Abstract
The persistent tetrodotoxin (TTX)-sensitive sodium current, detected in neurons of many regions of mammalian brains, is associated with many essential neuronal activities, including boosting of excitatory synaptic inputs, acceleration of firing rates, and promotion of oscillatory neuronal activities. However, the origin and molecular basis of the persistent current have remained controversial for decades. Here, we provide direct evidence that U-to-C RNA editing of an insect sodium channel transcript generates a sodium channel with a persistent current. We detected a persistent TTX-sensitive current in a splice variant of the cockroach sodium channel gene BgNav (formerly paraCSMA). Site-directed mutagenesis experiments revealed that an F-to-S change at the C-terminal domain of this variant was responsible for the persistent current. We demonstrated that this F-to-S change was the result of a U-to-C RNA editing event, which also occurred in the Drosophila para sodium channel transcript. Our work provides direct support for the hypothesis that posttranscriptional modification of a conventional transient sodium channel produces a persistent TTX-sensitive sodium channel.
The voltage-gated sodium channel is responsible for the rising phase of the action potential in the membranes of neurons and other excitable cells (1). It typically activates and then completely inactivates within a few milliseconds, the latter of which contributes to the rapid repolarization of action potentials. However, substantial evidence indicates the existence of persistent (i.e., noninactivating) tetrodotoxin (TTX)-sensitive sodium currents in the neurons of many regions of mammalian brains, such as hippocampal neurons (2), cortical pyramidal cells (3), suprachiasmatic nucleus (4), cerebellar Purkinje neurons (5), and tuberomammillary neurons (6), and in invertebrate neurons, such as squid giant axons (7, 8), spontaneously active heart interneurons of the leech (Hirudo medicinalis) (9), and terminal abdominal efferent dorsal unpaired median neurons of the cockroach (Periplaneta americana) (10). Persistent sodium current is believed to play an important role in the boosting of excitatory synaptic inputs, acceleration of firing rates, and promotion of oscillatory neuronal activities (11, 12). However, the origin of the persistent current has remained controversial for decades. Some studies support the notion that the persistent current is mediated by an unidentified sodium channel isoform distinct from the conventional sodium channels, whereas others suggest that this current reflects a noninactivating gating mode of conventional transient sodium channels (11–13).
RNA editing is an important posttranscriptional modification that has received increasing attention in the last decade (14–16). RNA editing occurs by changes in individual nucleotides, such as an A-to-I or C-to-U conversion, in gene transcripts, leading to changes in protein structure and function. Interestingly, A-to-I RNA editing occurs mainly in gene transcripts encoding ion channels, neurotransmitter receptors, or G protein-coupled receptors in the nervous system (16). The well characterized examples include A-to-I editing in mammalian glutamate-gated receptor channels (GluR) that mediate excitatory synaptic transmission in the CNS (17), mammalian serotonin 2C receptor (5HT2CR) (18, 19), and squid voltage-gated potassium channels (20, 21). In each case, edited channels or receptors exhibit distinct functional and/or pharmacological properties. Ten A-to-I RNA editing sites have been found in the para sodium channel transcripts of Drosophila melanogaster (22), and some of these sites are also conserved in Drosophila virilis (23). Drosophila mutants with deletion of the gene encoding the A-to-I editing enzyme, ADAR, exhibit abnormal behavior, demonstrating the biological importance of RNA editing (22). However, the functional consequence of A-to-I RNA editing of sodium channel gene transcripts remains unknown.
In this study, we identified a cockroach sodium channel splice variant that exhibited a persistent TTX-sensitive current and associated gating properties. We further showed that a U-to-C RNA editing event was responsible for generating this persistent TTX-sensitive sodium current, suggesting a major role of U-to-C RNA editing in regulating sodium channel function.
Materials and Methods
Molecular Analysis. Total RNA was isolated from tissues of German cockroaches (Blattella germanica), including head/thorax, leg, ovary, gut, and nerve cord, and adult D. melanogaster by using the TRIzol Reagent kit (Invitrogen). Methods for first-strand cDNA synthesis, RT-PCR, cloning of a full-length cDNA, and PCR-amplification of genomic DNA were identical to those described by Tan et al. (24).
A sodium channel splice variant, named BgNav4, was isolated from the German cockroach. It contains eight scattered amino acid changes compared with the published BgNav sequence (25) (GenBank accession no. U73583): E428G, I700V, Q843H, V975A, I1663M, M1823V, D1846G, and F1919S. Site-directed mutagenesis was performed by using the Altered Sites II In Vitro Mutagenesis System (Promega) to individually replace the amino acid changes in BgNav4 with the corresponding amino acid residues in the published sequence. Briefly, a 1.2-kb EcoRV fragment, which contains V700, H843, and A975, was excised from BgNav4 and subcloned into pAlter 1. Site-directed mutagenesis was performed according to the manufacturer's instructions. The resultant fragment containing each mutation, I700, Q843, or V975, was subcloned back to BgNav4. Clones containing H843 or A975 were isolated, but clones containing I700 were not recovered. By using the same strategy, a 1.6-kb KpnI/AccI fragment containing G428 and a 1.7-kb HindIII fragment containing M1663, V1823, G1846, and S1919 were used for site-directed mutagenesis. Mutants containing M1823, D1846, or F1919 were isolated, but clones containing I1663 were not recovered.
For identification of the U-to-C editing site at nucleotide 5756 in BgNav, which results in an F-to-S change at amino acid 1919, a 300-bp PCR fragment encoding part of the C-terminal domain (I1854 to S1950) was amplified by using sense primer 5561tttggcagcagtttgacccagatgg5585 and antisense primer 5851tagaggacacaggctcatagc5831. For identification of the same U-to-C editing site in the Drosophila para gene, a 264-bp PCR fragment was amplified by using sense primer 5699atcagctgtccgaattcctgg5719 and antisense primer 5941tagaggacacaggctcatagc5921, which encodes part of the C-terminal domain (D1900 to G1980) of the Para protein (26) (GenBank accession no. M32078). These PCR fragments were cloned into pCR2.1-TOPO by using an A-T cloning kit (Invitrogen). To determine the frequency of the U-to-C editing event in the cockroach terminal abdominal ganglion and ovary, 561 ganglia and 553 ovary cDNA clones were isolated and analyzed by using the single-nucleotide polymorphism (SNP) PCR detection method, described below, to determine the presence of C or T at nucleotide 5756. Briefly, a 93-bp fragment was amplified from each clone by using TaqMan Universal PCR Master Mix (Applied Biosystems) and two SNP PCR primers, 5719tgtgtggacattttggatgctt5740 (sense primer) and 5791tggtggcacttcacccaactc5811 (antisense primer). Two SNP probes were made at Applied Biosystems, VIC-5765ttccttgcaaagaagtc5749-MGB and FAM-5765ttccttgcagagaagtc5749, and were added in the PCR mix for the detection of the T/C polymorphism at nucleotide 5756. PCR results were analyzed by using the Prism 7700 Sequence Detection System (Applied Biosystems). For further confirmation, 15% of these clones were sequenced at the Genomic Technology Service Facility of Michigan State University.
Expression of BgNav Sodium Channels in Xenopus Oocytes. Procedures for oocyte preparation and cRNA injection were identical to those described by Tan et al. (24). Briefly, oocytes were obtained surgically from oocyte-positive female Xenopus laevis (Xenopus I, Ann Arbor, MI) and incubated with 1 mg/ml type IA collagenase (Sigma) in a Ca2+-free ND96 medium, which contained 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 5 mM Hepes (pH 7.5). Follicle cells remaining on the oocytes were removed with forceps. Isolated oocytes were incubated in ND96 medium containing 1.8 mM CaCl2 supplemented with 50 μg/ml gentamicin, 5 mM pyruvate, and 0.5 mM theophylline. Healthy stage V–VI oocytes were used for cRNA injection. For robust expression of the cockroach BgNav sodium channel, BgNav cRNA (0.2–2 ng) was coinjected into oocytes with D. melanogaster tipE cRNA (0.2–2 ng), which is known to enhance the expression of insect sodium channels in oocytes (27, 28).
Electrophysiological Recording and Analysis. Methods for electrophysiological recording and data analysis were similar to those described in ref. 24. Sodium currents were recorded by using standard two-electrode voltage clamping. Capacitive transient and linear leak currents were corrected by using the P/N subtraction technique or by subtraction of records obtained in the presence of 20 nM TTX, which completely blocks BgNav sodium channels. The voltage dependence of sodium channel conductance (G) was calculated by measuring the peak current at test potentials ranging from -80 mV to +65 mV in 5-mV increments and divided by (V - Vrev), where V is the test potential and Vrev is the reversal potential for sodium ion. Peak conductance values were normalized to the maximal peak conductance (Gmax) and fitted with a two-state Boltzmann equation of the form G/Gmax = [1 + exp (V - V1/2)/k]-1, in which V is the potential of the voltage pulse, V1/2 is the half maximal voltage for activation, and k is the slope factor.
The voltage dependence of sodium channel inactivation was determined by using 200-ms inactivating prepulses from a holding potential of -120–0 mV in 5-mV increments, followed by test pulses to -10 mV for 20 ms. The peak current amplitude during the test depolarization was normalized to the maximum current amplitude and plotted as a function of the prepulse potential. Data were fitted with a two-state Boltzmann equation of the form I/Imax = [1 + (exp(V - V1/2)/k)]-1, in which Imax is the maximal current evoked, V is the potential of the voltage pulse, V1/2 is the half maximal voltage for inactivation, and k is the slope factor.
To determine the recovery from fast inactivation, sodium channels were inactivated by a 200-ms depolarizing pulse to -10 mV, then repolarized to -120 mV for an interval of variable duration, followed by a 20-ms test pulse to -10 mV. The peak current during the test pulse was divided by the peak current during the inactivating pulse and plotted as a function of duration time between the two pulses. To determine the time constant for recovery, the curve was fitted by using double exponential function (I = 1 - [A1 × exp(-t/τ1) + A2 × exp(-t/τ2)], in which A1 and A2 are the relative proportions of current recovering with time constants τ1 and τ2, and t is the recovery interval.
Results
Identification of a Cockroach Sodium Channel Isoform. In a previous study, we identified three splice variants, BgNav1, BgNav2, and BgNav3 (formerly KD1, KD2, and KD3), that exhibit distinct gating and pharmacological properties (24). BgNav1, BgNav2, and BgNav3 each contain one of the three mutually exclusive exons (G1, G2, and G3) that encode IIIS3–4. In addition, BgNav1 contains optional exon J (24). In this study, we isolated a previously unrecognized splice variant, BgNav4, by using the same RT-PCR cloning strategy. Like BgNav1, BgNav4 contains exon G1 but lacks optional exon J. Furthermore, BgNav4 contains eight scattered amino acid changes compared with the published BgNav sequence (25) (GenBank accession no. U73583): E428G, I700V, Q843H, V975A, I1663M, M1823V, D1846G, and F1919S. None of these eight amino acid changes was present in BgNav1, -2, or -3.
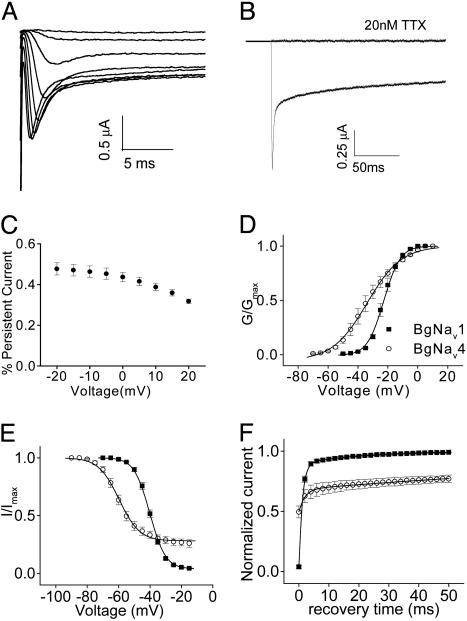
In contrast to previously characterized cockroach sodium channel splice variants BgNav1 and -2 (24), BgNav4 exhibited a noninactivating TTX-sensitive current (i.e., persistent current) during a 20-ms depolarization at test potentials from -60 to 10 mV (Fig. 1A). The persistent current was maintained during a long depolarization of 200 ms (Fig. 1B). This current was from sodium channel activity because it was completely blocked by the application of 20 nM TTX (Fig. 1B). Persistent currents were measured 15 ms into the 20-ms depolarization steps. The persistent current in percent of peak current was ≈50% at -20 mV and decreased gradually with increasing depolarizing voltages (Fig. 1C).
Fig. 1.
Unique gating properties of BgNav4. (A) Sodium current traces from a representative oocyte expressing BgNav4. Currents were recorded from a 20-ms depolarization of a series of depolarizing voltages (-65–10 mV) from the holding potential of -120 mV. (B) Sodium current recorded from a 200-ms depolarization to -10 mV from the holding potential of -120 mV. The current was completely blocked by 20 nM TTX. (C) Voltage dependence of the persistent current. The persistent current was measured 15 ms into a 20-ms depolarization, normalized by the amplitude of peak current, and plotted against the depolarizing voltages. (D) Voltage dependence of activation of BgNav4 compared with BgNav1 (18). (E) Voltage dependence of steady-state inactivation of BgNav4 compared with BgNav1. (F) Recovery from fast inactivation of BgNav4 compared with BgNav1. The protocols for recording these gating properties are described in Materials and Methods.
We next determined the voltage dependence of activation and steady-state inactivation and recovery from fast inactivation of BgNav4. The conductance curve was flatter than that of BgNav1, with a k value of 11 mV (Fig. 1D). Consequently, although BgNav4 can be activated at approximately -60 mV, 90% of activation was not reached until -10 mV (Fig. 1D). The voltage dependence of the inactivation curve revealed incomplete inactivation at positive potentials (Fig. 1E), consistent with the detection of the persistent current (Fig. 1 A). The recovery from fast inactivation exhibited two components, one with the same time constant as the fast component of other BgNav variants and the other with significantly slowed recovery that is minimal in other BgNav variants (Fig. 1F). Both persistent TTX-sensitive sodium current and these associated gating properties of BgNav4 are similar to characteristic features of persistent TTX-sensitive currents documented in many vertebrate and invertebrate neurons (12, 29).
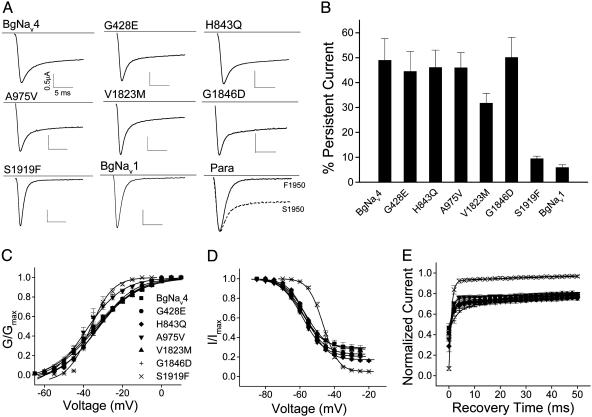
S1919 Is Responsible for the Persistent Current in BgNav4. BgNav4 contains eight scattered amino acid changes compared with the published BgNav sequence (25) (GenBank accession no. U73583): E428G, I700V, Q843H, V975A, I1663M, M1823V, D1846G, and F1919S. To identify the amino acid change(s) that contributed to the unique channel properties of BgNav4, we produced six recombinant channels by individually changing these residues to the corresponding residues in the published BgNav sequence by using site-directed mutagenesis. We were unable to construct full-length clones containing mutation V700I or M1663I. The other six mutants, G428E (i.e., G in BgNav4 to E in the published BgNav sequence), H843Q, A975V, V1823M, G1846D, and S1919F, produced sufficient sodium currents for further functional analysis. The gating properties of the six mutant channels were characterized and compared with those of the parental channel BgNav4 (Fig. 2 and Table 1). One mutation, S1919F, completely abolished the persistent current (Fig. 2 A and B). Furthermore, this mutation also converted the other unique gating properties associated with BgNav4 (Fig. 1) to those of the conventional sodium channels, such as BgNav1. The S1919F channel was activated at -50 mV instead of -60 mV for the BgNav4 channel (Fig. 2C). S1919F also restored the steepness of voltage dependence of activation from 11 to 4 mV (Table 1 and Fig. 2C). Moreover, the S1919F channel exhibited a faster rate of recovery from fast inactivation and an ≈10-mV depolarizing shift of the voltage dependence of inactivation compared with the BgNav4 channel (Table 1 and Fig. 2 D and E). None of the other mutations caused significant changes in the gating properties (Table 1 and Fig. 2). These results demonstrate that F1919S was responsible for the persistent current and associated gating properties of the BgNav4 channel.
Fig. 2.
S1919 at the C terminus is responsible for the persistent current and other unique properties identified in BgNav4. (A) Sodium current traces recorded from oocytes expressing BgNav4 and six single-amino-acid-substitution mutants, BgNav1 (containing F1919), Drosophila Para (containing F at 1950 equivalent to F1919 in BgNav1), and a Para mutant carrying S1950 for comparison. The current was measured by a 20-ms depolarization to -10 mV from the -120-mV holding potential. (B) Percentage of persistent currents. The persistent current was measured 15 ms into a 20-ms depolarization and was normalized by the amplitude of peak current. (C) Voltage dependence of activation of BgNav4 and six mutants. (D) Voltage dependence of steady-state inactivation of BgNav4 and six mutants. (E) Recovery from fast inactivation of BgNav4 and six mutants. The protocols for recording these gating properties are described in Materials and Methods.
Table 1. Gating properties of BgNav4 and its mutants.
| Activation
|
Fast inactivation
|
Recovery from inactivation
|
||||||
|---|---|---|---|---|---|---|---|---|
| V1/2, mV | k | V1/2, mV | k | τ1, ms | A1, % | τ2, ms | A2, % | |
| BgNav4 | -36.7 ± 3.9 | 10.6 ± 1.7 | -59.2 ± 1.8 | 5.5 ± 0.3 | 2.6 ± 1.2 | 70 ± 4 | 186 ± 67 | 30 ± 4 |
| G428E | -33.8 ± 3.4 | 8.9 ± 0.2* | -57.4 ± 1.1* | 5.6 ± 0.2 | 1.4 ± 0.3 | 72 ± 2 | 239 ± 47 | 28 ± 2 |
| H843Q | -34.6 ± 3.9 | 11.3 ± 0.9 | -56.7 ± 0.7† | 5.7 ± 0.1 | 1.2 ± 0.2 | 71 ± 4 | 168 ± 6 | 29 ± 4 |
| A975V | -36.5 ± 4.8 | 8.4 ± 0.7* | -58.4 ± 0.9 | 5.8 ± 0.3 | 1.0 ± 0.1 | 76 ± 2 | 283 ± 6 | 24 ± 2 |
| V1823M | -32.9 ± 2.8* | 9.8 ± 0.9 | -56.2 ± 1.5* | 5.1 ± 0.2* | 1.2 ± 0.2 | 69 ± 4 | 103 ± 14* | 31 ± 4 |
| G1846D | -35.9 ± 2.4 | 12.0 ± 1.4 | -58.1 ± 0.7 | 5.9 ± 0.3 | 3.2 ± 0.9 | 68 ± 1 | 209 ± 53 | 32 ± 1 |
| S1919F | -38.0 ± 5.2 | 4.0 ± 0.7† | -46.4 ± 1.2† | 3.7 ± 0.1† | 0.8 ± 0.2* | 91 ± 1† | 41 ± 10† | 9 ± 1† |
Values represent the mean ± SD for at least four oocytes.
Statistically significant difference compared with BgNav4 (P < 0.05)
Statistically significant difference compared with BgNav4 (P < 0.01)
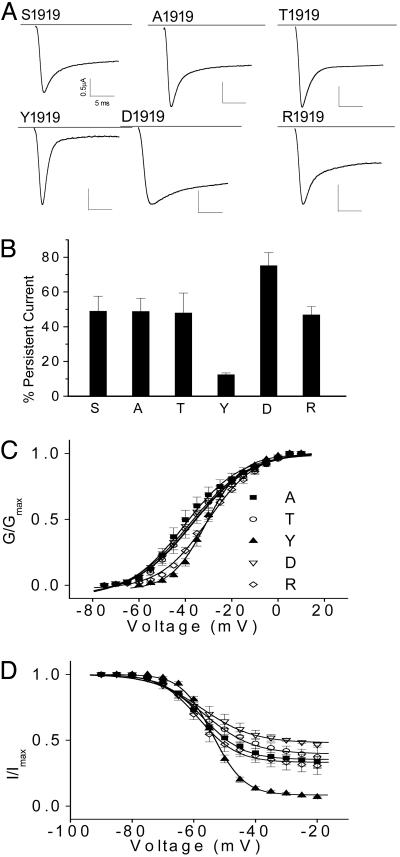
F/Y1919 at the C-Terminal Domain Is Essential for Fast Inactivation. Because the change from F to S at amino acid 1919 in BgNav4 introduced a consensus sequence for protein kinase C, we investigated the possibility of generation of persistent current by phosphorylation at S1919. Protein kinase C inhibitors calphostin C (500 nM), chelerythrine chloride (20 μM), or staurosporine (4 μM) were added to the oocyte recording solution 1 h before current recording. No significant change was observed in the amplitude of either peak or persistent current in the presence of any of these inhibitors (data not shown), suggesting that persistent current may not be generated by phosphorylation. To further characterize the nature of an amino acid at 1919 that is essential for the generation of persistent current, we replaced S1919 with another aromatic residue, tyrosine (Y), negatively charged aspartic acid (D), which mimics phosphorylation, positively charged arginine (R), neutral and hydrophilic alanine (A), or hydrophilic threonine (T). The mutant channels carrying A, D, R, or T produced persistent current comparable to that of BgNav4, whereas the Y1919 channel was able to almost completely inactivate (Fig. 3). These results indicated that S1919 was not essential for the generation of persistent current and that physophylation is not involved in the generation of persistent current. However, our analysis revealed that a phenyl-ring-containing bulky amino acid, F/Y1919, is essential for fast inactivation.
Fig. 3.
Effects of amino acid substitutions at S1919 on persistent current in BgNav4. (A) Sodium currents from five amino acid substitutions, A, T, Y, D, and R, at S1919 of BgNav4. The F substitution at 1919 is shown in Fig. 2. (B) Percentage of persistent current. The persistent current was measured 15 ms into a 20-ms depolarization and was normalized by the amplitude of peak current. (C) Voltage dependence of activation of single-amino-acid-substituted BgNav4 mutants. (D) Voltage dependence of steady-state inactivation of single-amino-acid-substituted BgNav4 mutants.
F1919S Is the Result of a U-to-C Editing Event. A nucleotide change from T to C at nucleotide 5756 results in the F (TTT) to S (TCT) change at the C-terminal domain at position 1919 in BgNav4. Sequence analysis of the corresponding genomic PCR fragment indicated that the nucleotide T was present in the genomic DNA. No alternative exon containing C was found. Therefore, the F1919S change results from a U-to-C editing event. By convention, this editing site is designated as the F/S site.
To determine the frequency of the U-to-C editing event, we isolated 561 partial cDNA clones from cockroach terminal abdominal ganglia because persistent current has been detected in dorsal unpaired median neurons in the terminal abdominal ganglion of the American cockroach (10). The 561 cDNA clones were analyzed by using the SNP PCR detection method described in Materials and Methods. Eighteen clones contained a C, whereas the remaining 543 clones contained a T (Table 2). Thus, 3.2% of BgNav transcripts were edited.
Table 2. Identification of the F/S editing site in BgNav and para.
| Source | Tissue* | F (TTT)† | S (TCT)† | Total clones |
|---|---|---|---|---|
| B. germanica | Ganglion | 543 | 18 | 561 |
| Ovary | 551 | 2 | 553 | |
| D. melanogaster | Whole body | 65 | 1 | 66 |
RNA isolated from the last abdominal ganglion or ovary from B. germanica or from the whole body of adult D. melanogaster
Number of clones containing T (underlined) or C (underlined)
Our previous RT-PCR analysis showed that the BgNav transcript was not only expressed abundantly in the nerve cord and leg but also detectable in ovary and gut (likely in neurons that innervate these tissues) (30). To determine whether the F-to-S change occurs in the ovary, we isolated 553 cDNA clones by using RNA from the ovary. We found that 551 ovary cDNA clones contained a T, whereas 2 clones contained a C (Table 2), indicating that the U-to-C editing event at the F/S site also occurs in the ovary but apparently at a much lower frequency than in terminal abdominal ganglia.
U-to-C Editing also Produces an F-to-S Change in Drosophila para. To determine whether the F/S editing event also occurs in other insect species, we isolated 65 cDNA clones encoding the corresponding region of the Drosophila para cDNA. One clone contained a nucleotide C corresponding to the F/S site in BgNav, and the remaining 64 clones contained a T (Table 2). A nucleotide T was present in the para genomic DNA at the corresponding position. Therefore, Drosophila para also undergoes the U-to-C editing at the same site. Furthermore, when this F-to-S change was introduced into a para cDNA clone, which does not contain any of the other seven amino acid changes detected in BgNav4 (Z.L. and K.D., unpublished data), a persistent current similar to that detected in BgNav4 also was observed (Fig. 2 A). Therefore, the U-to-C editing-mediated F-to-S change appears to confer the persistent current irrespective of the sodium channel backbone sequence.
Discussion
Although the U-to-C editing is observed frequently in mitochondria and plastids of lower plants (31), only a few examples were reported in animals before our study (32, 33). Here, we discovered a U-to-C editing event in the cockroach sodium channel transcript. Most significantly, the U-to-C editing in the BgNav transcript resulted in the F1919S change at the C-terminal domain of the cockroach sodium channel protein and transformed a transient sodium current to a persistent TTX-sensitive current. Furthermore, this U-to-C editing event also occurred in the Drosophila para transcript. Our findings demonstrate that RNA editing is a major regulatory mechanism that modulates sodium channel functions.
The persistent sodium current has been documented in well studied neurons from various regions of the mammalian brain, as well as in invertebrate neurons. It can contribute to a wide range of membrane depolarizations, such as subthreshold synaptic potentials and oscillations, somatic spike bursts, and depolarizing afterpotentials, which consequently enhance synaptic input, promote oscillatory activities, and accelerate firing frequency (11, 12). It has been debated for decades whether the persistent current can be attributed to noninactivating modes of conventional transient sodium channels or the expression of a novel sodium channel subtype not yet identified (11, 12). Supporting the first hypothesis, sodium channels can switch between two or more gating modes (34–36). The gating shift between fast and slow modes of transient sodium currents was proposed, but never proven, to be responsible for the persistent current in neocortical pyramidal neurons (3). It was also reported that modulation of the Nav1.2 channel by ectopic expression of the βγ subunits of a G protein induced a persistent sodium current in mammalian cell lines (37). More recently, Taddese and Bean (6) showed that persistent current detected in tuberomammillary neurons may be the result of an allosteric gating of the transient sodium channel. Rakowski et al. (7) also attributed the persistent current recorded from squid giant axons to the conventional transient sodium channel.
Reanalysis of Rakowski et al. (7) results by Clay (8), however, showed that the contribution of transient current to persistent current occurs in the -60 to -40 mV range, and outside of this range transient current appears an unlikely candidate, thus supporting the second hypothesis that the persistent current is attributable to a yet-to-be-identified subtype distinct from the conventional transient sodium channels. The most convincing results supporting the second hypothesis were from single-channel studies. Persistent current was detected in single-channel patches in cultured rat hippocampal neurons (38). Distinct persistent and transient gating properties were observed from single-channel recordings (39). However, the identity of the sodium channel that conducts persistent TTX-sensitive sodium current remained at large before this study.
Here, we found a sodium channel isoform that conducts persistent current. Our analysis showed that this previously uncharacterized sodium channel, BgNav4, was generated by a U-to-C editing event at the F/S site at the C-terminal domain. The persistent current detected in BgNav4 has two other hallmarks of the persistent currents identified in various vertebrate and invertebrate neurons: (i) it is TTX-sensitive, and (ii) it activates at more negative membrane potentials (at least 10 mV below the threshold of conventional sodium channels). Persistent TTX-sensitive current has been detected in various insect neurons, including cockroach terminal abdominal dorsal unpaired median neurons (10, 40–42). As expected, we detected this U-to-C editing event in the cockroach terminal abdominal ganglion where dorsal unpaired median neurons are located. Because the F/S editing event is found in both cockroach and Drosophila (Table 2), and F1919 is conserved in all sequenced sodium channel genes in arthropods including Drosophila, house fly, cockroach, and varroa mite sodium channels, we suggest that RNA-editing-mediated generation of persistent current is likely a conserved mechanism in arthropods.
Our finding that the F1919S change at the C terminus impairs the fast inactivation adds to the growing literature that supports a critical role of the C-domain in modulating sodium channel inactivation (43). For example, the C-terminal domain contributes to differences in inactivation kinetics between rat Nav1.2 and 1.5 channels (44) and rat Nav1.4 and 1.5 channels (45). Multiple disease-associated mutations at the C-terminal domain induce sustained sodium current (46–49). Both F1795C and F1795H mutations in the human Nav1.5 channel promote sustained sodium channel activity (48). How these mutations alter inactivation, however, is not clear.
Although the F1919S change introduces a potential phosphorylation site for protein kinase C, our kinase inhibitor experiments indicate that the persistent current is not generated by phosphorylation at S1919. Furthermore, S1919 is not essential for persistent current. BgNav4 derivatives, with negatively charged aspartic acid (D), positively charged arginine (R), neutral and hydrophilic alanine (A), or hydrophilic threonine (T) at position 1919, all induce the persistent current. Only another aromatic residue, tyrosine, can substitute for F without affecting fast inactivation. These results indicate that an aromatic residue at 1919 is essential for fast inactivation. The IFM motif in the short intracellular linker connecting domains III and IV is critical for fast inactivation and serves as the inactivation gate (1). The IFM motif is believed to interact with an inactivation receptor in or near the intracellular mouth of the pore during the inactivation process. Amino acid residues in the S4–S5 intracellular loops in domains III and IV may be part of the docking receptor, which is made available by channel activation (1). We speculate that the aromatic residue F1919 in the middle of the C-terminal domain could directly or indirectly interact with the IFM motif or the docking receptor to modulate the inactivation kinetics.
In conclusion, we identified U-to-C editing as a mechanism for increasing sodium channel functional plasticity, including the generation of a persistent TTX-sensitive current. Our study opens the possibility that RNA editing mediates generation of persistent TTX-sensitive current in other species, including mammals.
Note Added in Proof. While this paper was being reviewed, another paper on modulation of sodium channel gating properties by additional RNA editing events in the cockroach sodium channel gene was accepted for publication [Song, W., Liu, Z., Tan, J., Nomura, Y. & Dong, K. (2004) J. Biol. Chem., in press].
Acknowledgments
We thank Drs. Dalia Gordon, Noah Koller, and Vincent Salgado for critical review of an early version of this manuscript and Dr. Annette Thelen (Genomic Technology Service Facility, Michigan State University) for providing assistance with the SNP PCR detection method. This work was supported by National Science Foundation Grants IBN9808156 and IBN0224877.
This paper was submitted directly (Track II) to the PNAS office. Abbreviations: SNP, single-nucleotide polymorphism; TTX, tetrodotoxin.
References
- 1.Catterall, W. A. (2000) Neuron 26, 13-25. [DOI] [PubMed] [Google Scholar]
- 2.Hotson, J. R., Prince, D. A. & Schwartzkroin, P. A. (1979) J. Neurophysiol. 42, 889-895. [DOI] [PubMed] [Google Scholar]
- 3.Alzheimer, C., Schwindt, P. C. & Crill, W. E. (1993) J. Neurosci. 13, 660-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennartz, C. M. A., Bierlaagh, M. A. & Geurtsen, A. M. S. (1997) J. Neurophysiol. 78, 1811-1825. [DOI] [PubMed] [Google Scholar]
- 5.Raman, I. M. & Bean, B. P. (1997) J. Neurosci. 17, 4517-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taddese, A. & Bean, B. P. (2002) Neuron 33, 587-600. [DOI] [PubMed] [Google Scholar]
- 7.Rakowski, R. F., Gadsby, D. C. & DeWeer, P. (2002) J. Gen. Physiol. 119, 235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clay, J. R. (2003) J. Neurophysiol. 89, 640-644. [DOI] [PubMed] [Google Scholar]
- 9.Opdyke, C. A. & Calabrese, R. L. (1994) J. Comp. Physiol. 175, 781-789. [DOI] [PubMed] [Google Scholar]
- 10.Lapied, B., Malecot, C. O. & Pelhate, M. (1990) J. Exp. Biol. 151, 387-403. [Google Scholar]
- 11.Taylor, C. P. (1993) Trends Neurosci. 16, 455-460. [DOI] [PubMed] [Google Scholar]
- 12.Crill, W. E. (1996) Annu. Rev. Physiol. 58, 349-362. [DOI] [PubMed] [Google Scholar]
- 13.Huguenard, J. R. (2002) Neuron 33, 492-494. [DOI] [PubMed] [Google Scholar]
- 14.Bass, B. L., ed. (2001) RNA Editing (Oxford Univ. Press, Oxford).
- 15.Seeburg, P. H. (2000) Neuron 25, 261-263. [DOI] [PubMed] [Google Scholar]
- 16.Seeburg, P. H. (2002) Neuron 35, 17-20. [DOI] [PubMed] [Google Scholar]
- 17.Seeburg, P. H., Higuchi, M. & Sprengel, R. (1998) Brain Res. Rev. 26, 217-229. [DOI] [PubMed] [Google Scholar]
- 18.Burns, C. M., Chu, H., Rueter, S. M., Hutchison, L. K., Canton, H., Sanders-Bush, E. & Emeson, R. B. (1997) Nature 387, 303-308. [DOI] [PubMed] [Google Scholar]
- 19.Niswender, C. M., Copeland, S. C., Herrick-Davis, K., Emeson, R. B. & Sanders-Bush, E. (1999) J. Biol. Chem. 274, 9472-9478. [DOI] [PubMed] [Google Scholar]
- 20.Patton, D. E., Silva, T. & Bezanilla, F. (1997) Neuron 19, 711-722. [DOI] [PubMed] [Google Scholar]
- 21.Rosenthal, J. J. C. & Bezanilla, F. (2002) Neuron 34, 743-757. [DOI] [PubMed] [Google Scholar]
- 22.Palladino, M. J., Keegan, L. P., O'Connell, M. A. & Reenan, R. A. (2000) Cell 102, 437-449. [DOI] [PubMed] [Google Scholar]
- 23.Hanrahan, C. J., Palladino, M. J., Ganetzky, B. & Reenan, R. A. (2000) Genetics 155, 1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, J., Liu, Z., Nomura, Y., Goldin, A. L. & Dong, K. (2002) J. Neurosci. 22, 5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong, K. (1997) Insect Biochem. Mol. Biol. 27, 93-100. [DOI] [PubMed] [Google Scholar]
- 26.Loughney, K., Kreber, R. & Ganetzky, B. (1989) Cell 58, 1143-1154. [DOI] [PubMed] [Google Scholar]
- 27.Feng, G., Deak, P., Chopra, M. & Hall, L. M. (1995) Cell 82, 1001-1011. [DOI] [PubMed] [Google Scholar]
- 28.Warmke, J. W., Reenan, R. A., Wang, P., Qian, S., Arena, J. P., Wang, J., Wunderler, D., Liu, K., Kaczorowski, G. J., Van der Ploeg, L. H., et al. (1997) J. Gen. Physiol. 110, 119-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grolleau, F. & Lapied, B. (2000) J. Exp. Biol. 203, 1633-1648. [DOI] [PubMed] [Google Scholar]
- 30.Liu, Z., Chung, I. & Dong, K. (2001) Insect Biochem. Mol. Biol. 31, 703-713. [DOI] [PubMed] [Google Scholar]
- 31.Bock, R. (2001) in RNA Editing, ed. Bass, B. L. (Oxford Univ. Press, Oxford), pp. 38-60.
- 32.Sharma, P. M., Bowman, M., Madden, S. L., Rauscher, F. J., III, & Sukumar, S. (1994) Genes Dev. 8, 720-731. [DOI] [PubMed] [Google Scholar]
- 33.Nagalla, S. R., Barry, B. J. & Spindel, E. R. (1994) Mol. Endocrinol. 8, 943-951. [DOI] [PubMed] [Google Scholar]
- 34.Nilius, B. (1988) Biophys. J. 53, 857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moorman, J. R., Kirsch, G. E., VanDongen, A. M. J., Joho, R. H. & Brown, A. M. (1990) Neuron 4, 243-252. [DOI] [PubMed] [Google Scholar]
- 36.Zhou, J., Potts, J. F., Trimmer, J. S., Agnew, W. A. & Sigworth, F. J. (1991) Neuron 7, 775-785. [DOI] [PubMed] [Google Scholar]
- 37.Ma, J. Y., Catterall, W. A. & Scheuer, T. (1997) Neuron 19, 443-452. [DOI] [PubMed] [Google Scholar]
- 38.Masukawa, L. M., Hansen, A. J. & Shepherd, G. (1991) Cell. Mol. Neurobiol. 11, 231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magistretti, J., Ragsdale, D. S. & Alonso, A. (1999) J. Neurosci. 19, 7334-7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito, M. & Wu, C.-F. (1991) J. Neurosci. 11, 2135-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schafer, S., Rosenboom, H. & Menzel, R. (1994) J. Neurosci. 14, 4600-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kloppenburg, P. & Horner, M. (1998) J. Exp. Biol. 201, 2529-2541. [DOI] [PubMed] [Google Scholar]
- 43.Goldin, A. L. (2003) Curr. Opin. Neurobiol. 13, 284-290. [DOI] [PubMed] [Google Scholar]
- 44.Mantegazza, M., Yu, F. H., Catterall, W. A. & Scheuer, T. (2001) Proc. Natl. Acad. Sci. USA 98, 15348-15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deschenes, I., Trottier, E. & Chahine, M. (2001) J. Membr. Biol. 183, 103-114. [DOI] [PubMed] [Google Scholar]
- 46.Veldkamp, M. W., Viswanathan, P. C., Bezzina, C., Baartscheer, A., Wilde, A. A. M. & Balser, J. R. (2000) Circ. Res. 86, e91-e97. [DOI] [PubMed] [Google Scholar]
- 47.Baroudi, G. & Chahine, M. (2000) FEBS Lett. 487, 224-228. [DOI] [PubMed] [Google Scholar]
- 48.Rivolta, I., Abriel, H., Tateyama, M., Liu, H., Memmi, M., Vardas, P., Napolitano, C., Priori, S. G. & Hass, R. S. (2001) J. Biol. Chem. 276, 30623-30630. [DOI] [PubMed] [Google Scholar]
- 49.Lupoglazoff, J. M., Cheav, T., Baroudi, G., Berthet, M., Denjoy, I., Caushemez, B., Extramiana, F., Chahine, M. & Guicheney, P. (2001) Circ. Res. 89, e16-e21. [DOI] [PubMed] [Google Scholar]