Abstract
Objective(s):
Similar characteristics of molecular pathways between cellular reprogramming events and tumorigenesis have been accentuated in recent years. Reprogramming-related transcription factors, also known as Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC), are also well-known oncogenes promoting cancer initiation, progression, and cellular transformation into cancer stem cells. Long non-coding RNAs (lncRNAs) are a major class of RNA molecules with emerging roles in stem cell pluripotency, cellular reprogramming, cellular transformation, and tumorigenesis. The long intergenic non-coding RNA ROR (lincRNA-ROR, linc-ROR) acts as a regulator of cellular reprograming through sponging miR-145 that normally negatively regulates the expression of the stemness factors NANOG, OCT4, and SOX2.
Materials and Methods:
Here, we employed a real-time PCR approach to determine the expression patterns of linc-ROR and its two novel spliced variants (variants 2 and 4) in esophageal squamous cell carcinoma (ESCC).
Results:
The quantitative real-time RT-PCR results revealed a significant up-regulation of linc-ROR (P=0.0098) and its variants 2 (P=0.0250) and 4 (P=0.0002) in tumor samples of ESCC, compared to their matched non-tumor tissues obtained from the margin of same tumors. Our data also demonstrated a significant up-regulation of variant 4 in high-grade tumor samples, in comparison to the low-grade ones (P=0.04). Moreover, the ROC curve analysis demonstrated that the variant 4 of ROR has a potential to discriminate between tumor and non-tumor samples (AUC=0.66, P<0.05).
Conclusion:
Our data suggest a significant up-regulation of linc-ROR and its variants 2 and 4 in ESCC tissue samples.
Keywords: Esophageal squamous - cell carcinoma, Linc-ROR, Non-coding RNA, Spliced variants
Introduction
Esophageal cancer is the 6th most cause of cancer-related death, and also it is the 8th most common cancer in the word (1, 2). Two main subtypes of esophageal malignancies, esophageal squamous cell carcinoma (ESCC; a common form of the cancer with a high incidence in the East) and adenocarcinoma (ADC; a rare form of the cancer with a high incidence in the West) have a five-year survival rate of less than 10%, and both of them are highly lethal (3). Golestan province in Northeast of Iran is one of the highest risk areas of ESCC form in the world. In this area, ESCC subtype is accounting for ~90% of all types of esophageal cancers (4).
Forced ectopic expression of OCT4, SOX2, KLF4, and c-MYC (known as Yamanaka factors; OSKM) could reprogram somatic fully differentiated cells to an embryonic-like or pluripotent state (5, 6). Recent studies indicated OSKM factors in cancer initiation and progression; supporting the hypothesis that cellular reprogramming events and tumorigenesis processes share similar regulatory mechanisms (7). In line with the hypothesis, recent evidences suggest that cancer could be considered as a reprogramming-like process (8).
Long non-coding RNAs (lncRNAs) are a new class of RNA molecules which comprises the most proportion of human transcriptome. LncRNAs have emerged as main players in regulating key cellular pathways promoting proliferation, stem cells self-renewal, and cellular reprogramming events (9-13). Altered expression signatures of a large number of lncRNAs have been reported in several human malignancies (14, 15). The exact molecular mechanisms of lncRNAs functions have not yet been fully elucidated. However, some of their well-known functions in promoting cancer progression and maintaining stem cell pluripotency carry out through association with chromatin remodeling complexes and transcription factors (9, 13, 16).
Long intergenic ncRNA ROR (lincRNA-ROR, linc-ROR) is a multi-exon lncRNA gene located in the genomic region of human 18q21.3. Linc-ROR was initially introduced by Loewer and colleagues as a regulator of reprogramming (ROR) in human induced pluripotent stem cells (iPSc) (17). OCT4, SOX2, and Nanog transcription factors directly bind to the promoter region of linc-RoR gene to induce its expression. Interestingly, linc-RoR shares miR-145-response elements with that of the aforementioned transcription factors; and hence, it refrains microRNA-mediated suppression of OCT4, SOX2, and NANOG transcripts to maintain self-renewal and pluripotency status of iPSc (18).
Here, we explored a potential expression alteration of linc-ROR and its newly identified splice variants (2 to 5) in ESCC tissue samples. Since variants 3 and 5 had no expression levels in ESCC samples, we continued the work with linc-ROR variants 2 and 4.
Materials and Methods
Clinical ESCC samples
Fresh tumor and non-tumor tissue biopsies were obtained from the biobanks of Gastroenterology and Hepatology Research Center (Golestan University of Medical Sciences) and Buali (Avicenna) Research Institute (Mashhad University of Medical Sciences). The clinical samples were classified in two groups: 30 tumor biopsy samples obtained from patients with ESCC and 30 apparently normal tissue biopsies were taken from the margin of same tumor biopsies. Histopathological characteristic were evaluated according to the American Joint Committee on Cancer criteria and TNMG procedure for stage and grade classification (Table 1). The whole experimental procedure was approved by the Ethics Committee of Tarbiat Modares University and Golestan University of Medical Sciences. Written consents had been also obtained from all patients prior to surgical removal of their tumors.
Table 1.
The clinico-pathological characteristics of the patients with esophageal squamous cell carcinoma
| Case number | Clinic-pathological Characteristics |
|---|---|
| 65.26 | Age (median) |
| 16/14 | Sex (male/female) |
| 1/12/2/2/9/4 | Stage (I/IIA/IIB/ IIA-III/III/unknown) |
| 5/17/5/3 | Grade (I/II/III/unknown) |
| 16/10/4 | Location (middle/lower/unknown) |
| 1/1/3/1/20/1/3 | Tumor invasion (T1/T1a/T2/T2a/T3/T4/unknown) |
| 13/11/3/3 | Regional lymph node (N0/N1/Nx/unknown) |
RNA extraction and cDNA synthesis
Total RNA was extracted from both frozen tumor and non-tumor clinical samples, using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA) and according to the manufacturer’s instructions. To remove any potential contamination with DNA molecules, the extracted RNA was treated with a DNase enzyme (Takara, Japan) in an RNase-free condition. We then synthesized the first strand of complementary DNA (cDNA) by using the Hyperscript RT reagent Kit (GeneAll, South Korea) and random hexamer primers (Takara, Japan), as described by the manufacturers.
Quantitative real-time PCR
Bioinformatics analysis was used to design specific PCR primers (Figure 1A, Table 2) to amplify the main and the variants 2 and 4 of linc-ROR (with the GenBank accession numbers of NR_048536.1, AB844430.1, and AB844432.1, respectively), using Gene Runner (version 3.05), PerIPrimer (version 1.1.21), and Oligo (version 7.56) softwares. Real Q plus 2x master mix Green (Ampliqon, Denmark) supplemented with ROX dye was used for all quanti-tative real-time PCR reactions. As an endogenous control, the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH, NM_002046.4) transcript was quantified, and the expression of linc-ROR and its variants were normalized to its expression level. All real-time PCR reactions were carried out by the ABI STEP ONE real-time PCR systems (Applied Biosystems, Foster City, CA) using the following cycling conditions: initiation at 95°C for 15 min, amplification for 40 cycles with denaturation at 95°C for 15 sec, annealing and extending at 63°C for 55 sec. Moreover, we confirmed the authenticity of the PCR products by melt curve analysis and direct sequencing.
Figure 1.
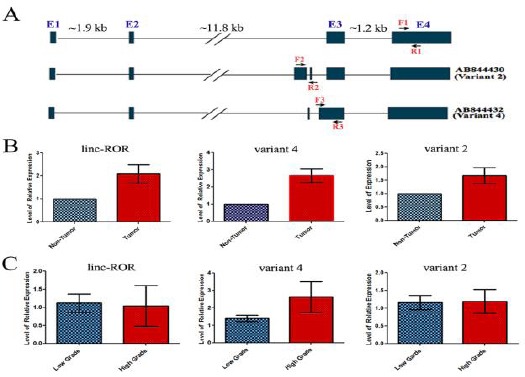
A) A representative schematic view of the genomic organization of linc-ROR and its spliced variants 2 and 4. The location of the forward and reverse primers used to amplify specifically each transcript is indicated with different set of arrows. B) Expression alteration of linc-ROR, and its splice variants 2 and 4 in ESCC tumor samples. The relative expression levels of linc-ROR and its splice variants 2 and 4are represented in ESCC tumors vs. their matched non-tumor samples obtained from the same patients. An up-regulation of linc-ROR, and its splice variants in ESCC tumor vs non-tumor samples is evident. C) Relative gene expressions in high and low grades esophageal tumors. The red and blue boxes represent high grade (GIII) and low grade (GI and GII) tumor samples, respectively. Note that there is a significant up-regulation just for the linc-ROR variant 4 in high-grade tumors of ESCC
ESCC: esophageal squamous cell carcinoma
Table 2.
The list of primers used to specifically amplify the main and variants 2 and 4 of linc-ROR as well as the internal control, GAPDH
| Transcript | Primer | sequence | PCR product (bp) |
|---|---|---|---|
| Linc-ROR | F R |
ACAAGGAGGAAAGGGCTGAC TTCTGGAAGCTAAGTGCACATG |
124 |
| AB844430 variant 2 | F R |
AAGCAGCTGTGACCTGGC TCTGGCACCTAGCATAGCAC |
179 |
| AB844432 variant 4 | F R |
CAGCCCAAAAATATCGTCAGAGT TCTTACTTAGCGACAATGCCATC |
265 |
| GAPDH | F R |
ATGGGGAAGGTGAAGGTCG GGGGTCATTGATGGCAACAATA |
108 |
Statistical analysis
All real-time PCR experiments were carried out in duplicates. The LinReg PCR software (version 11) was used to calculate the exact PCR efficiency, and the real-time PCR data was adjusted based on these efficiencies. In order to calculate the expression fold change of the interest genes, the expression level of them in each clinical sample was normalized to that of GAPDH gene, as an internal control. We then normalized the expression of candidate genes in tumor samples to the matched non-tumor ones (2^-ΔΔCT method). To discriminate between tumor and non-tumor clinical samples, as well as between different grades of tumor samples, receiver operating characteristic (ROC) curve analysis was plotted by Graphpad Prisom (version 5.0.0.288).
Results
Up-regulation of linc-ROR in ESCC tissue samples
To quantify linc-ROR variants transcripts in ESCC samples, we designed specific primers on regions that were unique for each RNA transcript (Figure 1A). Our quantitative real-time RT-PCR data demonstrated a significant up-regulation of linc-ROR in tumor samples of ESCC, compared to the marginal non-tumor tissues obtained from the same patients (fold change 1/61, P=0.0098, Figure 1B). Next, we investigated a potential association between linc-ROR expression and the grade of malignancy of ESCC tumors. The data analysis, however, failed to show any association between the expression level of linc-ROR with the grade of malignancy of samples (Figure 2C). Moreover, there was no statistically significant difference of linc-ROR expression in tumor samples between different genders (data not shown).
Figure 2.
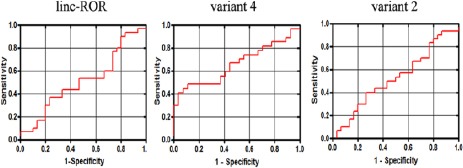
Analyzing the sensitivity and specificity of linc-ROR variants, as potential tumor markers of ESCC. ROC curve analysis demonstrated only a moderate sensitivity and specificity of linc-ROR spliced variant 4’sexpression level to discriminate between tumor and non-tumor states of ESCC tissue samples. Similarly, Area Under the Curve (AUC) was statistically significant only for variant 4 (AUC=0.65, P=0.049). An AUC>70 is considered as a good index for a biomarker with a high ability to correctly classify tumor and non-tumor groups of samples. On the other hand, the ROC curve analysis failed to discriminate between high and low grade ESCC tumor samples
ESCC: esophageal squamous cell carcinoma
Up-regulation of linc-ROR variants 2 and 4 in ESCC tissue samples
We next quantified the expression patterns of two novel variants of linc-ROR, variants 2 and 4, in ES CC tissue samples. Specific primers were designed in regions that were unique for each transcript (Figure 1A). The quantitative real-time PCR data revealed a significant up-regulation of variant 2 (fold change 1/67, P=0.025) and variant 4 (fold change 2/65, P=0.0002) in tumor specimens, in comparison to their paired non-tumor samples (Figure 1B). The data was unable to find any association between the expression levels of variant 2 with the grade of malignancy of the tumors (Figure 2B). In contrast, variant 4 had a significantly higher expression level in high grade (GIII) tumors, compared to the low grade (GI and GII) ones (P=0.04. Figure 1B). Moreover, there was no statistically significant difference between expression alteration of the linc-ROR variants 2 and 4 and the gender of patients (data not shown).
The expression level of linc-ROR spliced variant 4 could discriminate between tumor and non-tumor states of ESCC samples
We performed ROC curve analysis to evaluate the suitability of linc-ROR and its variants (2 and 4) expression levels to discriminate between tumor and non-tumor states of ESCC clinical samples. The data represent AUC=0.512 (P>0.05), AUC=0.522 (P>0.05), and AUC=0.656 (P=0.049) for linc-ROR, variant 2, and variant 4, respectively (Figure 2). In contrast to linc-ROR and variant 2, variant 4 showed a better suitability to classify tumor and non-tumor samples of esophagus. However, its AUC score, slightly fell below the score needed for a good biomarker (AUC>70). Moreover, in evaluation of different grades of malignancy and expression of the linc-ROR and its variants, ROC curve analysis failed to discriminate high and low grades tumor samples (data not shown).
Discussion
Up-regulation of several lncRNAs including HOTAIR, MALAT1, PCAT-1, UCA1, and SOX2OT have been reported in ESCC (19-24). Based on numerous similarities between molecular pathways of tumorigenesis and reprogramming events and the fact that linc-ROR promotes cellular reprogramming, we hypothesized that this non-coding RNA could have a part in cancer initiation and/or progression processes. For this reason, we aimed to investigate its expression pattern in ESSC clinical specimens. Furthermore, we examined a potential correlation between its expression pattern and histopathological characteristics of the ESCC tumor samples.
Our data revealed that linc-ROR and its spliced variants 2 and 4 were up-regulated in ESCC tumors samples, compared to their corresponding non-tumors ones obtained from the margin of same tumors. Moreover, our data demonstrated a significant up-regulation of variant 4 (but neither linc-ROR nor variant 2) in high-grade ESCC tumors, suggesting a potential role for variant 4 in ESCC tumor progression. Despite a statistically significant up-regulation of the main and spliced variants of linc-ROR in ESCC tissue samples, the data obtained from ROC curve analysis demonstrated that only variant 4 has a moderate sensitivity and specificity in discriminating between tumor and non-tumor ESCC samples.
Our data on elevated levels of linc-ROR transcripts in ESCC samples, is in agreement with recent studies reporting an up-regulation of linc-ROR in several other somatic cancers including breast cancer and hepatocellular carcinoma (25, 26). Overexpression of linc-ROR caused an epithelial to mesenchymal transition (EMT) process in immortalized human mammary epithelial cells, probably through preventing the degradation of miR-205 target genes, such as the inducer of EMT known as ZEB2 (25). Linc-ROR is also implicated in breast cancer cells metastasis, and the induction and maintenance of stemness properties to the pluripotent and cancer stem cells (25). Linc-ROR was also highly up-regulated in hepatocellular cancer cells. The extracellular linc-ROR derived from hepatocellular tumor cells could modulate the chemosensitivity of the tumor cells (26). Moreover, human linc-RoR is a strong negative regulator of P53 transcription factor, in response to DNA damage. In contrast to MDM2 that causes P53 degradation, linc-ROR suppresses P53 translation, and p53-mediated cell cycle arrest and apoptosis (27).
In contrast to the aforementioned studies claiming an up-regulation of linc-ROR in some cancers, Feng et al. reported a significant down-regulation of linc-ROR expression in glioma tumors. Accordingly, ectopic expression of linc-ROR diminished cell proliferation, inhibited KLF4 expression, and decreased the capacity of tumorosphere formation in U87 glioma cell line. The latter findings suggested that linc-ROR may act as a tumor suppressor gene in glioma (28).
Conclusion
Our data highlighted the up-regulation of linc-ROR and its splice variants 2 and 4 in ESCC. Our data also propose the existence of more similarities in molecular pathways common between tumorigenesis and reprogramming events. Altogether, our data suggest that linc-ROR transcripts might have a potential role in tumor initiation and/or progression of esophagus cancer. However, this conclusion needs further functional analysis for experimental validation.
Acknowledgment
This work is financially supported by research grants from research deputies of Golestan University of Medical Sciences and Tarbiat Modares University.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 3.Semnani S, Sadjadi A, Fahimi S, Nouraie M, Naeimi M, Kabir J, et al. Declining incidence of esophageal cancer in the Turkmen Plain, eastern part of the Caspian Littoral of Iran: A retrospective cancer surveillance. Cancer Detect Prev. 2006;30:14–19. doi: 10.1016/j.cdp.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Sadjadi A, Marjani H, Semnani S, Nasseri-Moghaddam S. Esophageal Cancer in Iran: A Review. Middle East J Cancer. 2010;1:5–14. [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- 8.Castellanos A, Vicente-Duenas C, Campos-Sanchez E, Cruz JJ, Garcia-Criado FJ, Garcia-Cenador MB, et al. Cancer as a reprogramming-like disease: implications in tumor development and treatment. Semin Cancer Biol. 2010;20:93–97. doi: 10.1016/j.semcancer.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2007;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 15.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-ROR modulates reprogramming of human induced pluripotent stem cell. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-ROR regulates Oct4, Nanog and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8:e63516. doi: 10.1371/journal.pone.0063516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, et al. Up-regulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, et al. Up-regulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36:2501–2507. doi: 10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 23.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;71:7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 24.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, et al. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32:126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 25.Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, et al. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng S, Yao J, Chen Y, Geng P, Zhang H, Ma X, et al. Expression and functional role of reprogramming-related long noncoding RNA (lincRNA-ROR) in Glioma. J Mol Neurosci. 2015;56:623–630. doi: 10.1007/s12031-014-0488-z. [DOI] [PubMed] [Google Scholar]


