Abstract
A portion of the nopaline synthase gene under the control of the cauliflower mosaic virus 35S promoter was used to transform a tobacco plant that had previously been transformed with a wild-type nopaline synthase (nos) gene. Unexpectedly, in all nine primary transformants tested the wild-type nos expression was virtually completely suppressed. In contrast, plants transformed with the control vector DNA, which differed only in the absence of the partial nos gene, did not show any inhibition of nos expression. Progeny plants were analyzed for the stability of the gene-silencing phenotype. All of the progeny that carried both the wild-type and partial nos genes had no detectable nopaline synthase activity. In addition, wild-type nos mRNA could not be detected in these plants. In most plants in which the wild-type gene was segregated away from the partial nos gene, wild-type levels of activity were detected. Although DNA methylation has been shown to be correlated with a decrease in promoter activity in plants, none of the progeny appeared to carry a methylated nos promoter. The underlying mechanism causing this gene suppression phenomenon is unclear at this time.
Full text
PDF



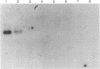
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chandler V. L., Walbot V. DNA modification of a maize transposable element correlates with loss of activity. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1767–1771. doi: 10.1073/pnas.83.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomet P. S., Wessler S., Dellaporta S. L. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 1987 Feb;6(2):295–302. doi: 10.1002/j.1460-2075.1987.tb04753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N. V. About maize transposable elements and development. Cell. 1989 Jan 27;56(2):181–191. doi: 10.1016/0092-8674(89)90891-x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- John M. C., Amasino R. M. Extensive changes in DNA methylation patterns accompany activation of a silent T-DNA ipt gene in Agrobacterium tumefaciens-transformed plant cells. Mol Cell Biol. 1989 Oct;9(10):4298–4303. doi: 10.1128/mcb.9.10.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Dunsmuir P., Bedbrook J. High level expression of introduced chimaeric genes in regenerated transformed plants. EMBO J. 1985 Oct;4(10):2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J. M., Shanklin J., Vierstra R. D., Hershey H. P. Expression of a functional monocotyledonous phytochrome in transgenic tobacco. EMBO J. 1989 Apr;8(4):1005–1012. doi: 10.1002/j.1460-2075.1989.tb03467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini L. M., Bradford S., Rothstein S. Peroxidase-Induced Wilting in Transgenic Tobacco Plants. Plant Cell. 1990 Jan;2(1):7–18. doi: 10.1105/tpc.2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A., Primig M., Trnovsky J., Matzke A. J. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989 Mar;8(3):643–649. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C., Lemieux C., Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990 Apr;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten L. A., Schilperoort R. A. A rapid micro scale method for the detection of lysopine and nopaline dehydrogenase activities. Biochim Biophys Acta. 1978 Dec 8;527(2):497–500. doi: 10.1016/0005-2744(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Dimaio J., Strand M., Rice D. Stable and heritable inhibition of the expression of nopaline synthase in tobacco expressing antisense RNA. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8439–8443. doi: 10.1073/pnas.84.23.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Lahners K. N., Lotstein R. J., Carozzi N. B., Jayne S. M., Rice D. A. Promoter cassettes, antibiotic-resistance genes, and vectors for plant transformation. Gene. 1987;53(2-3):153–161. doi: 10.1016/0378-1119(87)90003-5. [DOI] [PubMed] [Google Scholar]
- Yamada S., Itano H. Phenanthrenequinone as an analytical reagent for arginine and other monosubstituted guanidines. Biochim Biophys Acta. 1966 Dec 28;130(2):538–540. doi: 10.1016/0304-4165(66)90256-x. [DOI] [PubMed] [Google Scholar]
- van der Krol A. R., Mur L. A., Beld M., Mol J. N., Stuitje A. R. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990 Apr;2(4):291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]