. To assess directly the effects of Zantrins on GTP-dependent assembly of FtsZEc, we used negative-stain electron microscopy to visualize the polymers. Two distinct effects were observed: Zantrins either destabilized FtsZ assembly causing a diminution in PF length and abundance, or they induced PF pairing or, less frequently, PF bundling (Fig. 3), thereby increasing the amount of steady-state polymer mass (Fig. 4). The images of paired or bundled filaments together with enhanced polymer levels suggested that some of the Zantrins stabilized FtsZ polymers, although the relationship between PF pairing and stabilization turned out to be complex (see below).
In the presence of GTP and a pH 6.5 buffer, 2 μM FtsZEc underwent assembly into predominantly single PFs, 5–6 nm wide (Fig. 3A), similar to that observed previously by several laboratories (11, 12, 15, 22). It should be noted that at a higher FtsZ concentration, such as >5–20 μM, a significant percentage of PFs undergoes pairing (15, 16). In this report, assembly experiments were carried out with 2 μM FtsZEc. Zantrins Z1 (Fig. 3B) and Z4 (Fig. 3 G and H) caused a dose-dependent decrease in both the number and length of FtsZ PFs. Because a few PFs persisted in the presence of 30 μM Z1 and 75 μM Z4, we favor the possibility that these inhibitors act by destabilizing the longitudinal bonds between FtsZ subunits or by blocking the growing ends of the PFs, rather than by poisoning the FtsZ monomers to prevent de novo assembly.
In contrast to Z1 and Z4, the other three Zantrins appeared to stabilize FtsZ assembly. Z2 caused a dose-dependent increase in the number of PF doublets (Fig. 3 C and D). Similarly, as the concentration of Z3 was raised from 15 μM (Fig. 3E) to 45 μM (Fig. 3F), FtsZ PFs laterally associated into two-stranded structures that further associated to form bundles. PF pairing also occurred in the presence of Z5 (Fig. 3 I and J).
To validate the electron microscopy results, we quantified the effects of Zantrins on steady-state FtsZ polymer mass and structure by sedimentation. Although high concentrations (3–15× IC50) of Zantrins allowed PF pairing/bundling to be visible by electron microscopy (Fig. 3), we used the inhibitors at lower concentrations (2× IC50) to quantify their effects on steady-state assembly of FtsZEc. Although 40% of total FtsZ was recovered in the pellet fraction of the control reaction, Z1, Z2, and Z4 reduced sedimentation to ≈2–15% of total FtsZ in the reactions, with Z4 causing the most severe effect (Fig. 4). This indicates a destabilizing effect of these compounds on polymer assembly. In contrast, the remaining two Zantrins induced higher levels of FtsZ assembly; ≈85% of total FtsZ accumulated in the pellet in the presence of Z3, whereas Z5 allowed ≈50% of input FtsZ to be sedimented (Fig. 4). The increase in polymer mass seen with Z3 and Z5 indicates that these molecules stabilize FtsZ assembly to different extents, consistent with the electron microscopy data.
Z2 caused an ≈3-fold decrease in the amount of FtsZ in the pellet fraction at 20 μM (Fig. 4) but induced significant PF pairing at 60 μM (Fig. 3D). This suggests that besides inhibiting or destabilizing the longitudinal bonds between FtsZ subunits, Z2 also exerts a weaker stabilizing effect on the lateral bonds between the PFs. This behavior is broadly similar to the effects of vinblastine on microtubules (29). The overall effect of 20 μM Z2 on FtsZ assembly is, therefore, a net diminution in the steady-state polymer mass (Fig. 4) and an occasional stabilization of lateral bonds, causing infrequent PF pairing (Fig. 3C).
Zantrins Cause Bacterial Lethality. To test the effects of Zantrins in vivo, we determined the MICs of the compounds against a range of Gram-positive and -negative bacterial strains. Z1 and Z2 could kill the wild-type E. coli strain MC1000, whereas the other three inhibitors failed to abolish MC1000 growth up to 80 μM (Table 2). We hypothesized that E. coli could be pumping out the inactive molecules and that AcrAB, the major drug pump in E. coli (27, 30), could be involved in the postulated efflux. We therefore tested the acrAB-null strain DRC39 for its sensitivity against the inhibitors. DRC39 was markedly more sensitive to Z3 and Z5, but still insensitive toward Z4 (Table 2). Although Z2 killed MC1000 at 40 μM, its MIC against DRC39 decreased to 2.5 μM (Table 2), indicating that it is also a substrate of the AcrAB pump. The sensitivity toward Z1 was identical for the wild-type and acrAB-null strains (Table 2). The majority of Zantrins did not cause overt filamentation or minicell formation in DRC39 except Z5, which caused significant lethality and cell filamentation. After 14 h treatment of DRC39 with 80 μM Z5, a small residual turbidity persisted, and it consisted of filaments, filamentous ghosts, and some short nonmotile cells (Fig. 6, which is published as supporting information on the PNAS web site).
Table 2. MICs of Zantrins, μM.
| Organism | Z1 | Z2 | Z3 | Z4 | Z5 |
|---|---|---|---|---|---|
| E. coli MC 1000 | 20 | 40 | >80 | >80 | >80 |
| E. coli DRC 39 (ΔacrAB::Tn903kanr)* | 20 | 5 | 10 | >80 | 80‡ |
| E. coli DRC 40 (ΔacrAB::Tn903kanr ftsZ84-ts)* | 40 | 5 | 10 | >80 | 80§ |
| E. coli DRC 42 (ΔacrAB::Tn903kanr recA::cat)* | 20 | 5 | 5 | >80 | 80‡ |
| S. dysenteriae 60R | 10 | 10 | 20 | >80 | >80 |
| V. cholerae N16961 | 5 | 5 | 5 | >80 | >80 |
| P. aeruginosa PAK | 40 | >80 | >80 | >80 | >80 |
| P. aeruginosa (ΔmexAB)* | 40 | >80 | >80 | >80 | >80 |
| B. subtilis JH 642 | 1.25 | 2.5 | 2.5 | 2.5 | 40 |
| B. cereus CIP 3852 | 0.625 | 5 | 20 | 2.5 | 80 |
| S. aureus H | 2.5 | 1.25 | 5 | 10 | >80 |
| S. aureus MRSA† | 2.5 | 2.5 | 10 | 10 | >80 |
| S. pneumoniae TIGR 4 | 0.312 | 2.5 | 5 | 10 | >80 |
| C. perfringens Strain 13 | 5 | 10 | 80 | 5 | >80 |
DRC 39, DRC 40, and DRC 42 are derivatives of the wild-type E. coli strain MC 1000; ΔmexAB mutant of P. aeruginosa is a derivative of the wild-type strain PAK.
Methicillin-resistant S. aureus is a clinical isolate from Tufts—New England Medical Center.
Denotes residual turbidity due to unlyzed filaments, filament ghosts, and some short nonmotile cells.
Faint turbidity, less than in DRC 39; very long filaments, some with increased diameter, filament ghosts, and few short cells.
We tested Zantrins against DRC40 (MC1000 ΔacrAB::Tn903kanr ftsZ84-ts) to examine whether the presence of the thermosensitive ftsZ84 mutation may cause altered sensitivity under permissive growth conditions (30°C). DRC40 was 2-fold more resistant to Z1, whereas its sensitivity to the other four molecules was similar to that of its isogenic parent DRC39 (Table 2). This is consistent with a role for Z1–FtsZ interaction in the lethal effect of Z1 in vivo and does not rule out a role for FtsZ in the lethality of other compounds.
To rule out whether Zantrins might block DNA replication or cause DNA damage and thereby induce the FtsZ-inhibitor SulA as part of the SOS-checkpoint mechanism (31, 32), we transduced the null recA::cat allele from strain BW1074 (33) into DRC39 to obtain DRC42 as a tester strain. RecA is essential for the induction of SOS regulons in E. coli and, in its absence, SulA is not induced (34). Therefore, if Zantrins were acting indirectly through SulA induction to block cell growth and division, DRC42 should be resistant to these compounds. As shown in Table 2, DRC42 displayed similar sensitivity to Zantrins as its parent DRC39, indicating that these inhibitors are unlikely to cause SulA-mediated FtsZ inhibition.
We next tested the effects of Zantrins on pathogenic Gram-negative bacteria. The Shiga toxin-producing Shigella dysenteriae 60R and the seventh-pandemic (El Tor biotype) Vibrio cholerae strain N16961 were sensitive to Zantrins Z1–Z3 but failed to be killed by Z4 and Z5 (Table 2). Although V. cholerae culture treated with 80 μM Z5 showed substantial turbidity, the cells appeared nonmotile with a subpopulation that were longer, fatter, and markedly curved (data not shown). In contrast, Z5-treated S. dysenteriae cells retained apparently normal morphology (not shown). Of all organisms tested, Pseudomonas aeruginosa showed the highest resistance toward Zantrins, and the absence of the MexAB drug pump did not enhance its sensitivity to the majority of the inhibitors (Table 2). However, 40 μM Z1 abolished growth of P. aeruginosa, confirming that it is a highly cell-permeable molecule and is not subject to drug pump-mediated efflux (Table 2).
The Gram-positive organisms were significantly more sensitive to Z1 and Z4 than Gram-negative bacteria but showed resistance to Z5, exceptions being Bacillus subtilis and Bacillus cereus (Table 2). Strikingly, the MICs of Z1 against the virulent Streptococcus pneumoniae type 4 TIGR4 strain (0.312 μM) and the opportunistic food-borne pathogen B. cereus (0.625 μM) are comparable to the known MICs of penicillin, vancomycin, and ciprofloxacin against streptococci (35). Zantrins caused bulging of cells in S. pneumoniae chains and increased the proportion of short chains or isolated cells compared to control cultures (Fig. 7, which is published as supporting information on the PNAS web site).
B. cereus is the closest relative of Bacillus anthracis, the causative agent of anthrax (36), and the sensitivity of B. cereus to all five Zantrins suggests that FtsZ inhibitors could be effective antidotes against virulent bacteria belonging to Category A priority pathogen group (www.niaid.nih.gov/biodefense/bandc_priority.htm). Furthermore, the majority of Zantrins could kill a methicillin-resistant clinical isolate of Staphylococcus aureus (Table 2), suggesting that FtsZ antagonists may provide protection against antibiotic-resistant pathogens. Besides causing lethality to aerobic organisms, Zantrins also were effective against the Gram-positive toxigenic anaerobe Clostridium perfringens (Table 2). This suggests that Zantrins may not require redox modifications within bacterial cells to cause lethality.
Zantrins Perturb Z Ring Assembly. Although Zantrins other than Z5 did not induce filamentation in E. coli, perhaps because of secondary effects on metabolism, we tested whether these molecules may nonetheless target FtsZ in vivo to perturb Z ring assembly. We analyzed the effects of Zantrins (except Z4, which does not kill E. coli; Table 2) on Z ring assembly in growing cultures of DRC39 by immunofluorescence microscopy (Fig. 5). Control cultures treated with 0.66% DMSO for 30 min (Fig. 5 a and a′) or for 60 min (Fig. 5 b and b′) showed distinct equatorial Z rings between segregated nucleoids in ≈90% cells. In contrast, after 30- or 60-min treatment of DRC39 with Zantrins (0.5× or 1× MIC), Z rings were either absent or dimly visible in a large percentage of cells or appeared as polar foci in a smaller fraction of the population (Fig. 5 c–g; Table 3, which is published as supporting information on the PNAS web site). There was no major chromosome segregation defect in the presence of Zantrins, exemplified with Z2 (Fig. 5 d and d′ and Table 3), except that Z1 caused ≈33% DRC39 cells to display an unsegregated nucleoid mass between polar FtsZ foci (Fig. 5 c and c′). Because Z1 affected nucleoid partitioning only in a fraction of cells, we consider it a kinetic defect rather than a permanent block. Although Z3 stabilized FtsZ assembly in vitro (Figs. 3 and 4), it perturbed Z ring assembly and integrity significantly (Fig. 5f and Table 3). Z5 also affected Z ring assembly with the caveat that the delocalized fluorescence seen in the majority of treated cells could be due partly to Z5 autofluorescence that may have caused a high background and masked the Z rings (Fig. 5g and Table 3). Overall, we conclude that Zantrins perturb Z ring assembly by directly targeting FtsZ in vivo, without exerting any major effect on chromosome segregation.
Discussion
Zantrins as Tools to Study FtsZ Polymerization Dynamics. We have shown that Zantrins Z1, Z2, and Z4 destabilize FtsZ assembly, whereas Z3 stabilizes FtsZ assembly significantly and Z5 is a weak stabilizer (Figs. 3 and 4 and Table 3). The persistence of a small number of short PFs in the presence of the destabilizers suggests that these molecules may not inhibit de novo FtsZ assembly but probably induce polymer lability through other mechanisms. Based on microtubule–colchicine interaction (37), we envisage a scenario where a destabilizing Zantrin may bind to a pocket between FtsZ subunits to push them apart such that the synergy loop T7 in one FtsZ monomer fails to make optimum contact with the GTP bound to loops T1–T6 in the neighboring monomer, an interaction essential for polymerization and for stimulating nucleotide hydrolysis (13, 17). Interestingly, the SOS-inducible division inhibitor SulA has recently been shown to bind to the T7 loop surface of FtsZ to block polymer formation (32), revealing an effective evolutionary strategy for targeting FtsZ. It would be of interest to examine whether any of the small molecule inhibitors may also target the T7 loop.
The stabilizing Zantrins could inhibit FtsZ depolymerization by opposing the movement of the switch loop T3 that has been proposed to cause a bend in the filament upon GTP hydrolysis (38). If a Zantrin were to counteract this structural transition, it could inhibit GTP hydrolysis and allow lateral association of the FtsZ PFs. Alternatively, Z3 or Z5 may simply crosslink the PFs and thus stabilize the lateral contacts.
Interestingly, contrary to their in vitro stabilizing effects, Z3 and Z5 caused a significant reduction in Z ring assembly in E. coli (Fig. 5 and Table 3). Because the integrity of Z rings depends, in part, on the stabilizing effects of ZipA and FtsA (3, 22, 39, 40), it is conceivable that the binding of Zantrins to the FtsZ PFs in vivo may adversely affect the recruitment of ZipA and/or FtsA to the septum, thus causing lability to the Z rings. Alternatively, these inhibitors may promote FtsZ polymerization or aggregation at inappropriate sites, thus using up the pool of FtsZ in the cell and preventing ring assembly indirectly. Such effects may underlie vinblastine and Taxol inhibition of microtubule function (37).
Zantrins as Potential Therapeutic Leads. Zantrins Z1–Z5 are neither sufficiently potent nor sufficiently specific to be considered as antibiotics in their current form (Tables 2 and 3). However, several Zantrins were effective against a range of organisms in broth cultures, including antibiotic-resistant and virulent pathogens (Table 2). In particular, Z1 was broadly cell-permeable and displayed significant potency against Gram-positive organisms (Table 2). Therefore, it is possible that one or more Zantrins may serve as leads for antibiotic development through systematic chemical modifications to optimize inhibitor potency and specificity.
An approach to optimize FtsZ inhibitors will be to identify the atomic contacts between the ligands and the protein by solving cocrystal structures of FtsZ–Zantrin complexes. A crystallization trial with FtsZEc had previously revealed conditions where crystals formed, but they were thin and failed to diffract (D.R.C., N. Nassar, and A. Wittinghofer, unpublished results). The majority of crystallization conditions caused FtsZ polymerization, even in the presence of GDP. Because the cocrystal structure of P. aeruginosa FtsZ in a complex with the inhibitor SulA has been solved recently (32), we expect FtsZEc to also crystallize in the presence of chemical antagonists. If successful, this effort could facilitate chemical derivatization of Zantrins to design potent analogs.
Acknowledgments
We thank A. Camilli, D. Hava, N. Mani, J. T. Park, A. Serio, A. L. Sonenshein, and M. Waldor (all at Tufts University School of Medicine); A. Kane [Center for Gastroenterology Research on Absorptive and Secretory Processes (GRASP) at Tufts–New England Medical Center]; S. Lory (Harvard Medical School) for bacterial strains; and D. Hava and N. Mani for help with culturing streptococci and clostridia, respectively. We also thank the High Throughput Screening Group at the Harvard Institute of Chemistry and Cell Biology for help with robotics and screening. This work was supported by the Defense Advanced Research Projects Agency (Grant N65236-98-1-5408), by the Charles A. Dana Foundation, and by the GRASP Center at Tufts–New England Medical Center (National Institute of Diabetes and Digestive and Kidney Diseases, P30 DK-34928).
Abbreviations: PF, protofilament; FtsZEc, untagged Escherichia coli FtsZ; MIC, minimum inhibitory concentration; FtsZMt, FtsZ from Mycobacterium tuberculosis.
† Present address: Harvard Medical School, Boston, MA 02115.
References
- 1.Bi, E. & Lutkenhaus, J. (1991) Nature 354, 161–164. [DOI] [PubMed] [Google Scholar]
- 2.Buddelmeijer, N. & Beckwith, J. (2002) Curr. Opin. Microbiol. 5, 553–557. [DOI] [PubMed] [Google Scholar]
- 3.Romberg, L. & Levin, P. A. (2003) Annu. Rev. Microbiol. 57, 125–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhardt, T. G. & de Boer, P. A. (2003) Mol. Microbiol. 48, 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt, K. L., Peterson, N. D., Kustusch, R. J., Wissel, M. C., Graham, B., Phillips, G. J. & Weiss, D. S. (2004) J. Bacteriol. 186, 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Löwe, J. & Amos, L. A. (1998) Nature 391, 203–206. [DOI] [PubMed] [Google Scholar]
- 7.RayChaudhuri, D. & Park, J. T. (1992) Nature 359, 251–254. [DOI] [PubMed] [Google Scholar]
- 8.de Boer, P. A., Crossley, R. E. & Rothfield, L. I. (1992) Nature 359, 254–256. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee, A., Dai, K. & Lutkenhaus, J. (1993) Proc. Natl. Acad. Sci. USA 90, 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson, H. P., Taylor, D. W., Taylor, K. A. & Bramhill, D. (1996) Proc. Natl. Acad. Sci. USA 93, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu, X.-C. & Margolin, W. (1997) EMBO J. 16, 5455–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee, A. & Lutkenhaus, J. (1998) EMBO J. 17, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löwe, J. & Amos, L. (1999) EMBO J. 18, 2364–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White, E. L., Ross, L. J., Reynolds, R. C., Seitz, L. E., Moore, G. D. & Borhani, D. W. (2000) J. Bacteriol. 182, 4028–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romberg, L., Simon, M. & Erickson, H. P. (2001) J. Biol. Chem. 276, 11743–11753. [DOI] [PubMed] [Google Scholar]
- 16.Oliva, M. A., Huecas, S., Palacios, J. M., Martin-Benito, J., Valpuesta, J. M. & Andreu, J. M. (2003) J. Biol. Chem. 278, 33562–33570. [DOI] [PubMed] [Google Scholar]
- 17.Scheffers, D.-J., de Wit, J. G., den Blaauwen, T. & Dreissen, A. J. M. (2002) Biochemistry 41, 521–529. [DOI] [PubMed] [Google Scholar]
- 18.Peterson, J. R. & Mitchison, T. J. (2002) Chem. Biol. 9, 1275–1285. [DOI] [PubMed] [Google Scholar]
- 19.Crews C. M. & Mohan, R. (2000) Curr. Opin. Chem. Biol. 4, 47–53. [DOI] [PubMed] [Google Scholar]
- 20.White, E. L., Suling, W. J., Ross, L. J., Seitz, L. E. & Reynolds, R. C. (2002) J. Antimicrob. Chemother. 50, 111–114. [DOI] [PubMed] [Google Scholar]
- 21.Wang, J., Galgoci, A., Kodali, S., Herath, K. B., Jayasuriya, H., Dorso, K., Vicente, F., Gonzalez, A., Cully, D., Bramhill, D., et al. (2003) J. Biol. Chem. 278, 44424–44428. [DOI] [PubMed] [Google Scholar]
- 22.RayChaudhuri, D. (1999) EMBO J. 18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergmeyer, H. U., Gawehn, K. & Grassl, M. (1974) Methods of Enzymatic Analysis, ed. Bergmeyer, H. U. (Academic, New York), Vol. I, pp. 501–510. [Google Scholar]
- 24.RayChaudhuri, D. & Park, J. T. (1994) J. Biol. Chem. 269, 22941–22944. [PubMed] [Google Scholar]
- 25.Lee, E., Taussig, R. & Gilman, A. G. (1992) J. Biol. Chem. 267, 1212–1218. [PubMed] [Google Scholar]
- 26.Akiyama, Y., Kihara, A., Tokuda, H. & Ito, K. (1996) J. Biol. Chem. 271, 31196–31201. [DOI] [PubMed] [Google Scholar]
- 27.Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H. & Hearst, J. E. (1995) Mol. Microbiol. 16, 45–55. [DOI] [PubMed] [Google Scholar]
- 28.Mingorance, J., Rueda, S., Gomez-Puertas, P., Valencia, A. & Vicente, M. (2001) Mol. Microbiol. 41, 83–91. [DOI] [PubMed] [Google Scholar]
- 29.Khan, I. A. & Luduena, R. F. (2003) Invest. New Drugs 21, 3–13. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido, H. (2001) Semin. Cell Dev. Biol. 12, 215–223. [DOI] [PubMed] [Google Scholar]
- 31.Bi, E. & Lutkenhaus, J. (1993) J. Bacteriol. 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordell, S. C., Robinson, E. J. H. & Löwe, J. (2003) Proc. Natl. Acad. Sci. USA 100, 7889–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wanner, B. L. & Boline, J. A. (1990) J. Bacteriol. 172, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 35.Gordon, K. A., Beach, M. L., Biedenbach, D. J., Jones, R. N., Rhomberg, P. R. & Mutnick, A. H. (2002) Diagn. Microbiol. Infect. Dis. 43, 157–162. [DOI] [PubMed] [Google Scholar]
- 36.Read, T. D., Peterson, S. N., Tourasse, N., Baillie, L. W., Paulsen, I. T., Nelson, K. E., Tettelin, H., Fouts, D. E., Eisen, J. A., Gill, S. R., et al. (2003) Nature 423, 81–86. [DOI] [PubMed] [Google Scholar]
- 37.Downing, K. H. (2000) Annu. Rev. Cell Dev. Biol. 16, 89–111. [DOI] [PubMed] [Google Scholar]
- 38.Diaz, J. F., Kralicek, A., Mingorance, J., Palacios, J. M., Vicente, M. & Andreu, J. M. (2001) J. Biol. Chem. 276, 17307–17315. [DOI] [PubMed] [Google Scholar]
- 39.Pichoff, S. & Lutkenhaus, J. (2002) EMBO J. 21, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geissler, B., Elraheb, D. & Margolin W. (2003) Proc. Natl. Acad. Sci. USA 100, 4197–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]