Abstract
Molecular factors that contribute to the emergence of new virulent bacterial subclones and epidemics are poorly understood. We hypothesized that analysis of a population-based strain sample of serotype M3 group A Streptococcus (GAS) recovered from patients with invasive infection by using genome-wide investigative methods would provide new insight into this fundamental infectious disease problem. Serotype M3 GAS strains (n = 255) cultured from patients in Ontario, Canada, over 11 years and representing two distinct infection peaks were studied. Genetic diversity was indexed by pulsed-field gel electrophoresis, DNA–DNA microarray, whole-genome PCR scanning, prophage genotyping, targeted gene sequencing, and single-nucleotide polymorphism genotyping. All variation in gene content was attributable to acquisition or loss of prophages, a molecular process that generated unique combinations of proven or putative virulence genes. Distinct serotype M3 genotypes experienced rapid population expansion and caused infections that differed significantly in character and severity. Molecular genetic analysis, combined with immunologic studies, implicated a 4-aa duplication in the extreme N terminus of M protein as a factor contributing to an epidemic wave of serotype M3 invasive infections. This finding has implications for GAS vaccine research. Genome-wide analysis of population-based strain samples cultured from clinically well defined patients is crucial for understanding the molecular events underlying bacterial epidemics.
Keywords: population genetics, evolution, phage, subclone
All species of pathogenic microbes are composed of genetically diverse strains that differ in gene content and allelic diversity (1). These genetic differences can produce variation in pathogen–host interactions, resulting in changes in disease frequency and character (2). Hence, understanding the contribution that an infecting organism makes to the outcome of pathogen–host interactions requires detailed knowledge of microbial gene content and clinical disease characteristics. Various techniques have been used to index genetic diversity among bacterial isolates for study of strain genotype–disease phenotype relationships, population genetics, and evolution (2–5). Although many insights have been obtained, these studies have substantially underestimated genetic diversity among isolates due to limitations in the resolving power of the techniques applied, such as pulsed-field gel electrophoresis (PFGE), multilocus enzyme electrophoresis, and multilocus sequence typing (2–5). Moreover, convenience rather than population-based strain sampling generally has been used, thereby further limiting our understanding of changes in disease frequency and severity.
Group A Streptococcus (GAS) is a human-adapted pathogen that causes diseases ranging in severity from superficial lesions to fulminating invasive infections with high morbidity and mortality (6, 7). GAS can cause localized disease outbreaks that fluctuate in frequency, disease manifestation, and the predominant M protein serotype (6) (M protein is a highly polymorphic surface protein that is antiphagocytic and forms the basis of a commonly used classification scheme for GAS strains). Although no single M type or virulence determinant is uniquely associated with a specific disease, strains expressing certain M proteins have long been associated with certain infection types (6, 7). For example, in most patient populations studied, serotype M3 strains cause a disproportionate number of invasive disease cases, including necrotizing fasciitis, bacteremia, and streptococcal toxic shock syndrome (6, 8–16, ¶). In addition, large prospective population-based studies conducted in the U.S. and Canada have found that serotype M3 strains cause a higher rate of lethal infections than strains of other M types (11–13, ¶). Moreover, serotype M3 and other GAS strains can undergo very rapid shifts in disease frequency and exhibit epidemic behavior. The molecular basis for these phenomena is unknown.
We recently sequenced the genome of a serotype M3 strain (MGAS315) that is genetically representative of the principal clone of M3 isolates causing contemporary episodes of human disease in the U.S., Canada, western Europe, and Japan (8, 17). To gain new insight into the molecular genetic basis of subclone emergence and disease epidemics and to study the relationship between bacterial strain genotype and patient disease phenotype on a genome-wide level, we analyzed 255 serotype M3 invasive isolates collected in an 11-year population-based surveillance study conducted in Ontario, Canada (9–11, 16). The results provided understanding of the molecular events underlying bacterial epidemics.
Materials and Methods
Detailed protocols are provided as Supporting Text, which is published as supporting information on the PNAS web site.
Bacterial Strains. The study was based on 255 M3 strains (Table 1, which is published as supporting information on the PNAS web site) recovered in a prospective population-based surveillance study of GAS invasive infections conducted in Ontario, Canada (population ≈11.4 million), from January 1, 1992, to December 31, 2002. Serotype M3 strain MGAS315 has been well described (8, 17, 18).
emm3 Gene Sequencing. The region of the emm gene encoding amino acids 1–98 of the mature (after cleavage of the secretion signal sequence) M3 protein was sequenced in all 255 strains (19).
PFGE Profile. The PFGE profile was determined for all 255 strains with SmaI (20).
Prophage Genotyping. PCR (Table 2, which is published as supporting information on the PNAS web site) was used to screen all 255 invasive GAS strains for known serotype M3 prophages, the virulence genes encoded by these prophages, and their chromosomal context (21). Previous studies have found speC present in many ET2/M3 strains (8). Therefore, we also screened for the presence of speC and spd1 encoded by Φ370.1 and Φ8232.2 of the sequenced serotype M1 and M18 strains, respectively (22, 23). Cluster analysis of the phage profiles was accomplished with cluster (http://rana.lbl.gov/EisenSoftware.htm) (24).
DNA–DNA Microarray Hybridization. DNA–DNA microarray hybridization (23, 25) was used to assess variation in gene content among a representative subset (n = 33) of the 255 serotype M3 strains from Ontario (Table 3, which is published as supporting information on the PNAS web site).
Whole-Genome PCR Scanning (WGPS). WGPS was recently described as a method to identify previously undetected genome diversity in serotype O157 strains of Escherichia coli causing enterohemorrhagic infections (26). WGPS was used to assess variation in gene content among a representative subset of 19 serotype M3 strains from Ontario (Table 3) by analogous procedures using 634 PCR primer pairs.
Analysis of Variation in the Gene (sclB) Encoding Streptococcal Collagen-Like Protein B. Nucleotide variation in sclB was assessed by PCR amplification and DNA sequence analysis (27).
Single-Nucleotide Polymorphism (SNP) Genotyping. SNPs potentially present in serotype M3 strains were identified by in-silico comparison of the genome sequence of strains MGAS315 and SSI-1 (28) using blastn. Seventy-three putative SNPs (55 SNPs in coding sequences and 18 SNPs in intragenic regions; Table 4, which is published as supporting information on the PNAS web site), located in the core chromosome, were sequenced (Table 5, which is published as supporting information on the PNAS web site) in strain MGAS315 and a representative subset of nine serotype M3 strains from Ontario (Table 3). This analysis identified 15 SNPs (13 in CDS and 2 intergenic) that were polymorphic in the 9 test strains. These 15 SNPs, plus 5 additional putative SNPs in virulence regulatory genes, found to be invariant in the 10 test strains, were analyzed in all 255 invasive serotype M3 Ontario strains by the SNaPshot primer extension method (Applied Biosystems) (Table 6, which is published as supporting information on the PNAS web site) (29).
PCR Analysis of Chromosomal Inversions. PCR was used to test for two large chromosomal inversions present in the genome of some serotype M3 strains (28). Orientation of the chromosomal segments altered by these inversions was determined by PCR amplification of products spanning the inversion recombination junctions (Table 7, which is published as supporting information on the PNAS web site).
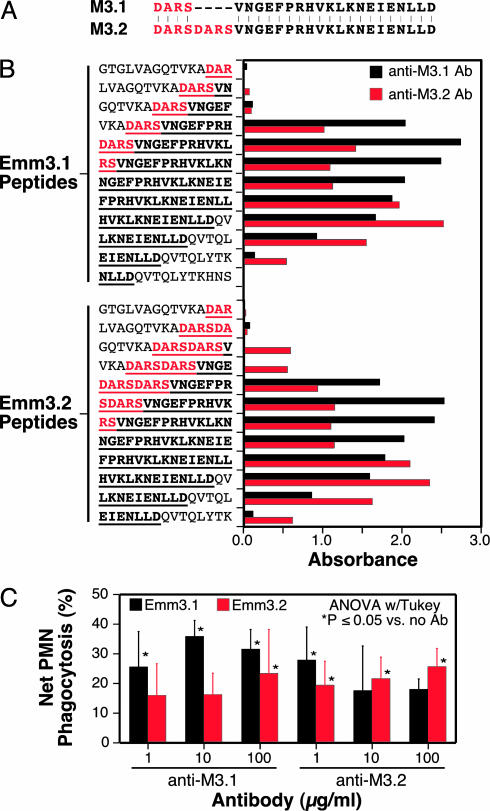
Serologic Analysis of M3 Variants. Thirty-three overlapping 15-mer synthetic peptides (Chiron) spanning the N-terminal variable region of the GAS M3 protein were used. The peptides correspond to amino acids 1–99 of the mature M3 protein, variant Emm3.0. Overlapping 15 mers, corresponding to regions of variation in Emm3.1 and Emm3.2, also were used. Analysis of the reactivity of sera from rabbits immunized with synthetic peptides M3.1 and M3.2 was done in 96-well streptavidin-coated plates (30).
Phagocytosis Assay. Polymorphonuclear leukocytes (PMNs) were isolated from venous blood (31) obtained from healthy donors in accordance with a protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases. Phagocytosis of GAS by human PMNs was assessed as described, with minor modifications (32). Data were analyzed for statistical significance with a one-way ANOVA with Tukey's posttest (instat, GraphPad, San Diego).
Statistical Analysis. Associations among strain molecular genetic characteristics, disease category, and peaks of infection were assessed by using contingency tables and χ2 or Fisher's exact tests of independence. Probabilities calculated with χ2 tests are given as P values, and probabilities calculated with Fisher's exact tests are given as α values.
Results
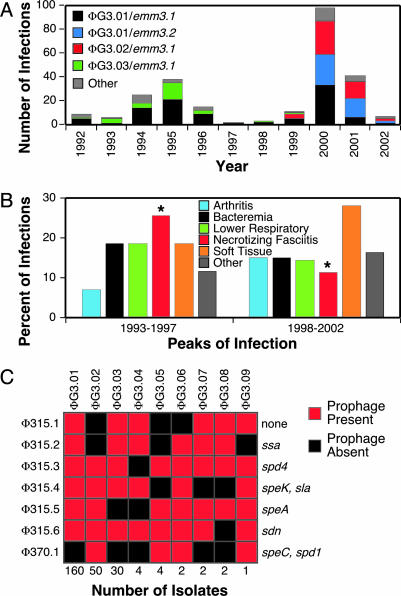
Epidemiologic Overview. Between 1992 and 2002, population-based surveillance identified 255 invasive infections caused by serotype M3 isolates (9–11, 16). The frequency of occurrence of invasive episodes in the 2000 peak (i.e., years 1998–2002) was twice that of the 1995 peak (i.e., years 1993–1997) (Fig. 1A). Five clinical disease categories (soft tissue, lower respiratory tract, necrotizing fasciitis, bacteremia, and arthritis) accounted for 85% of the cases (Table 1). A significantly greater proportion (P = 0.008) of necrotizing fasciitis cases occurred in the 1995 peak compared to the 2000 peak of infection (Fig. 1B).
Fig. 1.
Characteristics of M3 strains studied and infection type. (A) Epidemiologic curve of GAS serotype M3 invasive infections in Ontario, Canada. Stacked columns are color-coded to indicate prophage genotype and emm3 allele. (B) Occurrence of invasive disease types. Illustrated is the percent of the most abundant disease types in the epidemic peaks centered around 1995 and 2000. Significantly more necrotizing fasciitis infections occurred in the 1995 peak than in the 2000 peak (P = 0.008). (C) Prophage and prophage-encoded virulence factor gene content of the isolates. Indicated is the arbitrarily designated prophage genotype (on the top), number of isolates in each prophage genotype (on the bottom), and the prophage content (on the left) and corresponding prophage-encoded virulence factor genes (on the right).
Variation in Genomic PFGE Profile. Seven distinct PFGE patterns were identified and assigned a two-letter designation (Table 1). Most (97%) of the isolates were pattern AA (n = 157, 63%), AX (n = 50, 20%), or AB (n = 34, 14%). The PFGE profiles were distributed nonrandomly over time (P < 0.0001), with virtually all AB strains present in the peak of infection centered around 1995, and all AX strains in the infection peak centered in 2000.
Prophage and Prophage-Encoded Virulence Factor Gene Profiling. All 255 strains had between four and seven prophages, and nine distinct prophage genotypes (ΦGs) were identified by PCR (21) (Fig. 1C and Table 1). ΦG3.01, ΦG3.02, and ΦG3.03 accounted for 94% of the isolates. These three genotypes had unique combinations of the speC, spd1, ssa, and speA genes (Fig. 1C). Each of the prophage-encoded virulence factor genes was integrated adjacent to the chromosomal loci described for reference strain MGAS315 (ssa and speA) and serotype M1 strain SF370 (speC and spd1).
Three major findings were revealed. First, prophage profile ΦG3.01, characterized by the presence of all six prophages present in strain MGAS315, was abundantly represented in both peaks of infections. Second, virtually all ΦG3.03 and ΦG3.02 strains were limited to the peaks of infection centered around 1995 and 2000, respectively (Fig. 1 A). Third, prophage genotypes correlated strongly with PFGE patterns (Table 1).
Sequence Analysis of the emm3 Gene. The N-terminal variable region of M protein is the portion of the molecule against which type-specific immunity is generated (7). Amino acid sequence variation in this region has been identified among isolates of the same M protein serotype (19, 33, 34) and has been associated with variation in opsonophagocytosis and killing of GAS by human PMNs (35–38).
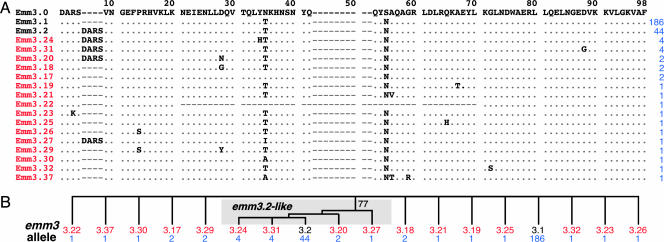
To test the hypothesis that the two peaks of disease in Ontario were linked to variation in the amino acid sequence of the N terminus of the M3 protein, we sequenced the part of the emm3 gene encoding the first 98 aa of the extracellular mature form of the protein in all 255 isolates. Eighteen distinct M3 protein variants were identified (Fig. 2 and Table 1), virtually all explainable by single molecular events such as point mutation or insertion or deletion of short regions of the gene. Emm3.1 and Emm3.2 accounted for 73% and 17% of the isolates, respectively, and were differentiated from each other by a duplication of the first four amino acid residues (D-A-R-S) of the mature M3 protein (Fig. 2 and Table 1).
Fig. 2.
M3 protein variants. (A) The inferred N-terminal amino acid sequences of the 18 emm3 alleles found in this study are shown aligned with the prototype Emm3.0 sequence. The designation of the M3 protein variants (on the left), variants identified in this study (red), and the number of isolates comprising each variant (blue) are indicated. (B) Relationships among emm3 alleles. Phylogenetic reconstruction by the method of neighbor joining was used to generate an unrooted tree by using the emm3 nucleotide sequence encoding amino acids 1–98 of the mature M3 protein. Only alleles encoding Emm3.2-like variants with the D-A-R-S duplication diverged as a genetically related group.
Emm3.1 isolates were present in each year of the study and were proportionally distributed between the two peaks of infection. In striking contrast, isolates with the Emm3.2 variant and related variants Emm3.24 and Emm3.31 (Fig. 2) were not present in the sample until 2000 and consequently were disproportionately distributed between the two epidemic peaks (P < 0.0001) (Fig. 1 A).
Analysis of Variation in Chromosomal Gene Content. DNA–DNA microarray analysis was used to assess the extent of variation in chromosomal gene content among the serotype M3 strains. Because DNA–DNA microarray is labor intensive, and the results of the other genomic variation studies suggested the presence of a relatively limited number of distinct clones, we analyzed 33 of the 255 strains (Table 3). These 33 strains were selected to represent broad temporal distribution and variation in PFGE pattern, disease type, and prophage-encoded virulence gene content. All strains had a core-gene content identical to serotype M3 strain MGAS315. All differences in gene content were located in regions of the genome that contain prophages in the sequenced serotype M1, M3, or M18 strains. There was complete concordance between the serotype M3 subclones identified by prophage PCR profiling and DNA–DNA microarray analysis (data not shown).
WGPS of Serotype M3 GAS Strains. A limitation of DNA–DNA microarray analysis is that it fails to reveal genes or gene segments present in the test strain but absent from the genomes of strains used to formulate the microarray. Thus, it is possible that strains studied by microarray contain previously uncharacterized DNA segments that may contribute to pathogenesis but would not be identified by DNA–DNA microarray analysis. The problem can be circumvented with WGPS (26).
To test the hypothesis that uncharacterized genetic content contributed to genome diversity among the Ontario serotype M3 strains, we analyzed 19 of the 255 isolates (Table 3). These 19 strains were selected from the 33 strains examined by DNA–DNA microarray and represent the major subclones identified by the other molecular genetic methods. Although PCR size differences as small as 200 bp were detected, very little additional genetic diversity was detected by WGPS. Size variation was identified in 1 of the 634 PCR products, in the region corresponding to sclB (Fig. 7, which is published as supporting information on the PNAS web site).
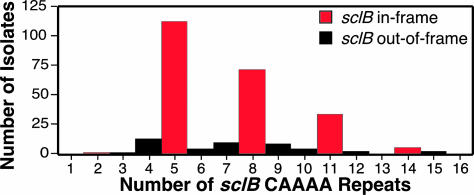
SclB is a collagen-like surface protein that has been implicated in host–pathogen interactions (27, 39). Sequence variation at the 5′ end of sclB was due to differences in the number of CAAAA nucleotide repeats (range, 2–15) located in the gene region directly following the start codon (Fig. 7). Most (84%) strains had 5, 8, 11, or 14 CAAAA repeats (Fig. 3), numbers that result in in-frame alleles of sclB, and the capacity to produce full-length SclB. These results suggest that strains with the potential to express full-length SclB have a selective advantage over strains making a truncated SclB, consistent with a role in host–pathogen interactions (27, 39). The sequence of sclB located 3′ of the collagen structural motif (CSM)-encoding domain was virtually invariant among the 255 isolates. Most of the variation in sclB amplicon size was attributable to variation in the region of the gene encoding the CSM domain. The number of Gly-X-Y repeats in the CSM domain varied from 10 to ≈220 among the 255 serotype M3 isolates (Table 1).
Fig. 3.
Distribution of sclB CAAAA nucleotide repeats in the 255 isolates. The 5′ end of the sclB gene was sequenced in all 255 isolates, and the number of CAAAA pentanucleotide repeats was determined.
There was no simple association between occurrence of in-frame or out-of-frame sclB alleles and infection peak, prophage genotype, emm3 allele, or disease phenotype (data not shown). This result is consistent with the idea that a transition between in-frame and out-of-frame alleles occurs very rapidly in natural populations. The lack of nucleotide sequence variation in the 5′ and 3′ ends of sclB also is consistent with this idea. In contrast, the distribution of strains with 5, 8, or 11 CAAAA repeats varied significantly across peaks of infection, prophage genotype, emm3 allele, but not with disease phenotype (P = 0.81). For example, strains with eight CAAAA repeats were overrepresented among organisms with Emm3.2 variants (P < 0.001).
PCR-Based Analysis of Large Chromosomal Inversions. The genome sequences of serotype M3 strains MGAS315 and SSI-1 are very closely related (17, 28). The most prominent difference was a rearrangement of the genome of strain SSI-1 caused by two large chromosomal inversions (28) (Fig. 8, which is published as supporting information on the PNAS web site). It was speculated that the resurgence of rheumatic fever and severe invasive infections in Japan was associated with the emergence of strains with this genome configuration (28).
To determine whether these genome rearrangements were associated with distinct M3 subclones, a PCR-based strategy was used. Three amplicon patterns were identified with the first inversion, arbitrarily designated pattern A, B, and AB (Fig. 8A). Among the 255 strains studied, 47 (19%) had pattern A, and 142 (57%) had pattern B. Importantly, 59 (24%) strains had an AB pattern, indicating a mixture of both genome arrangements. Most of these 59 strains had a dominant pattern, that is, the pattern was primarily A or B. Taken together, these data suggest that this chromosomal inversion occurs relatively frequently during in vitro growth. There was no significant association of inversion pattern and infection category (χ2 = 15.4, P = 0.12).
PCR amplification of products spanning the chromosomal junctions demarcating the second inversion was performed on all 198 isolates for which prophage PCR screening indicated the presence of both Φ315.1 and Φ315.2. Three amplicon patterns were obtained, arbitrarily designated C, D, and CD (Fig. 8B). Virtually all strains (n = 193) had pattern D, the configuration present in strain SSI-1. Hence, the results do not support the contention (28) that the first chromosomal inversion induced the second. We believe it is more likely that the two processes are independent events that occur at different frequencies.
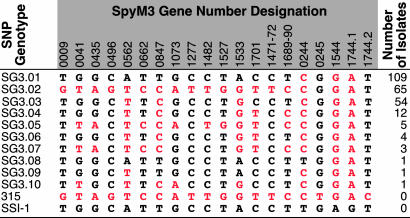
SNP Analysis. Genetic relationships among strains can be inferred on the basis of analysis of SNPs (29). Twenty SNPs were analyzed in all 255 serotype M3 isolates, and 10 distinct SNP genotypes, designated SG3.01–SG3.10 in order of abundance, were identified (Fig. 4). SG3.01, SG3.02, and SG3.03 accounted for 89% of the isolates. SG3.01 and SG3.02 strains were present in both 1995 and 2000 epidemic peaks, but virtually all SG3.03 strains were found only in the 2000 peak. SG3.01 and SG3.02 strains were predominately ΦG3.01 or ΦG3.03, whereas virtually all SG3.03 strains were ΦG3.02. SG3.02 strains were significantly overrepresented in necrotizing fasciitis infections (P = 0.010). SG3.03 strains were overrepresented in soft tissue infections (P = 0.015) but underrepresented in lower respiratory tract infections (P = 0.014). Thus, SNP genotypes were significantly associated with epidemic peaks, prophage genotypes, and infection categories.
Fig. 4.
SNP genotypes identified among the 255 M3 isolates. SNP genotypes (SGs) based on nucleotides present at 20 sites are shown. 315 refers to strain MGAS315, and SSI-1 refers to strain SSI-1.
Analysis of Variation in Immune Recognition and Phagocytosis Between Emm3.1 and Emm3.2. In principle, the Emm3.2 protein could represent an escape variant that arose from an Emm3.1 precursor by host immune selection. If this were the case, we expect that Emm3.1 and Emm3.2 would differ in immunologic properties, such as serologic reactivity. Consistent with this idea, linear epitope mapping with rabbit antisera raised against synthetic peptides revealed differential reactivity to the peptides representing the extreme N terminus of Emm3.1 and Emm3.2 (Fig. 5A). Anti-M3.1 antibody reacted with an epitope located toward the N terminus of the immunizing peptides (Fig. 5B). In contrast, anti-M3.2 antibodies reacted with an epitope located toward the C terminus of the immunizing peptides (Fig. 5B). In addition, anti-M3.1 and anti-M3.2 antibodies differed in reactivity to the duplicated D-A-R-S sequence. Only anti-M3.2 antibodies reacted with the first 12 aa of Emm3.2 (Fig. 5B).
Fig. 5.
Immunologic analysis of Emm3.1 and Emm3.2. (A) Emm3 synthetic peptides used to immunize rabbits. M3.1 and M3.2 peptides correspond to the first 24 and 28 aa of mature Emm3.1 and Emm3.2, respectively. The first four amino acids of mature Emm3.1, which are duplicated in Emm3.2, are shown in red. (B) ELISA reactivity of rabbit antibodies with Emm3.1 and Emm3.2 peptides. Affinity-purified rabbit anti-Emm3 peptide antibodies were diluted 1:80,000. Underlined amino acids correspond to the peptides used to immunize rabbits. (C) Human PMN phagocytosis studies. Strains MGAS3392 (Emm3.1) and MGAS9887 (Emm3.2) were opsonized with either rabbit anti-M3.1 or anti-M3.2 antibodies at the indicated concentrations and incubated with human PMNs. Values are the mean of five to six independent assays using PMNs obtained from different donors. Error bars show the standard error.
Next, we compared the ability of human PMNs to phagocytosize strain MGAS3392 (Emm3.1) or strain MGAS9887 (Emm3.2) opsonized with anti-M3.1 or anti-M3.2 antibodies (Fig. 5C). In the aggregate, phagocytosis of strain MGAS3392 was greater than strain MGAS9887 at all concentrations of anti-M3.1 antibody tested (P = 0.003 at 10 μg/ml, t test). In contrast, phagocytosis of strain MGAS9887 was not consistently higher than strain MGAS3392. Taken together, the data suggest that the N termini of Emm3.1 and Emm3.2 differ sufficiently in immunologic character such that anti-M3.1 antibodies recognize Emm3.2 less well than Emm3.1.
Conclusion
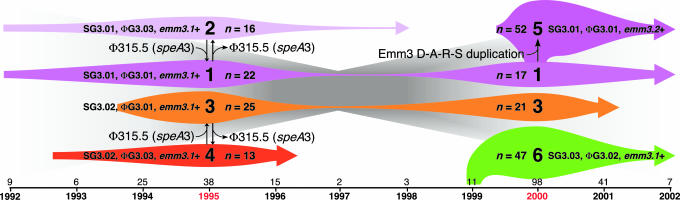
The primary goal of our study was to gain new insight into the molecular genetic factors that bear on the emergence of virulent subclones and epidemics using the model pathogen GAS. A population-based strain sample was used that was composed of virtually all serotype M3 GAS strains causing invasive episodes in Ontario from 1992 to 2002. Our data implicate three contributory factors (Fig. 6). First, acquisition and loss of prophages is the major generator of distinct genotypes with novel combinations of proven and putative virulence factor genes. These distinct genotypes can undergo very rapid population expansion and cause infections that differ significantly in character (Fig. 9 and Table 8, which are published as supporting information on the PNAS web site).
Fig. 6.
Schematic showing summary of temporal changes in serotype M3 subclones. The six major serotype M3 subclones defined by difference in distribution of SNPs, prophage content, and/or emm3 allele, identified among the isolates are shown (large font). Colored arrow length reflects the temporal distribution, and colored arrow height reflects the relative abundance of the subclones. The number of isolates of each subclone in the 1995 and 2000 epidemic peaks are given, and the total annual number of isolates is shown above the time line (on the bottom).
Second, a critical observation was that two subclones (2 and 4 in Fig. 6) lacking the prophage encoding the speA gene were recovered only in the first epidemic peak centered around 1995. In contrast, two subclones (1 and 3 in Fig. 6) containing this prophage were prominent causes of disease in both epidemic peaks. Subclones 5 and 6 (Fig. 6) that increased greatly in frequency in the epidemic peak centered around 2000 also had the SpeA-encoding prophage. Taken together, the data support the hypothesis that serotype M3 isolates with this prophage are more fit than organisms that lack this prophage. The data are consistent with a model in which loss of the speA-containing prophage results in a less fit (and potentially less virulent) organism that is more prone to undergo clonal extinction, presumably because it is less abundant in natural populations. In this regard, the model is consistent with data indicating that speA-containing GAS are significantly more likely to cause recurrent pharyngitis than are GAS lacking this gene (40).
Third, duplication of four amino acids located at the extreme N terminus of the M protein was the only molecular change we identified in all strains representing an abundantly occurring M3 subclone that rose to great prominence in the peak of invasive episodes centered around 2000. This 4-aa duplication conferred altered immune recognition to M protein. This fact, together with the observation of extreme underrepresentation of synonymous (silent) nucleotide changes in M protein in natural populations (19, 33, 34), rapid change in M protein structure in epidemiologically linked patients (41, 42), and ability of sequence changes in the N terminus of the M protein to alter the efficiency of phagocytosis and killing of GAS by human PMNs (35–38), strongly suggests that the Emm3.2 variant rose to prominence as a consequence of host selective pressure rather than by chance alone. Inasmuch as GAS initially interacts with many hosts in the oral cavity and epithelial surfaces in the posterior pharynx, we believe that the selection occurs in the upper respiratory tract. Hence, these findings have implications for GAS vaccines that are based on N-terminal M protein antigens.
In conclusion, our genome-wide analysis revealed a hitherto unknown complexity of the molecular population genetics of strains of a single GAS M protein serotype. Distinct serotype M3 genotypes experienced rapid population expansion and caused infections that differed significantly in character and severity. The molecular genetic analysis, combined with immunologic studies, implicated a 4-aa duplication in the extreme N terminus of M protein as a factor contributing to a new epidemic wave of serotype M3 invasive infections. Study of other microbial pathogens by the general strategy we used will be a very fruitful line of investigation.
Acknowledgments
We thank A. Henion and A. Mora for assistance with statistical analysis and graphics, respectively.
Abbreviations: GAS, group A Streptococcus; WGPS, whole-genome PCR scanning; SNP, single-nucleotide polymorphism; PFGE, pulsed-field gel electrophoresis; PMN, polymorphonuclear leukocyte.
Footnotes
Cowgill, K. D., Van Beneden, C., Wright, C., Beall, B. & Schuchat, A. (2003) 41st Annual Meeting of the Infectious Disease Society of America, p. 238 (abstr.).
References
- 1.Blaser, M. J. & Musser, J. M. (2001) J. Clin. Invest. 107, 391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musser, J. M. (1996) Emerg. Infect. Dis. 2, 1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selander, R. K., Caugant, D. A., Ochman, H., Musser, J. M. Gilmour, M. N. & Whittam, T. S. (1986) Appl. Environ. Microbiol. 51, 873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenover, F. C., Arbiet, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H. & Swaminathan, B. (1995) J. Clin. Microbiol. 33, 2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright, M. C., Spratt, B. G., Kalia, A., Cross, J. H. & Bessen, D. E. (2001) Infect. Immun. 69, 2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musser, J. M. & Krause, R. M. (1998) in Emerging Infections, ed. Krause, R. M. (Academic, New York), pp. 185-218.
- 7.Cunningham, M. W. (2000) Clin. Microbiol. Rev. 13, 470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musser, J. M., Hauser, A. R., Kim, M. H., Schlievert, P. M., Nelson, K. & Selander, R. K. (1991) Proc. Natl. Acad. Sci. USA 88, 2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, H. D., McGeer, A., Schwartz, B., Green, K., Cann, D., Simor, A. & Low, D. E. (1996) N. Engl. J. Med. 335, 547-554. [DOI] [PubMed] [Google Scholar]
- 10.Kaul, R., McGeer, A., Low, D. E., Green, K. & Schwartz, B. (1997) Am. J. Med. 103, 18-24. [DOI] [PubMed] [Google Scholar]
- 11.Sharkawy, A., Low, D. E., Saginur, R., Gregson, D., Schwartz, B., Jessamine, P., Green, K., McGeer, A. & Ontario Group A Streptococcal Study Group (2002) Clin. Infect. Dis. 34, 454-460. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien, K. L., Beall, B., Barrett, N. L., Cieslak, P. R., Reingold, A., Farley, M. M., Danila, R., Zell, E. R., Facklam, R., Schwartz, B., et al. (2002) Clin. Infect. Dis. 35, 268-276. [DOI] [PubMed] [Google Scholar]
- 13.Li, Z., Sakota, V., Jackson, D., Franklin, A. R. & Beall, B. (2003) J. Infect. Dis. 188, 1587-1592. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz, F.-J., Beyer, A., Charpentier, E., Normark, B. H., Schade, M., Fluit, A. C., Hafner & Novak, R. (2003) J. Infect. Dis. 188, 1578-1586. [DOI] [PubMed] [Google Scholar]
- 15.Moses, A. E., Hidalgo-Grass, C., Dan-Goor, M., Jaffe, J., Shetzigovsky, I., Ravins, M., Korenman, Z., Cohen-Poradosu, R. & Nir-Paz, R. (2003) J. Clin. Microbiol. 41, 4655-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller, M. P., Low, D. E., Green, K. A., Simor, A. E., Loeb, M., Gregson, D., McGeer, A. & Ontario Group A Streptococcal Study (2003) Arch. Intern. Med. 163, 467-472. [DOI] [PubMed] [Google Scholar]
- 17.Beres, S. B., Sylva, G. L., Barbian, K. D., Lei, B., Hoff, J. S., Mammarella, N. D., Liu, M.-Y., Smoot, J. C., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banks, D. J., Lei, B. & Musser, J. M. (2003) Infect. Immun. 71, 7079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musser, J. M., Kapur, V., Szeto, J., Pan, X., Swanson, D. S. & Martin, D. R. (1995) Infect. Immun. 63, 994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Single, L. A. & Martin, D. R. (1992) FEMS Microbiol. Lett. 91, 85-90. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto, M., Hoe, N. P., Liu, M., Beres, S. B., Sylva, G. L., Brandt, C. M., Haase, G. & Musser, J. M. (2003) J. Infect. Dis. 187, 604-612. [DOI] [PubMed] [Google Scholar]
- 22.Ferretti, J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., Primeaux, C., Sezate, S., Suvorov, A. N., Kenton, S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smoot, J. C., Barbian, K. D., Van Gompel, J. J., Smoot, L. M., Chaussee, M. S., Sylva, G. L., Sturdevant, D. E., Ricklefs, S. M., Porcella, S. F., Parkins, L. D., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald, J. R., Sturdevant, D. E., Mackie, S. M., Gill, S. R. & Musser, J. M. (2001) Proc. Natl. Acad. Sci. USA 98, 8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi, M., Terajima, J., Kurokawa, K., Nakayama, K., Murata, T., Tamura, K., Ogura, Y., Watanabe, H. & Hayashi, T. (2002) Proc. Natl. Acad. Sci. USA 99, 17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukomski, S., Nakashima, K., Abdi, I., Cipriano, V. J., Shelvin, B. J., Graviss, E. A. & Musser, J. M. (2001) Infect. Immun. 69, 1729-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, I., Kurokawa, K., Yamashita, A., Nakata, M., Tomiyasu, Y., Okahashi, N., Kawabata, S., Yamazaki, K., Shiba, T., Yasunga, T., et al. (2003) Genome Res. 13, 1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutacker, M. M., Smoot, J. C., Lux Migliaccio, C. A., Ricklefs, S. M., Hua, S., Cousins, D. V., Graviss, E. A., Shashkina, E., Kreiswirth, B. N. & Musser, J. M. (2002) Genetics 162, 1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoe, N. P., Kordari, P., Cole, R., Liu, M., Palzkill, T., Huang, W., McLellan, D., Adams, G., Hu, M., Vuopio-Varkila, J., Cate, T. R., et al. (2000) J. Infect. Dis. 182, 1425-1436. [DOI] [PubMed] [Google Scholar]
- 31.Boyum, A. (1968) Scand. J. Clin. Lab. Invest. Suppl. 97, 77-89. [PubMed] [Google Scholar]
- 32.Lei, B., DeLeo, F. R., Hoe, N. P., Graham, M. R., Mackie, S. M., Cole, R. L., Liu, M., Hill, H. R., Low, D. E., Federle, M. J., et al.. (2001) Nat. Med. 7, 1298-1305. [DOI] [PubMed] [Google Scholar]
- 33.Hoe, N., Nakashima, K., Grigsby, D., Pan, X., Dou, S. J., Naidich, S., Garcia, M., Kahn, E., Bergmire-Sweat, D. & Musser, J. M. (1999) Emerg. Infect. Dis. 5, 254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoe, N.P., Nakashima, K., Lukomski, S., Grigsby, D., Liu, M., Kordari, P., Dou, S.-J., Pan, X., Vuopio-Varkila, J., Salmelinna, S., et al. (1999) Nat. Med. 5, 924-929. [DOI] [PubMed] [Google Scholar]
- 35.Harbaugh, M. P., Podbielski, A., Hugl, S. & Cleary, P. P. (1993) Mol. Microbiol. 8, 981-991. [DOI] [PubMed] [Google Scholar]
- 36.de Malmanche, S. A. & Martin D. R. (1994) Med. Microbiol. Immunol. 183, 299-306. [DOI] [PubMed] [Google Scholar]
- 37.Villasenor-Sierra, A., McShan, W. M., Salmi, D., Kaplan, E. L., Johnson, D. R. & Stevens, D. L. (1999) J. Infect. Dis. 180, 1921-1928. [DOI] [PubMed] [Google Scholar]
- 38.Eriksson, B. K. G., Villasenor-Sierra, A., Norgren, M. & Stevens, D. L. (2001) Clin. Infect. Dis. 32, e24-e30. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen, M. & Bjorck, L. (2001) Mol. Microbiol. 40, 1427-1438. [DOI] [PubMed] [Google Scholar]
- 40.Musser, J. M., Gray, B. M., Schlievert, P. M. & Pichichero, M. E. (1992) J. Clin. Microbiol. 30, 600-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischetti, V. A., Jarymowycz, M., Jones, K. F. & Scott, J. R. (1986) J. Exp. Med. 164, 971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollingshead, S. K., Fischetti, V. A. & Scott, J. R. (1987) Mol. Gen. Genet. 207, 196-203. [DOI] [PubMed] [Google Scholar]