Abstract
Murine leukemia viruses (MLVs) have been classified as N-tropic (N-MLV) or B-tropic (B-MLV), depending on their ability to infect particular mouse strains. The early phase of N-MLV infection is blocked in the cells of several mammalian species, including humans. This block is mediated by a dominant host factor that targets the viral capsid soon after virus entry into the cell has been achieved. A similar block to HIV-1 in rhesus monkey cells is mediated by TRIM5α. Here we show that human TRIM5α is both necessary and sufficient for the restriction of N-MLV in human cells. Rhesus monkey TRIM5α, which potently blocks HIV-1 infection, exhibited only modest inhibition of N-MLV infection. B-MLV was resistant to the antiviral effects of both human and rhesus monkey TRIM5α; susceptibility to TRIM5α-mediated restriction was conferred by alteration of residue 110 of the B-MLV capsid protein to the amino acid found in the N-MLV capsid. Our results demonstrate that species-specific variation in TRIM5α governs its ability to block infection by diverse retroviruses.
Mammals have been subjected to retrovirus infections repeatedly over millions of years of evolution, as documented by the presence of endogenous retroviral elements in the germ lines of these species (1–3). Retroviral epidemics likely have exerted selective pressure for optimal innate and adaptive immune responses. Innate cellular resistance to the early phase of retroviral infection can influence retroviral tropism (4, 5). Retroviral infection of host cells is initiated by virus attachment, fusion of the viral and cellular membrane and entry of the viral core into the cytoplasm. Several retroviruses encounter blocks in the target cells of certain hosts at early, postentry stages. Some subtypes of murine leukemia virus (MLV), designated N-tropic (N-MLV) or B-tropic (B-MLV) are blocked in specific mouse strains by the product of the Fv1 gene of mice (6–8). This block occurs after reverse transcription of the viral genome but before its integration into the host cell genome (9–11). The Fv1 gene is present only in mice but is related to murine and human endogenous retroviruses and encodes a protein similar to retroviral core (Gag) proteins (12). The expression of Fv1 occurs at only very low levels, hampering the elucidation of a mechanism of the Fv1-mediated block. However, it has been shown that the viral target for Fv1 restriction is the viral core (13–15). Indeed, a single amino acid change at position 110 in the MLV capsid protein determines the susceptibility of the virus to the blocking effects of different Fv1 alleles (15).
In addition to governing the ability of retroviruses to infect particular mouse strains, early postentry restrictions can also determine retroviral tropism at the species level. N-MLV, for example, inefficiently infects human cells and certain cell lines from African green monkeys (16, 17). By contrast, B-MLV and dual-tropic (NB-tropic) MLV efficiently infect these cells. HIV-1 encounters a postentry block in Old World monkeys, whereas simian immunodeficiency virus (SIVmac) infection is blocked in most New World monkey cells (18–20). These species-specific restrictions share several features:
The blocks occur before or concurrently with reverse transcription; levels of reverse transcripts are reduced in restricted cells (16–19, 21–23).
The viral determinant of susceptibility to the block is the capsid protein (16, 21, 24–26). Of note, the same changes in residue 110 of the MLV capsid that determine susceptibility to Fv1-mediated restrictions also specify susceptibility to the block of N-MLV in human cells (15, 16). Other capsid-binding proteins, such as cyclophilin A in the case of HIV-1 (27), can modify the degree of the restriction.
The restriction is mediated by dominant host factors, the activity of which can be titrated by the introduction of virus-like particles containing proteolytically processed capsid proteins of the restricted viruses (21–23, 28). The N-MLV restriction factor in human cells has been called REF1 (16). Competition studies with virus-like particles suggested that some of the host factors restricting infection by different retroviruses might be related (28).
Recently, a genetic screen identified a major restriction factor in Old World monkey cells that acts on HIV-1 and, to a lesser extent, on SIVmac (29). The factor, TRIM5α, is a member of the tripartite motif (TRIM) family of proteins. TRIM proteins contain a RING domain, a B-box 2 domain, and a coiled coil domain (30). Some TRIM proteins, like TRIM5α, also possess a C-terminal B30.2 or SPRY domain (30). TRIM5α from rhesus monkeys was shown to be sufficient to confer potent resistance to HIV-1 infection in otherwise susceptible cells (29). Interference with TRIM5α expression in Old World monkey cells relieved the early block to HIV-1 infection (29). Cells expressing rhesus monkey TRIM5α (TRIM5αrh) exhibited partial inhibition of SIVmac infection but were as susceptible as control cells to infection by Moloney MLV, a dual-tropic MLV (29). The human orthologue of TRIM5α (TRIM5αhu), is 87% identical in amino acid sequence to TRIM5αrh (29). When expressed at comparable levels, TRIM5αhu was less potent at suppressing HIV-1 and SIVmac infection than TRIM5αrh (29). Thus, each TRIM5α protein, TRIM5αhu or TRIM5αrh, specifies an intermediate level of resistance to the naturally infecting cognate virus, HIV-1 or SIVmac, respectively. Here we test the hypothesis that TRIM5αhu contributes to the restriction of N-MLV infection in human cells.
Methods
Creation of Cells Stably Expressing TRIM5α Variants. Retroviral vectors encoding TRIM5αhu or TRIM5αrh were created by using the pLPCX vector (29). The pLPCX-TRIM5αhu and pLPCX-TRIM5αrh vectors contain only the amino acid-coding sequence of the TRIM5α cDNA. In both constructs, the TRIM5α proteins possess C-terminal epitope tags derived from influenza hemagglutinin (HA). Recombinant viruses were produced in 293T cells by cotransfecting these pLPCX plasmids with the pVPack-GP and pVPack-VSV-G packaging plasmids (Stratagene). The pVPack-VSV-G plasmid encodes the vesicular stomatitis virus (VSV) G envelope glycoprotein, which allows efficient entry into a wide range of vertebrate cells. The resulting virus particles were used to transduce ≈1 × 105 NIH 3T3, 293T, or HeLa cells in the presence of 5 μg/ml polybrene. Cells were selected in 1 μg/ml puromycin (Sigma).
Infection with Viruses Expressing GFP. Moloney MLV-GFP (MoMLV-GFP) viruses were prepared by cotransfecting 293T cells with 15 μg of pFB-hrGFP encoding humanized Renilla reniformis GFP, 15 μg of pVPack-GP, and 4 μg of pVPack-VSV-G (all from Stratagene). A similar protocol was used to generate N-MLV and B-MLV expressing GFP, except that pVPack-GP-N and pVPack-GP-B, respectively, were used in place of pVPack-GP (16, 17). The NBNN-MLV and BNBB-MLV GFP-expressing vectors were created by site-directed mutagenesis of pVPack-GP-N and pVPack-GP-B, respectively, by using the following primers: NBNN:5′-GATTACACCACCCAAGAAGGTAGGAACC-3′ and BNBB:5′-GATTACACCACTACAAGAGGTAGGAACC-3′.
Viral stocks were titrated on 293T cells and normalized based on a dilution of virus that yielded 50% infected cells. For infections, 4 × 104 NIH 3T3, HeLa, or 293T cells seeded in 24-well plates were incubated with recombinant viruses for 24 h. Cells were washed and returned to culture for 48 h and then subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScan (Becton Dickinson).
Quantitative Real-Time PCR. Virus stocks derived from the transfection of 293T cells were treated with 50 units/ml Turbo DNase (Ambion, Austin, TX) for 60 min at 37°C. NIH 3T3 cells (2 × 105) were infected with VSV G-pseudotyped N-MLV-GFP or control viruses lacking envelope glycoproteins. Genomic DNA was isolated from the cells at various time points (0–24 h). Primers specific for the GFP gene in the vector were used as described in refs. 16 and 17. Reaction mixtures contained TaqMan universal master mix (PE Biosystems), 300 nM primers, 100 nM probe, and 500 ng of genomic DNA. The PCR conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 min at 60°C, on an ABI Prism 7700 (Applied Biosystems).
Immunoblotting. HA-tagged TRIM5αhu and TRIM5αrh expressed stably in NIH 3T3 cells or 293T cells were detected in whole-cell lysates (PBS plus 1% Nonidet P-40) by Western blotting with horseradish peroxidase-conjugated 3F10 antibody (Roche). β-actin was detected with A5441 antibody (Sigma).
RNA Interference. The short interfering RNAs (siRNA) used correspond to siRNAs 3 and 7 and the control siRNA described by Stremlau et al. (29). HeLa or TE671 cells (1.5 × 105) were seeded in six-well plates and transfected with 120 nM siRNA by using 10 μl of Oligofectamine (Invitrogen). After 48 h, cells were reseeded for infection by N-MLV-GFP, B-MLV-GFP, and MoMLV-GFP.
Results
Expression of TRIM5αhu in Permissive Cells Specifically Blocks N-MLV Infection. To examine whether TRIM5αhu contributes to the block to N-MLV in human cells, two cell lines permissive for N-MLV infection were transduced with a retroviral vector expressing HA-tagged TRIM5αhu or with an empty pLPCX vector. NIH 3T3 cells are mouse cells that are able to be efficiently infected by N-MLV; B-MLV infection is less efficient in these cells because of expression of the Fv1n allele (7, 8). Atypically, 293T cells are human cells that do not exhibit a block to N-MLV (17). NIH 3T3 and 293T cells transduced with the pLPCX vector expressing TRIM5αhu or with the empty vector were incubated with different amounts of recombinant N-MLV-GFP, B-MLV-GFP, and MoMLV-GFP. Expression of TRIM5αhu specifically inhibited N-MLV-GFP infection in both cell lines compared with cells transduced with the empty pLPCX vector (Fig. 1 A–C). Infection by B-MLV-GFP and MoMLV-GFP was not significantly affected by the expression of TRIM5αhu. We conclude that TRIM5αhu specifically and efficiently inhibits N-MLV infection.
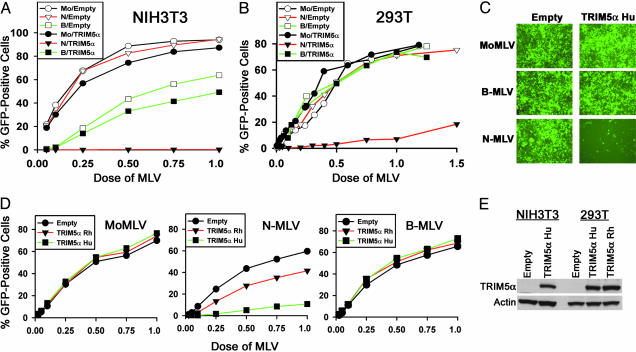
Fig. 1.
Human TRIM5α blocks N-MLV infection more potently than TRIM5αrh. (A–C) TRIM5αhu specifically blocks N-MLV infection. NIH 3T3 cells (A) and 293T cells (B and C) transduced with the empty pLPCX vector or the pLPCX vector expressing TRIM5αhu were incubated with various amounts of N-MLV-GFP (N), B-MLV-GFP (B), or MoMLV-GFP (Mo). Infected, GFP-positive cells were counted by FACS. In A and B, the results of a typical experiment are shown. Similar results were obtained in three independent experiments. (D) We incubated 293T cells transduced with the empty pLPCX vector or the pLPCX vectors expressing TRIM5αrh or TRIM5αhu with various amounts of MoMLV-GFP, N-MLV-GFP, or B-MLV-GFP. Infected, GFP-positive cells were counted by FACS. The results shown represent the means from two independent experiments. (E) Expression of TRIM5αhu and TRIM5αrh in cells. Lysates from NIH 3T3 and 293T cells expressing HA-tagged TRIM5α protein or transduced with the empty pLPCX vector were subjected to Western blotting with an antibody against HA. The lysates were also Western blotted with an antibody directed against actin.
Next, we examined whether TRIM5αrh would similarly block N-MLV infection. Expression of TRIM5αrh in 293T cells resulted in a modest inhibition of N-MLV but did not affect the efficiency of infection of B-MLV or MoMLV (Fig. 1D). TRIM5αrh inhibited N-MLV less efficiently than TRIM5αhu. The expression levels of TRIM5αhu and TRIM5αrh were equivalent in the transduced 293T cells (Fig. 1E). We conclude that TRIM5αrh mediates a specific but much less potent block to N-MLV infection than that mediated by TRIM5αhu.
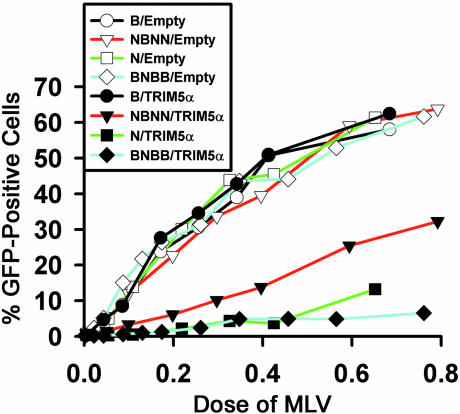
Capsid Is the Viral Target for TRIM5αhu Restriction. To investigate the viral target of the TRIM5αhu-mediated restriction, 293T cells expressing TRIM5αhu or control cells transduced with the empty pLPCX vector were incubated with recombinant N-MLV-GFP, B-MLV-GFP, NBNN-MLV-GFP, or BNBB-MLV-GFP. NBNN-MLV-GFP is identical to N-MLV except that residue 110 in the capsid protein has been changed from arginine to glutamic acid, the residue found in B-MLV. BNBB-MLV-GFP is identical to B-MLV-GFP except that capsid residue 110 has been changed from glutamic acid to arginine. These single amino acid changes have been shown to alter N-MLV susceptibility to postentry restrictions in human cells (17). N-MLV-GFP and BNBB-MLV-GFP infections were blocked to a similar extent in cells expressing TRIM5αhu (Fig. 2). In independent experiments, NBNN-MLV-GFP was less sensitive to the TRIM5αhu-mediated restriction than N-MLV-GFP but was more sensitive than B-MLV-GFP, which efficiently infected the TRIM5αhu-expressing cells. All of the viruses efficiently infected the control cells containing the empty pLPCX vector. We conclude that the MLV capsid, specifically residue 110, is an important determinant of susceptibility to TRIM5αhu-mediated restriction.
Fig. 2.
MLV capsid residue 110 influences viral susceptibility to TRIM5αhu antiviral effects. We exposed 293T cells transduced with the empty pLPCX vector or the pLPCX vector expressing TRIM5αhu to the indicated GFP-expressing viruses. Infected, GFP-positive cells were counted by FACS. The results shown are typical of those obtained in three independent experiments.
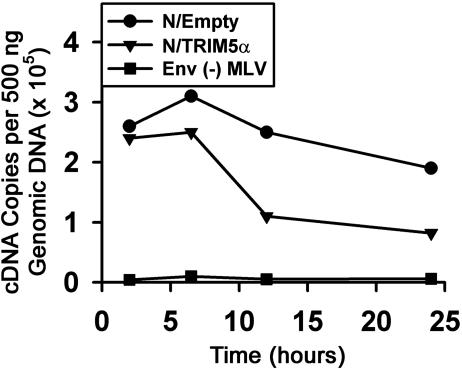
Reverse Transcription of N-MLV Is Less Efficient in Cells Expressing TRIM5αhu. To determine the level at which N-MLV infection is blocked by TRIM5αhu, we used a real-time PCR assay to detect viral cDNA at various times after incubating VSV G-pseudotyped N-MLV-GFP with either NIH 3T3 or 293T cells expressing TRIM5αhu or control cells. The levels of reverse transcripts detected for N-MLV-GFP were reduced in NIH 3T3 cells expressing TRIM5αhu, in comparison with those in cells containing the empty pLPCX vector (Fig. 3). Similar results were obtained in 293T cells (data not shown). We conclude that at least part of the block to N-MLV in TRIM5αhu-expressing cells involves a decrease in the efficiency of early events before the completion of reverse transcription.
Fig. 3.
Reverse transcription of N-MLV is less efficient in target cells expressing TRIM5αhu. NIH 3T3 cells stably transduced with the empty pLPCX vector or the pLPCX vector expressing TRIM5αhu were incubated with VSV G-pseudotyped, DNase-treated N-MLV-GFP. Approximately 50% of the cells with the empty vector became GFP-positive compared with 0.72% of the cells expressing TRIM5αhu. NIH 3T3 cells transduced with the empty pLPCX vector were also incubated with N-MLV-GFP viruses lacking envelope glycoproteins [Env (-) MLV]. Target cell DNA was isolated at the indicated times and used to detect reverse transcripts. Similar results were obtained in two independent experiments.
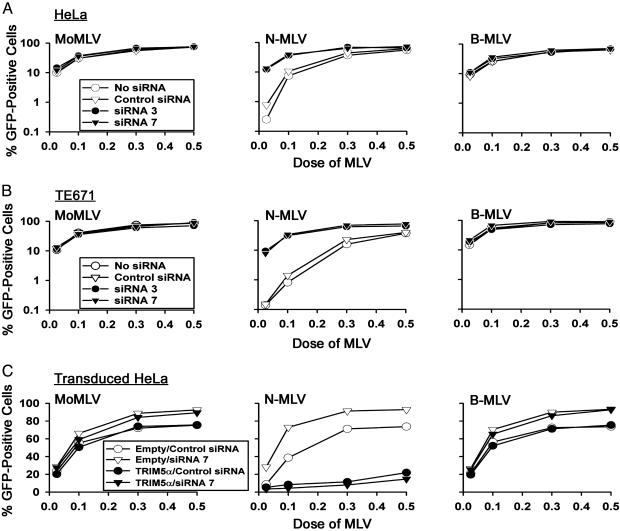
TRIM5αhu Expression Is Necessary for the Block to N-MLV Infection in Human Cells. To determine whether TRIM5αhu is necessary for the restriction to N-MLV infection in human cells, siRNAs directed against TRIM5αhu were used to down-regulate its expression. These siRNAs were previously shown to decrease TRIM5αrh protein levels significantly (29). Transfection of HeLa and TE671 cells with TRIM5αhu-specific siRNA 3 and siRNA 7, but not control siRNA, resulted in a marked increase in the efficiency of N-MLV infection (Fig. 4 A and B). In the cells that were incubated with siRNA 3 or siRNA 7, the efficiency of infection by N-MLV was comparable with that seen for B-MLV-GFP and MoMLV-GFP. Infections by B-MLV-GFP and MoMLV-GFP were unaffected by any of the siRNAs. As an additional control for specificity, the effects of siRNA 7 were rescued by expression of TRIM5αhu by a vector resistant to the effects of this siRNA. The siRNA 7 targets a 3′ noncoding sequence on the natural trim5 message that is not included in the TRIM5αhu-expressing pLPCX vector; therefore, siRNA 7 is expected to down-regulate endogenous TRIM5αhu expression in human cells but not the expression of vector-derived TRIM5αhu in the transduced HeLa cells (29). In HeLa cells stably transduced with the vector expressing TRIM5αhu, transfection of siRNA 7 did not relieve the restriction to N-MLV infection (Fig. 4C). We conclude that TRIM5αhu is necessary for the postentry block to N-MLV in human cells.
Fig. 4.
Expression of TRIM5αhu is essential for the block to N-MLV in human cells. (A and B) HeLa (A) and TE671 (B) cells were transfected with 120 nM of control siRNA, siRNA 3, or siRNA 7. Cells were then incubated with the indicated GFP-expressing viruses. Infected, GFP-positive cells were counted by FACS. The results shown are typical of those obtained in three independent experiments. (C) HeLa cells stably transduced with the empty pLPCX vector or the pLPCX vector expressing TRIM5αhu were transfected with 120 nM control siRNA, siRNA 3, or siRNA 7. Cells were then incubated with the indicated GFP-expressing viruses. Infected, GFP-positive cells were counted by FACS. The results shown are typical of those obtained in two independent experiments.
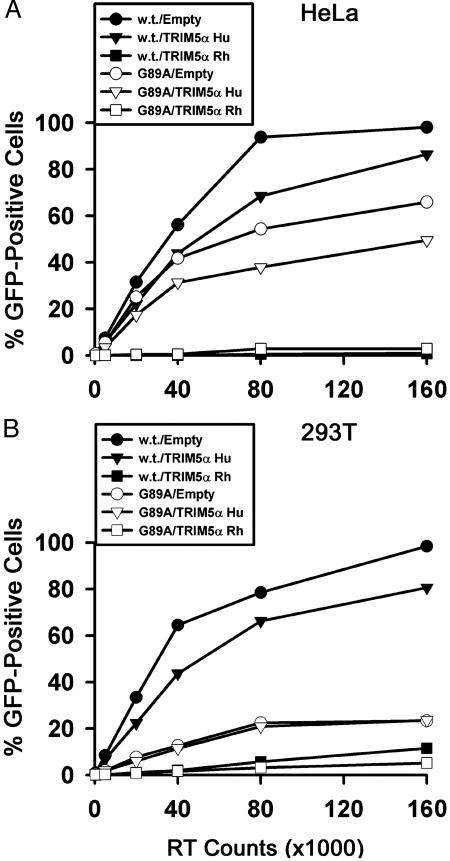
Effect of TRIM5αhu on Infection by an HIV-1 Mutant Defective for Cyclophilin A Binding. Cyclophilin A binding to the HIV-1 capsid protein results in an enhancement of virus infectivity (31–33). It has been reported that when cyclophilin A binding is interrupted, HIV-1 capsids can interact with and be inhibited by the N-MLV-restricting factor in human cells (27). The above studies identify TRIM5αhu as the major N-MLV-restricting factor in human cells and provide an opportunity to investigate the relationship of cyclophilin A and TRIM5αhu interaction with HIV-1. In particular, we wished to examine whether the interaction with TRIM5αhu might contribute to the poor infectivity of an HIV-1 mutant that does not bind cyclophilin A. The infectivities of wild-type HIV-1-GFP and the G89A mutant, which contains a single-residue alteration in the HIV-1 capsid protein that disrupts cyclophilin A binding (26, 34) were examined. The target cells for these assays were HeLa cells, which exhibit an N-MLV restriction, and 293T cells, which do not (17). HeLa and 293T cells stably overexpressing TRIM5αhu and TRIM5αrh were also used as target cells. The infectivity of the G89A mutant was lower than that of the wild-type HIV-1, and this decrease in relative infectivity was even more pronounced in the 293T cells than in HeLa cells (Fig. 5). The expression of TRIM5αrh in both cell types dramatically inhibited the infectivity of wild-type HIV-1 and the G89 mutant. In HeLa cells, TRIM5αhu expression resulted in a modest level of inhibition of both viruses. A moderate level of inhibition was also observed for wild-type HIV-1 infection of 293T cells expressing TRIM5αhu. This inhibitory effect was not evident for the infection of 293T cells by the G89A mutant virus. These results indicate that virus-specific and host cell-specific variables may influence the modest anti-HIV-1 effect associated with TRIM5αhu expression. The results in 293T cells also suggest that the negative effect of the G89A change in capsid on HIV-1 replication does not depend on the presence of a functional TRIM5αhu-mediated restriction in the target cell. Finally, cyclophilin A–capsid interaction is not an absolute requirement for HIV-1 susceptibility to the suppressive effects of TRIM5αrh or TRIM5αhu.
Fig. 5.
Infection of HeLa and 293T cells by wild-type HIV-1 or a mutant defective in cyclophilin A binding. HeLa (A) or 293T (B) cells stably transduced with the empty pLPCX vector or pLPCX vectors expressing TRIM5αhu or TRIM5αrh were incubated with the wild-type (w.t.) or G89A HIV-1-GFP viruses. Infected, GFP-positive cells were counted by FACS. The results shown are typical of those obtained in three independent experiments.
Discussion
In this report, we demonstrate that TRIM5αhu can potently and specifically block infection by N-MLV. TRIM5αhu is both necessary and sufficient for the establishment of this restriction in both human and murine cell lines. These results indicate that any cofactors required for the block to N-MLV infection must be functionally conserved between humans and rodents.
The results of this study emphasize the species-specific nature of the ability of TRIM5α proteins to establish blocks to particular viruses. TRIM5αhu potently restricted N-MLV, whereas TRIM5αrh exhibited only modest inhibition of N-MLV infection. Conversely, TRIM5αrh more potently blocked the primate lentiviruses HIV-1 and SIVmac than did TRIM5αhu (29). We can only speculate on the kinds of virus infections that might have influenced the evolution of the human and Old World monkey TRIM5α proteins in the ≈25 million years since the divergence of these primate lineages. The current epidemic of HIV-1 in humans is thought to have originated 60–90 years ago (35–37) and thus is far too limited in duration and extent to represent a significant selective force in TRIM5αhu evolution. It is possible that previous lentivirus epidemics afflicted the hominoids without leaving evidence of proviruses in the germline. However, the modest antiviral activity of TRIM5αhu against HIV-1 and the potent antiviral activity of this protein against N-MLV raise the possibility that past infections with gammaretroviruses like MLV may have exerted greater selective pressure on human ancestors. The presence in the human genome of human endogenous retroviruses (e.g., HERV-C, HERV-L, HERV-W, HERV-H, HERV-I, and ERV-9), which resemble the gammaretroviruses such as MLV (38–42), supports this model. Because gammaretroviruses and lentiviruses are only distantly related, our observations hint that resistance to a broad range of retroviruses and perhaps other viruses could be mediated by vertebrate TRIM5 or other TRIM proteins.
TRIM5αhu and TRIM5αrh exhibit ≈13% variation in primary sequence (29). Some subset of these amino acid differences accounts for the significant difference in the ability of TRIM5αhu and TRIM5αrh to restrict N-MLV. It will be of interest to determine whether the same amino acid differences that specify the ability to inhibit N-MLV infection negatively influence potency for the inhibition of lentivirus infection.
A single amino acid change in residue 110 of the B-MLV capsid protein, which has been shown to limit the ability of the virus to infect human cells (16), was sufficient to confer susceptibility to TRIM5αhu-mediated inhibition. The reciprocal chimera, NBNN-MLV, exhibited decreased susceptibility to, but not complete escape from, the TRIM5αhu-mediated block. Partial escape from TRIM5αhu may suffice to allow NBNN-MLV to infect natural human cell lines (16), which express lower levels of TRIM5αhu than the transduced cells used in our study (data not shown). Capsid residues other than 110 that differ between NBNN-MLV and B-MLV may contribute to the more complete escape of the latter virus from TRIM5αhu suppression. These results identify the viral capsid as an important determinant of susceptibility to TRIM5αhu-mediated restriction. It is intriguing that residue 110 of the MLV capsid also determines susceptibility to Fv1 restriction (15). This coincidence raises the possibility that trim5 orthologues in the mouse may encode factors that contribute to Fv1 restriction.
One difference between Fv1 restriction and the species-specific retroviral restrictions is that the former occurs after the completion of reverse transcription (9–11). A recent study in which Fv1n-mediated restriction was studied in human cells suggests that the level of the block is determined by the restriction factor, not by other host cell variables (17). Our results indicate that the level of N-MLV reverse transcripts is lower in TRIM5αhu-expressing cells than in control cells. This reduction in reverse transcripts was less dramatic than that observed for HIV-1 in TRIM5αrh-expressing cells, although the potencies of the infection blocks were comparable (29). Thus, TRIM5αhu may continue to inhibit early events in N-MLV infection even after some reverse transcription has occurred. If TRIM5α plays a role in MLV restriction mediated by Fv1, the presence of the capsid-like product of Fv1 (12) might influence the level at which the block occurs. Thus, the apparent dissimilarity between the retroviral restrictions mediated by TRIM5α and Fv1 may reflect contextual variation rather than intrinsic mechanistic differences.
It has been reported that if cyclophilin A binding to the HIV-1 capsid is inhibited by cyclosporin A treatment or by changes in the capsid protein, the HIV-1 capsid can interact with and be inhibited by the N-MLV-restricting factor in human cells (27). These observations have led to the proposal that cyclophilin A binding may decrease the binding of this restricting factor, herein identified as TRIM5αhu, to HIV-1 capsids (27). Our results in 293T cells indicate that the negative consequences to HIV-1 replication of the G89A capsid change, which eliminates cyclophilin A binding (35), do not depend on the presence of a functional TRIM5αhu-mediated restriction in the target cell. Our previous studies indicated that cyclosporin A treatment of HeLa target cells resulted in a slight increase in wild-type HIV-1 infection (26). These results are consistent with a model in which cyclophilin A binding results in an increase in TRIM5αhu interaction with HIV-1 capsids. Cyclophilin A binding to the HIV-1 capsid also increases the antiviral effects of the rhesus monkey restriction factor (26, 27), which has been identified as TRIM5αrh (29). Thus, our data support a modest, positive role for cyclophilin A–capsid binding in the antiviral effects of TRIM5α from both humans and monkeys. The study of the G89A mutant indicates that cyclophilin A binding by HIV-1 is not necessary for susceptibility to the restriction imposed by either TRIM5αhu or TRIM5αrh.
An appreciation that the antiviral effect of primate TRIM5α proteins can extend to a diverse set of retroviruses should provide additional incentive to understand the molecular basis for virus recognition and restriction by these proteins. Such understanding could reveal means to enhance this innate intracellular immunity to our advantage.
Acknowledgments
We thank Ms. Yvette McLaughlin and Sheri Farnum for manuscript preparation. This work was supported by National Institutes of Health Grant HL54785, Center for AIDS Research Grant P30 AI28691, the International AIDS Vaccine Initiative, the Bristol–Myers Squibb Foundation, and the late William F. McCarty-Cooper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MLV, murine leukemia virus; MoMLV, Moloney MLV; N-MLV, N-tropic MLV; B-MLV, B-tropic MLV; TRIM5αrh, rhesus monkey TRIM5α; TRIM5αhu, human TRIM5α; SIVmac, simian immunodeficiency virus; HA, hemagglutinin; VSV, vesicular stomatitis virus; FACS, fluorescence-activated cell sorting; siRNA, short interfering RNA.
References
- 1.Boeke, J. D. & Stoye, J. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-436. [PubMed]
- 2.Herniou, E., Martin, J., Miller, K., Cook, J., Wilkinson, M. & Tristem, M. (1998) J. Virol. 72, 5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin, J. M. (1996) in Fundamental Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott–Raven, Philadelphia), pp. 763-844.
- 4.Bieniasz, P. D. (2003) Trends Microbiol. 11, 286-291. [DOI] [PubMed] [Google Scholar]
- 5.Stoye, J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 11549-11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lilly, F. (1970) J. Natl. Cancer Inst. 45, 163-169. [PubMed] [Google Scholar]
- 7.Pincus, T., Hartley, J. W. & Rowe, W. P. (1971) J. Exp. Med. 133, 1219-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartley, J. W., Rowe, W. P. & Huebner, R. J. (1970) J. Virol. 5, 221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sveda, M. M. & Soeiro, R. (1976) Proc. Natl. Acad. Sci. USA 73, 2356-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolicoeur, P. & Rassart, E. (1980) J. Virol. 33, 183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prysiak, P. M. & Varmus, H. E. (1992) J. Virol. 66, 5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Best, S. P., LeTissier, P., Towers, G. & Stoye, J. P. (1996) Nature 382, 826-829. [DOI] [PubMed] [Google Scholar]
- 13.Ou, C.-Y., Boone, L. R., Koh, C.-K., Tennant, R. W. & Yang, W. K. (1983) J. Virol. 48, 779-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DesGroseillers, L. & Jolicoeur, P. (1983) J. Virol. 48, 685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, C. A. & Chakraborti, A. (1996) Virology 425, 300-306. [DOI] [PubMed] [Google Scholar]
- 16.Towers, G., Bock, M., Martin, S., Takeuchi, Y., Stoye, J. P. & Danos, O. (2000) Proc. Natl. Acad. Sci. USA 97, 12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besnier, C., Ylinen, L., Strange, B., Lister, A., Takeuchi, Y., Goff, S. P. & Towers, G. J. (2003) J. Virol. 77, 13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K. & Adachi, A. (1995) J. Gen. Virol. 76, 2723-2730. [DOI] [PubMed] [Google Scholar]
- 19.Himathongkham, S. & Luciw, P. A. (1996) Virology 219, 485-488. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, W., Schubert, D., LaBonte, J., Munson, L., Gibson, S., Scammell, J., Ferrigno, P. & Sodroski, J. (1999) J. Virol. 73, 10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowan, S., Hatziiannou, T., Cunningham, T., Muesing, M., Gottlinger, H. & Bieniasz, P. (2002) Proc. Natl. Acad. Sci. USA 99, 11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munk, C., Brandt, S. M., Lucero, G. & Landau, N. R. (2002) Proc. Natl. Acad. Sci. USA 99, 13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besnier, C., Takeuchi, Y. & Towers, G. (2002) Proc. Natl. Acad. Sci. USA 99, 11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owens, C. M., Yang, P. C., Gottlinger, H. & Sodroski, J. (2003) J. Virol. 77, 726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kootstra, N. A., Munk, C., Tonnu, N., Landau, N. R. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens, C. M., Song, B., Perron, M. J., Yang, P. C., Stremlau, M. & Sodroski, J. (2004) J. Virol. 78, 5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towers, G. J., Hatziioannou, T., Cowan, S., Goff, S. P., Luban, J. & Bieniasz, P. D. (2003) Nat. Med. 9, 1138-1143. [DOI] [PubMed] [Google Scholar]
- 28.Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D. & Towers, G. J. (2003) EMBO J. 22, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848-853. [DOI] [PubMed] [Google Scholar]
- 30.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thali, M., Bukovsky, A., Kondo, E., Rosenwirth, B., Walsh, C. T., Sodroski, J. & Gottlinger, H. G. (1994) Nature 372, 363-365. [DOI] [PubMed] [Google Scholar]
- 32.Franke, E. K., Yuan, H. E. & Luban, J. (1994) Nature 372, 359-362. [DOI] [PubMed] [Google Scholar]
- 33.Braaten, D., Franke, E. K. & Luban, J. (1996) J. Virol. 70, 3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bukovsky, A., Weimann, A., Accola, M. & Gottlinger, H. (1997) Proc. Natl. Acad. Sci. USA 94, 10943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korber, B., Muldoon, M., Theiler, J., Gao, F., Gupta, Lapedes, A., Hahn, B. H., Wolinsky, S. & Bhattacharya, T. (2000) Science 288, 1789-1796. [DOI] [PubMed] [Google Scholar]
- 36.Sharp, P. M., Bailes, E., Chaudhuri, R. R., Rodenburg, C. M., Santiago, M. I. & Hahn, B. H. (2001) Philos. Trans. R. Soc. London B 356, 867-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp, P. M., Bailes, E., Gao, F., Beer, B. E., Hirsch, V. M. & Hahn, B. H. (2000) Biochem. Soc. Trans. 28, 275-282. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson, D. A., Mager, D. L. & Leong, J.-A. (1994) in The Retroviridae, ed. Levy, J. A. (Plenum, New York), Vol. 3, pp. 465-525. [Google Scholar]
- 39.Martin, J., Herniou, E., Cook, J., O'Neill, R. W. & Tristem, M. (1997) J. Virol. 71, 437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tristem, M. (2000) J. Virol. 74, 3715-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benit, L., DeParseval, N., Casella, J.-F., Callebaut, I., Cordonnier, A. & Heidmann, T. (1997) J. Virol. 71, 5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blond, J. L., Beseme, F., Duret, L., Bouton, O., Bedin, F., Perron, H., Manbrand, B. & Mallet, F. (1999) J. Virol. 73, 1175-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]