Abstract
In higher plants, very-long-chain fatty acids (VLCFAs) are the main constituents of hydrophobic polymers that prevent dessication at the leaf surface and provide stability to pollen grains. Of the 21 genes encoding VLCFA elongases (VLCFAEs) from Arabidopsis thaliana, 17 were expressed heterologously in Saccharomyces cerevisiae. Six VLCFAEs, including three known elongases (FAE1, KCS1, and KCS2) and three previously uncharacterized gene products (encoded by At5g43760, At1g04220, and At1g25450) were found to be enzymatically active with endogenous yeast fatty acid substrates and to some extent with externally supplied unsaturated substrates. The spectrum of VLCFAs accumulated in expressing yeast strains was determined by gas chromatography/mass spectrometry. Marked specificity was found among elongases tested with respect to their elongation products, which encompassed saturated and monounsaturated fatty acids 20–30 carbon atoms in length. The active VLCFAEs revealed highly distinct patterns of differential sensitivity to oxyacetamides, chloroacetanilides, and other compounds tested, whereas yeast endogenous VLCFA production, which involves its unrelated elongase (ELO) in sphingolipid synthesis, was unaffected. Several compounds inhibited more than one VLCFAE, and some inhibited all six active enzymes. These findings pinpoint VLCFAEs as the target of the widely used K3 class herbicides, which have been in commercial use for 50 years, provide important clues as to why spontaneous resistance to this class is rare, and point to complex patterns of substrate specificity and product spectrum among members of the Arabidopsis VLCFAE family.
The Herbicide Resistance Action Committee (HRAC, www.plantprotection.org/HRAC) classifies herbicides into different groups according to their target sites, modes of action, similarity of induced symptoms, or chemical classes. The target sites for most commercial herbicides are known. For the HRAC class K3 and N herbicides, including the well known oxyacetamides and chloroacetanilides, the site of action is unknown. K3 and N herbicides are grouped by their similarity of induced physiological symptoms. K3 herbicides are described by HRAC as inhibitors of cell division or inhibitors of very-long-chain fatty acid (VLCFA, fatty acids >C18) synthesis, whereas N herbicides are described as inhibitors of lipid synthesis. Chloroacetanilides and chloroacetamides belong to group K3 and inhibit early plant development. These compounds have been widely used for 50 years (1); in 1998, this group accounted for 50% of all herbicides used in U.S. corn fields.
K3 and N herbicides lead to inhibition of VLCFA biosynthesis in plant and algal cells (reviewed in ref. 2). In higher plants, VLCFAs serve as components or precursors of wax, suberin, and cutin, which form the leaf cuticle (reviewed in ref. 3); as storage lipids in seeds; as periderm and endodermis components; as glycosylphosphatidyl-inositol anchors in plasma membrane proteins; and as sphingolipid components in various membranes. VLCFAs are formed by membrane-bound, multienzyme acyl-CoA elongase systems that catalyze a series of biochemical reactions analogous to those of de novo fatty acid synthase (4). The first step is the condensation of an acyl-CoA primer (>16 carbons in length) with malonyl-CoA to form β-ketoacyl-CoA followed by reduction, dehydration, and a second reduction, resulting in an acyl-CoA that is extended by two carbons (4). The elongase is the key activity determining whether fatty acids will be elongated and the amount and the chain lengths of the VLCFAs produced (5). Recent work (6) has shown that treatment of mono- and dicotyledonous plants with representatives of the K3 and N group phenocopies the VLCFA elongase (VLCFAE) mutant fiddlehead (7, 8) in Arabidopsis thaliana, which exhibits an organ-fusion phenotype. Although inhibition of fiddlehead gene product elongase activity is assumed to underlie the phenocopy (6), the specifically targeted enzymes within VLCFA biosynthesis remain unknown.
Several plant VLCFAEs have been characterized, including A. thaliana FAE1 (5, 9–11), KCS1 (12), KCS2 (13), CUT1 (14), Brassica napus FAE1 (15), Lesquerella fendleri KCS3 (16), Simmondsia chinensis KCS (17), and FAE2 from the moss Marchantia polymorpha (18). These VLCFAEs differ in terms of their tissue- and ontogeny-specific expression as well as their substrate specificity. Progress into VLCFAE biochemistry has been impaired because of the difficulty of heterologous expression, solubilization, and purification of these membrane-associated enzymes.
To identify the target of oxyacetamide, chloroacetanilide, and other herbicides of the HRAC groups K3 and N, we cloned, expressed, and characterized putative and known VLCFAEs from A. thaliana in Saccharomyces cerevisiae and examined the spectrum of VLCFAs produced by the expressing strains in the presence of various herbicides.
Materials and Methods
Cloning of Putative Elongases. Total RNA was isolated from 100 mg of 28-day-old A. thaliana leaves with the RNeasy plant minikit (Qiagen, Valencia, CA) according to manufacturer instructions and used to synthesize first-strand cDNA with NotI-d(T)18 primers and Moloney murine leukemia virus reverse transcriptase (Amersham Pharmacia); 2 μl of this cDNA was used directly for PCR with the appropriate primers. PCR products were cloned blunt into the pCR-Blunt II-TOPO vector (Invitrogen), cleaved with the appropriate restriction enzymes, and ligated in-frame with the C-terminal His-tag to the pYES2 vector (Invitrogen) digested with the same enzymes.
Yeast Transformation and Cultivation. S. cerevisiae strain INVSc1 (Invitrogen) was transformed by the CaCl2 method (19), and transformants were selected on minimal medium agar plates lacking uracil. Transformed cells were grown at 28°C in complete minimal-dropout-uracil medium supplemented with 2% glucose as the only carbon source. Expression of heterologous protein was induced by transferring log-phase cells to medium containing 1% raffinose and 2% galactose.
Expression of VLCFAEs and Inhibition of VLCFA Elongation in Yeast. A 50-ml culture of yeast transformed with the appropriate elongase was induced and grown for 18–20 h, and cells were harvested and freeze-dried. Fatty acid composition was investigated (see Analysis of Fatty Acids) and compared with that of cells transformed with the empty pYES2 vector. Where elongase activity was observed, inhibition was assayed in a new 50-ml culture by adding aK3 or N herbicide to a final concentration of 100 μM at the time of induction; after 18–20 h of incubation time, cells were processed as described above, and fatty acid composition was compared with that of cells expressing the appropriate elongase grown in the absence of herbicide.
Feeding Experiments. To examine the possible utilization of various substrates by different elongases, fatty acids were added to a final concentration of 500 μM in the culture medium of transformed yeasts 5 h after induction and 15 h before harvest. Fatty acid composition was investigated as described below.
Analysis of Fatty Acids. To determine total fatty acid composition, fatty acid methyl esters were prepared according to a modified protocol by Browse et al. (20). In brief, 50 mg of freeze-dried yeast cells was resuspended in 1 ml of 1 M methanolic HCl (Supelco) with 5% (vol/vol) 2,2-dimethoxypropane and incubated under nitrogen at 80°C for 1 h. After the samples had cooled, 1 ml of 0.9% NaCl was added, and fatty acid methyl esters were extracted into 300 μl of heptane and analyzed by gas chromatography/mass spectrometry (GC/MS).
GC/MS Analysis. Fatty acid methyl esters were identified by using a Hewlett–Packard 5890 gas chromatograph (HP 7673 MSD). Components were separated on a 0.2-mm methyl silicone capillary column with a 0.35-μm film thickness. The column was operated with helium carrier gas and 30 ml/min split injection (injection temperature, 250°C; detector temperature, 320°C). The oven temperature was increased from 50 to 320°C at 12°C min-1 and held for an additional 5 min.
Arabidopsis Cultivation and Herbicide Treatment. Cultivation and herbicide treatment of A. thaliana (L.) Heynhoe cv. Columbia (Col-0) were carried out as described (6).
Results
Cloning of Putative Elongases from Arabidopsis. A blastp (21) search of the Arabidopsis Institute for Genomic Research database (www.tigr.org/tdb/e2k1/ath1/ath1.shtml) with the protein sequence of A. thaliana FAE1 (11) revealed 20 genes with strong similarity to FAE1 (6), among them the well characterized VLCFAEs CUT1 (14), KCS1 (12), and KCS2 (13). This unexpectedly high number of Arabidopsis VLCFAEs suggests that their products may have a variety of specific physiological functions. Notably, fatty acid elongase (FAE)-like elongases are unique to plants (6). In animals and fungi, ELO-type elongases synthesize the fatty acids specific to sphingolipid synthesis (22–25). All 21 Arabidopsis elongase genes were amplified by PCR using leaf and inflorescence cDNA as a template.
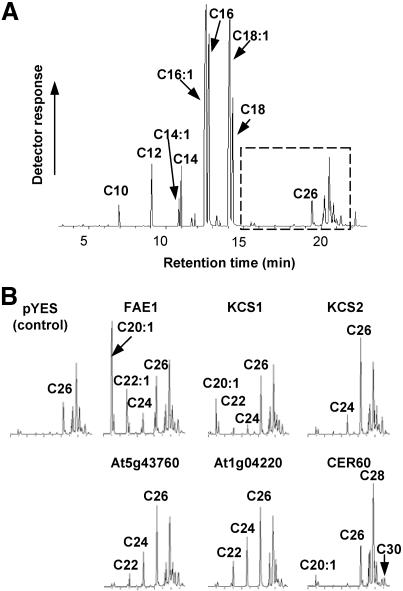
Expression in S. cerevisiae. PCR products for 17 of the 21 FAE1-like genes were cloned into the pYES2 vector for expression in Saccharomyces strain INVSc1, sequenced for verification, and expressed under a galactose-inducible promoter. After induction with galactose for 18–20 h, elongase expression was verified by immunodetection in Western blots with His-tag antibodies combined with secondary antibodies coupled to alkaline phosphatase detection. Elongase activity was determined by analyzing the total fatty acid composition of the yeast strains by using GC/MS. Control cells containing the empty pYES2 vector showed the typical fatty acid composition of yeast in which saturated and monounsaturated C16 and C18 fatty acids predominate (Fig. 1). Significant amounts of fatty acids longer than 18 carbon atoms were not observed in control cells except for the C26 fatty acid, which is a component of wild-type yeast sphingolipids. The extraction procedure does not exclude sterols, which elute later than C26:0.
Fig. 1.
Gas-chromatographic analyses of fatty acid methyl esters prepared from VLCFAE transgenic yeast cells compared with cells containing the empty vector as the control. (A) Gas-chromatographic analysis of total fatty acids from yeast cells transformed with the empty vector. The dashed box shows the VLCFA segment of the chromatogram displayed in B. Note the presence in the control of methyl esters of endogenous C26:0 and the later eluting minor peaks (sterols), which coextract with VLCFA methyl esters. (B) Gaschromatographic analysis of VLCFAs from transgenic yeast cells expressing the indicated elongase. Only the C20–C30 part of the gas chromatograms is shown. Elongation of yeast endogenous fatty acids to products from 20 up to 30 carbons in chain length can be observed in cells transformed with the corresponding elongases.
Fig. 1 shows the gas-chromatographic analyses of fatty acid methyl ester composition of transgenic lines compared with that from control cells transformed with the empty vector. Six transgenic yeast strains accumulated saturated and monounsaturated VLCFAs from 20 to 30 carbons in length (Fig. 1). Three of these elongation activities were represented by the known elongases FAE1, KCS1, and KCS2. The other three VLCFAE activities were encoded by the genes At5g43760, At1g04220, and At1g25450, annotated in the database as putative elongases with yet-unknown functions, although At1g25450 had been described as CER60, a gene that Fiebig et al. (34) presumed to be involved in VLCFA biosynthesis because of its high sequence similarity to CUT1.
As summarized in Table 1, yeasts expressing the known elongases FAE1, KCS1, and KCS2 displayed a spectrum of elongated products in agreement with previous findings (5, 9–13). Cells expressing the At5g43760 construct accumulated saturated VLCFAs from 22 to 26 carbons in length. The same result was observed with yeast containing the At1g04220 construct. Expression of CER60 resulted mostly in an increase of C26, C28, and C30 saturated fatty acids, with a small peak of monounsaturated C20 fatty acid accumulating as well. These data show that At5g43760, At1g04220, and At1g25450 (CER60) were functionally expressed in yeast cells, that they catalyze VLCFA elongation by cooperating with the dehydratase and two reductases of yeast elongase complexes, and that they are able to elongate endogenous Saccharomyces fatty acids.
Table 1. Proteins with elongation activity in S. cerevisiae.
| Elongase | Products |
|---|---|
| At1g01120 (KCS1) | C20:1/C20, C22, C24, C26 |
| At4g34520 (FAE1) | C20:1/C20, C22:1/C22, C24:1/C24, C26 |
| At5g43760 | C22, C24, C26 |
| At4g34510 (KCS2) | C20:1/C20, C24, C26 |
| At1g04220 | C22, C24, C26 |
| At1g25450 (CER60) | C20:1, C26, C28, C30 |
Different products of the elongation reaction for each elongase are shown. Main elongation products are underlined.
Several elongases produced distinctively different spectra of elongated fatty acids, indicating that different elongases catalyze different elongation steps. Furthermore, almost identical elongase functions can be observed when comparing the gaschromatographic analyses of yeasts expressing At5g43760 and At1g04220 (Table 1), which raises the possibility that different elongases might possess the same biochemical function but in different tissues.
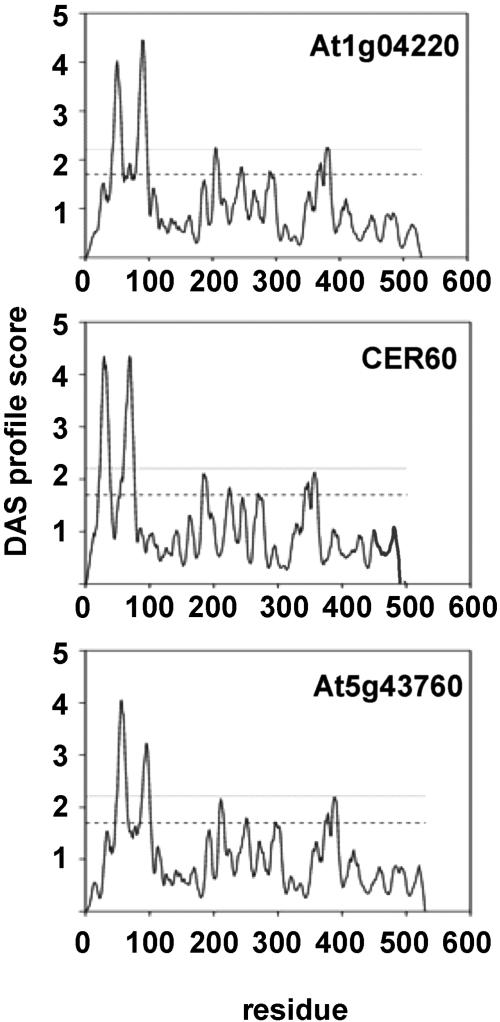
To determine whether the three previously uncharacterized condensing enzymes possess membrane-spanning domains, as suggested for FAE1 and CUT1, the DAS transmembrane prediction server (www.sbc.su.se/~miklos/DAS) was used (25). In hydropathy plots of At5g43760, At1g04220, and At1g25450 amino acid sequences, the N terminus of each sequence reveals two regions of high hydrophobicity, suggesting that each protein contains at least two transmembrane domains (Fig. 2), and FAE1 and CUT1 reveal a similar profile (data not shown). After centrifugation of disrupted transgenic yeast cells, At5g43760, At1g04220, and CER60 proteins did not accumulate in the soluble supernatant (data not shown), indicating that the expressed proteins are indeed membrane-anchored.
Fig. 2.
Hydropathy plots of the amino acid sequences of the three previously uncharacterized elongation activities according to Cserzo et al. (26). The lower dashed line represents the loose transmembrane region cutoff, and the upper dashed line represents the strict transmembrane region cutoff.
Feeding Experiments. Some expressed elongases might convert fatty acid substrates that are not available at sufficient concentrations in yeast or that are plant-specific, hence altogether lacking in yeast. To test this possibility, we fed a variety of fatty acids to transformed yeast cells and examined the resulting products (Table 2). FAE1- and At5g43760-expressing strains accumulate C20:2 and C22:2 after 15 h in medium supplemented with 500 μM C18:2, and At5g43760 strains accumulate no unsaturated VCLFAs when fed C18:3. FAE1 strains extend C18:3 to C20:3, and KCS2 strains extend neither C18:2 nor C18:3. No elongation of these substrates was observed with FDH.
Table 2. Results of feeding experiments with exogenous substrates.
| Elongase | Added fatty acid | Products |
|---|---|---|
| FAE1 | C18:2 | C20:2, C22:2 |
| C18:3 | C20:3 | |
| KCS2 | C18:2 | - |
| C18:3 | - | |
| At5g43760 | C18:2 | C20:2, C22:2 |
| C18:3 | - | |
| FDH | C20:0-C30:0 | - |
| C20:1-C26:1 | - | |
| C18:3 | - | |
| C20:4 | - | |
| C18:1-12OH | - | |
| C16-ω-diacid | - | |
| CUT1 | C26 | - |
| C28 | - |
Yeast strains expressing the VLCFAE indicated were supplemented to a 500-μM final concentration of each corresponding fatty acid individually in the culture medium 15 h before product analysis.
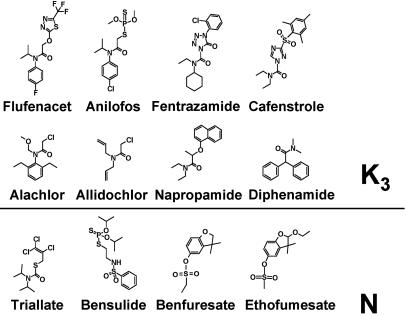
Mode of Action of K3 and N Herbicides. To investigate the herbicidal mode of action of chloroacetamides and other herbicides from HRAC classes K3 and N, the six transgenic Saccharomyces strains expressing the active elongases were incubated in the presence of different K3 and N herbicides (Fig. 3) at an initial concentration of 100 μM. In the presence of herbicide, cells no longer accumulated VLCFAs. The effects on VLCFA elongation by individual herbicides are shown in Table 3.
Fig. 3.
Structures of K3 and N herbicides tested as potential VLCFAE inhibitors in plant elongase-expressing yeast strains.
Table 3. Inhibition pattern of different elongases expressed in S. cerevisiae by group K3 and N herbicides.
| HRAC group | Inhibitor | FAE1 | KCS1 | KCS2 | CER60 | At5g43760 | At1g04220 | Phenotype |
|---|---|---|---|---|---|---|---|---|
| K3 | Alachlor | - | + | + | + | + | - | - |
| K3 | Allidochlor | + | + | + | + | + | + | - |
| K3 | Anilofos | - | + | + | + | + | + | - |
| K3 | Cafenstrole | + | + | + | + | + | + | + |
| K3 | Diphenamide | - | - | - | - | - | - | - |
| K3 | Fentrazamide | - | + | + | - | + | - | + |
| K3 | Flufenacet | + | + | + | + | + | + | + |
| K3 | Napropamide | - | - | - | - | - | - | - |
| N | Benfuresate | - | - | - | - | - | - | + |
| N | Bensulide | - | - | - | - | - | - | - |
| N | Ethofumesate | - | - | - | - | - | - | + |
| N | Triallate | - | - | - | (+) | - | - | - |
Inhibition of VLCFAE with the respective inhibitor at a concentration of 100 μM is indicated by +. In all cases, inhibition was complete at this concentration except for the inhibition of CER60 with triallate. A + in the last column indicates that a fiddlehead phenotype was induced after herbicide treatment of Arabidopsis plants (6).
Yeast cell growth was not affected during the incubation time by any of the herbicides tested here, as monitored by optical density in cultures with and without the inhibitor. Furthermore, formation of endogenous VLCFAs by Saccharomyces ELO genes (21, 22) was not inhibited, because the endogenous level of C26 fatty acids, presumably components of sphingolipids, was not changed by any herbicide added. As an additional control, yeast cultures grown in the continuous presence of herbicides rather than during the induction period only also showed no differences in C26 accumulation (data not shown) relative to untreated control cells, indicating that yeast ELO activity is indeed unaffected by the herbicides tested.
As shown in Table 3, each of the expressed elongases was inhibited by several herbicides, thereby revealing its individual pattern of active inhibitors. Conversely, some compounds inhibited a spectrum of several elongases. Flufenacet and cafenstrole inhibited all elongase activities tested here. Interestingly, inhibition was observed only with HRAC class K3 herbicides; none of the class N herbicides inhibited elongase activities in yeast except for a low inhibition that could be observed on CER60 with triallate, although treatment of Arabidopsis with ethofumesate, for example, resulted in a phenotype resembling fiddlehead, a mutant with a defect in VLCFA elongation (7, 8, 27). We previously attributed ethofumesate action to inhibition of VLCFA elongation (6).
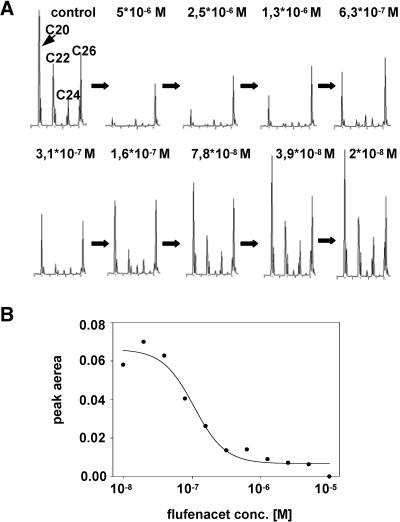
By gradually decreasing inhibitor concentrations from the initial value of 100 μM, which in each case led to a complete inhibition of elongation activity, we were able to generate dose–response curves (shown in Fig. 4). The example shown represents the inhibition of FAE1 with flufenacet. We found that up to a herbicide concentration of 10-5 M, elongation of fatty acids by FAE1 was inhibited completely in this case. The corresponding pI50 value, which designates the negative logarithm of the test compound concentration required to achieve 50% inhibition, can be determined from this curve at 7. The determination of pI50 values of each elongase with each herbicide is tedious; only some examples shown in Table 4 were measured.
Fig. 4.
Inhibition of VLCFA elongation and corresponding dose–response curve. (A) Gas-chromatographic analyses of VLCFAs from yeast cells expressing FAE1, inhibited with the indicated concentrations of flufenacet in the medium. (B) Dose–response curve generated from the peak areas of chromatograms shown in A. The peak area of C24 fatty acid at each concentration was divided by the area of the C16:1 peak as a standard.
Table 4. pI50 values [-log(M)] from inhibition of FAE1 and At5g43760 with allidochlor, cafenstrole, and flufenacet.
| Allidochlor | Cafenstrole | Flufenacet | |
|---|---|---|---|
| FAE1 | 7.1 | 6.7 | 7 |
| At5g43760 | 5.7 | 6.4 | - |
Discussion
The cuticle covers the outer surface of the leaf and stem epidermis and is the interface between the plant and the atmosphere. It has pivotal functions for the plant, including limiting the diffusion of water and solutes while permitting a controlled release of volatiles. It provides protection from attack by herbivores and pathogens and helps the plants resist drought and frost. The brunt of plant VLCFA synthesis occurs in epidermal cells. At the level of the whole plant, the main function of VLCFAs in terms of carbon mass partitioned is their role as precursors for various components of the cuticle such as cutin, epicuticular waxes, and other waxes that are embedded in suberin and cutin layers (3).
However, VLCFAs are also essential components of waxes in the tryphine layer of the extracellular pollen coat (28). They are required for proper pollen–stigma signaling and fertilization (29). Furthermore, VLCFAs accumulate as storage lipids in the seed oil of some plants, in which they are incorporated into triacylglycerols or wax esters (30). In plants, as in all eukaryotes studied thus far, an additional requirement for VLCFAs is in sphingolipid synthesis, in particular synthesis of ceramides, as components of membranes (31). In Arabidopsis, which we have studied here, VLCFAs constitute only a minor component of the ceramides (32).
As shown by a variety of phenotypes generated from mutations of VLCFAEs in plants, functions of VLCFAs are manifold and essential, making VLCFAEs interesting herbicide targets. The Arabidopsis mutants fiddlehead, hic, kcs1, fae1, and cut1 all possess defects in VLCFA elongation. Although hemizygotic fiddlehead plants display an organ-fusion phenotype (27), in hic mutants the stomatal density is affected in response to elevated CO2 (33). The kcs1 mutant phenotype has thinner stems compared with that of the wild type, and young mutants are more sensitive to low-humidity stress (12). Because of seed-specific elongase expression, the phenotype of mutant fae1 plants does not differ from wild-type plants (9). The cut1 phenotype involves sense suppression; these plants completely lack epicuticular waxes on stems and siliques and are conditionally male-sterile because of the lack of VLCFAs in the extracellular pollen coat (14). Keeping in mind that the CUT1 amino acid sequence is very similar to that of CER60, sense-suppression plants are possibly deficient in mRNAs for both genes, resulting in a stronger phenotype (34). These dysfunctions and their phytotoxic consequences show the broad spectrum of possible herbicide action caused by VLCFAE inhibition, exemplified by plants that, when treated with low concentrations of flufenacet, produce a phenocopy of the fiddlehead mutant organ-fusion phenotype.
The FAE-type elongases of plants share no sequence similarity with the ELO-type elongases that extend polyunsaturated and saturated fatty acids for sphingolipid synthesis in yeast, fungi, and animals (6, 16, 22–24). However, plants possess both FAE-type and ELO-type elongases (6, 35), the latter of which are presumably involved in sphingolipid synthesis as in yeast. However, the FAE pathway is unique to plants (6) and hence a favored herbicide target.
Three previously uncharacterized VLCFAEs identified here through heterologous expression of the Arabidopsis genes in yeast in addition to the three known elongases FAE1, KCS1, and KCS2 were specifically inhibited by K3 herbicides, identifying the condensing reaction of VLCFA biosynthesis as the activity that is inhibited by K3 herbicides. Together with the findings of Böger et al. (2) and our previous findings on the fiddlehead phenocopy (6), this result identifies VLCFAEs as the molecular targets of K3 herbicides.
Whereas 6 VLCFAEs showed clear elongation activity in yeast, 11 putative elongases did not, although they were produced as protein in yeast. Their inactivity could be caused by faulty folding, lack of cofactors, inability to interact with yeast enzymes catalyzing subsequent steps, interference by the C-terminal His tag, or a mere lack of their specific fatty acid substrates in Saccharomyces. Excluding the latter possibility (a prerequisite for excluding the others) is currently difficult, because the spectrum of fatty acid substrates that various Arabidopsis VLCFAEs might elongate is not known. Plants synthesize, elongate, and modify a much more complex spectrum of fatty acids (3) than yeast, in line with the presence of 21 different VLCFAE genes in Arabidopsis. The active VLCFAEs studied here convert endogenous yeast substrates into a spectrum of easily identified and largely gene-specific elongated products (Table 1). Identifying specificity at the substrate side of the reaction is more problematic, as shown by our feeding experiments (Table 2). Substrate specificity may bear decisively on the lack of activity observed for the 11 other VLCFAEs tested, because their natural substrates may be lacking in yeast. Conversely, the ability of At5g43760, At1g04220, At1g25450, FAE1, KCS1, and KCS2 to convert additional plant-specific substrates not present in yeast or not tested here cannot currently be excluded.
Because At3g10280 is identical to HIC except for a lack of the active-site Cys, it seems to be a nonfunctional copy of HIC (6), similar to the case of At3g52160, that lacks a His residue essential for elongation activity (6, 36). A role for covalent Cys binding by herbicides was shown recently for chalcone synthase and stilbene synthase (37), which are thought to have a reaction mechanism similar to the condensing step in VLCFA elongation. The C-terminal region of VLCFAEs is highly conserved (data not shown), suggesting a crucial role in catalytic function.
Very few VLCFAEs are well characterized concerning their substrates, products, and tissue-specific expression. In A. thaliana, these VLCFAEs are FAE1 (5, 9–11), KCS1 (12), and KCS2 (13). Arabidopsis FAE1 was shown recently to be inhibited by metolachlor through expression studies in yeast (38). The previously uncharacterized elongases from A. thaliana, At1g25450 (CER60), At5g43760, and At1g04220, expressed in S. cerevisiae under the control of a galactose-inducible promoter catalyzed accumulation of VLCFAs from 20 up to 30 carbons in length. At5g43760, At1g04220, and At1g25450 thus are condensing enzymes involved in VLCFA synthesis and furthermore are able to interact with endogenous yeast reductase and dehydratase activities to enable multiple rounds of VLCFA elongation. Yeast endogenous fatty acids, presumably C16 and C18 saturated and monounsaturated fatty acids, served as substrates. Strains transformed with At5g43760 and At1g04220 accumulated saturated C22, C24, and C26 fatty acids as products of the elongation reaction. Strains with the CER60 construct accumulated even longer-chain fatty acids from 26 to 30 carbon atoms and also seemed to have little activity with monounsaturated fatty acids, as indicated by accumulation of the C20:1 product. Because of its sequence similarity to CUT1, CER60 was presumed by Fiebig et al. (34) to be involved in wax synthesis; our findings that CER60 catalyzes the synthesis of VLCFAs 26- to 30-carbons-long support that view.
Yeast cells expressing At5g43760, At1g04220, At1g25450, FAE1, KCS1, and KCS2 grown in the presence of different K3 and N herbicides revealed that each expressed elongase was inhibited by more than one K3 herbicide, uncovering individual patterns of specific inhibitors. Conversely, each herbicide acted on more than one elongase, and some inhibited all six elongases tested. Because the herbicides showed a marked elongase preference, the condensing activity is the first and rate-limiting step in VLCFA elongation, and endogenous yeast VLCFA synthesis by ELO was not affected, the plant VLCFAE activity is clearly the target of these herbicides.
In weeds, K3 and N herbicides inhibit early development. Seeds usually germinate, but growth is inhibited and seedlings remain stunted (2). The first leaves to emerge from the coleoptile of monocots and the cotyledons of dicots are small and misformed. These phenotypes likely result from inhibition of several different elongases. The inhibition of a broad spectrum of elongases, each with different functions for the plant, might characterize the effective K3 herbicides. Furthermore, inhibition of several different elongases may explain why, despite 50 years of use, resistance to these herbicides is extremely rare.
No significant inhibition of VLCFAE activity in yeast could be observed with N class herbicides. Because earlier observations in plants demonstrated inhibition of VLCFA biosynthesis by N herbicides (39, 40), it was surprising that the elongases active in yeast were unaffected by the N class in this study, which might be because of lack of possible bioactivation of the thiocarbamates to the corresponding sulfoxides (41) or of ethofumesate and benfuresate to the corresponding semiacetales (42). Another possible reason could be low bioavailability of these inhibitors in yeast cells, but this remains to be shown. It also cannot be excluded that VLCFAEs for which no activity data are currently available are indeed inhibited by N herbicides in yeast. In earlier work, we observed the fiddlehead phenotype in Arabidopsis and other plants treated with N herbicides and attributed this to inhibition of the VLCFAE FDH, hence inducing a fiddlehead phenocopy (6). The inhibition of FDH, which was not found to be active in yeast, might therefore be of particular importance to herbicide action. Determining the role of FDH and other VLCFAEs yet to be functionally characterized in yeast requires activity assays using substrates that are accepted by these enzymes. Analysis of compounds accumulated in plants treated with K3 and N herbicides, in comparison with those accumulated in the fiddlehead mutant, may identify natural substrates to enable additional characterization of Arabidopsis VLCFAEs.
Abbreviations: HRAC, Herbicide Resistance Action Committee; VLCFA, very-long-chain fatty acid; VLCFAE, VLCFA elongase; FAE, fatty acid elongase.
References
- 1.Hamm, P. C. (1974) Weed Sci. 22, 541-545. [Google Scholar]
- 2.Böger, P., Matthes, B. & Schmalfuss, J. (2000) Pest Manag. Sci. 56, 497-508. [Google Scholar]
- 3.Post-Beittenmiller, D. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 405-430. [DOI] [PubMed] [Google Scholar]
- 4.Fehling, E. & Mukherjee, K. D. (1991) Biochim. Biophys. Acta 1082, 239-246. [DOI] [PubMed] [Google Scholar]
- 5.Millar, A. A. & Kunst, L. (1997) Plant J. 12, 121-131. [DOI] [PubMed] [Google Scholar]
- 6.Lechelt-Kunze, C., Meissner, R., Drewes, M. & Tietjen, K. (2003) Pest Manag. Sci. 59, 847-856. [DOI] [PubMed] [Google Scholar]
- 7.Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K. & Saedler, H. (1999) Plant Cell 11, 2187-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruitt, R. E., Vielle-Calzada, J.-P., Ploense, S. E., Grossniklaus, U. & Lolle, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 1311-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James, D. W. & Dooner, H. K. (1990) Theor. Appl. Genet. 80, 241-245. [DOI] [PubMed] [Google Scholar]
- 10.Kunst, L., Taylor, D. C. & Underhill, E. W. (1992) Plant Physiol. Biochem. (Paris) 30, 425-434. [Google Scholar]
- 11.James, D. W., Jr., Lim, E., Keller, J., Plooy, I., Ralston, E. & Dooner, H. K. (1995) Plant Cell 7, 309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Todd, J., Post-Beittenmiller, D. & Jaworski, J. (1999) Plant J. 17, 119-130. [DOI] [PubMed] [Google Scholar]
- 13.Clemens, S. & Kunst, L. (2001) Patent Cooperation Treaty Int. Patent Appl. WO 0107586.
- 14.Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C. & Kunst, L. (1999) Plant Cell 11, 825-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, J., Lühs, W., Sonntag, K., Zähringer, U., Borchard, D. S., Wolter, F. P., Heinz, E. & Frentzen, M. (2001) Plant Mol. Biol. 46, 229-239. [DOI] [PubMed] [Google Scholar]
- 16.Moon, Y. A., Shah, N. A., Mohapatra, S., Warrington, J. A. & Horton, J. D. (2001) J. Biol. Chem. 276, 45358-45366. [DOI] [PubMed] [Google Scholar]
- 17.Lassner, M. W., Lardizabal, K. & Metz, J. G. (1996) Plant Cell 8, 281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajikawa, M., Yamaoka, S., Yamato, K. T., Kanamaru, H., Sakuradani, E., Shimizu, S., Fukuzawa, H. & Ohyama, K. (2003) Biosci. Biotechnol. Biochem. 67, 605-612. [DOI] [PubMed] [Google Scholar]
- 19.Dohmen, R. J., Strasser, A. W., Honer, C. B. & Hollenberg, C. P. (1991) Yeast 7, 691-692. [DOI] [PubMed] [Google Scholar]
- 20.Browse, J., McCourt, P. J. & Somerville, C. R. (1986) Anal. Biochem. 152, 141-145. [DOI] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 22.Toke, D. A. & Martin, C. E. (1996) J. Biol. Chem. 271, 18413-18422. [DOI] [PubMed] [Google Scholar]
- 23.Oh, C.-S., Toke, D. A., Mandala, S. & Martin, C. E. (1997) J. Biol. Chem. 272, 17376-17384. [DOI] [PubMed] [Google Scholar]
- 24.Parker-Barnes, J. M., Das, T., Bobik, E., Leonard, A. E., Thurmond, J. M., Chaung, L.-T., Huang, Y.-S. & Mukerji, P. (2000) Proc. Natl. Acad. Sci. USA 97, 8284-8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon, H., Smith, M. A. & Kunst, L. (2001) Plant Physiol. 127, 1635-1643. [PMC free article] [PubMed] [Google Scholar]
- 26.Cserzo, M., Wallin, E., Simon, I., von Heijne, G. & Elofsson, A. (1997) Protein Eng. 10, 673-676. [DOI] [PubMed] [Google Scholar]
- 27.Lolle, S. J., Cheung, A. Y. & Sussex, I. M. (1992) Dev. Biol. 152, 383-392. [DOI] [PubMed] [Google Scholar]
- 28.Mariani, C. & Wolters-Arts, M. (2000) Plant Cell 12, 1795-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preuss, D., Lemieux, B., Yen, G. & Davis, R. W. (1993) Genes Dev. 7, 974-985. [DOI] [PubMed] [Google Scholar]
- 30.Harwood, J. L. (1980) in The Biochemistry of Plants, eds. Stumpf, P. K. & Conn, E. (Academic, New York), pp. 2-56.
- 31.Lynch, D. V. (1993) in Lipid Metabolism in Plants, ed. Moore, T. S., Jr. (CRC, Boca Raton, FL), pp. 286-308.
- 32.Uemura, M., Joseph, R. A. & Steponkus, P. L. (1995) Plant Physiol. 109, 15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray, J. E., Holroyd, G. H., van der Lee, F. M., Bahrami, A. R., Sijmons, P. C., Woodward, F. I., Schuch, W. & Hetherington, A. M. (2000) Nature 408, 713-716. [DOI] [PubMed] [Google Scholar]
- 34.Fiebig, A., Mayfield, J. A., Miley, N. L., Chau, S., Fischer, R. L. & Preuss, D. (2000) Plant Cell 12, 2001-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zank, T. K., Zähringer, U., Beckmann, C., Pohnert, G., Boland, W., Holtorf, H., Reski, R., Lerchl, J. & Heinz, E. (2002) Plant J. 31, 255-268. [DOI] [PubMed] [Google Scholar]
- 36.Ghanevati, M. & Jaworski, J. G. (2001) Biochim. Biophys. Acta 1530, 77-85. [DOI] [PubMed] [Google Scholar]
- 37.Eckermann, C., Matthes, B., Nimtz, M., Reiser, V., Lederer, B., Böger, P. & Schröder, J. (2003) Phytochemistry 64, 1045-1054. [DOI] [PubMed] [Google Scholar]
- 38.Böger, P. (2003) J. Pesticide Sci. 28, 324-329. [Google Scholar]
- 39.Abulnaja, K. O. & Harwood, J. L. (1991) Phytochemistry 30, 2883-2887. [Google Scholar]
- 40.Abulnaja, K. O., Tighe, C. R. & Harwood, J. L. (1992) Phytochemistry 31, 1155-1159. [Google Scholar]
- 41.Kern, A. J., Jackson, L. L. & Dyer, W. E. (1997) Pestic. Sci. 51, 21-26. [Google Scholar]
- 42.Kawahigashi, H., Hirose, S., Hayashi, E., Ohkawa, H. & Ohkawa, Y. (2003) Pestic. Biochem. Physiol. 74, 139-147. [Google Scholar]