Abstract
Circadian clocks control daily rhythms in physiology and behavior across all phyla. These rhythms are intrinsic to individual cells that must synchronize to their environment and to each other to anticipate daily events. Recent advances in recording from large numbers of cells for many circadian cycles have enabled researchers to begin to evaluate the mechanisms and consequences of intercellular circadian synchrony. Consequently, methods have been adapted to estimate the period, phase and amplitude of individual circadian cells and calculate synchrony between cells. Stable synchronization requires that the cells share a common period. As a result, synchronized cells maintain constant phase relationships to each (e.g. with cell 1 peaking an hour before cell 2 each cycle). This chapter reviews how circadian rhythms are recorded from single mammalian cells and details methods for measuring their period and phase synchrony. These methods have been useful, for example, in showing that specific neuropeptides are essential to maintain synchrony among circadian cells.
Keywords: circadian, Fourier transform, Period gene, vasoactive intestinal polypeptide, Rayleigh plot, synchronization index
Introduction
What is synchrony?
When a good marching band enters the field, the players step at exactly the same moment. The drummers keep time so that each band member synchronizes their paces to their neighbors’. The musicians perform with the same period. As they march across the field, the line of trumpeters might arrive at midfield first followed by, perhaps, the trombonists. The trombonists share the same period as the trumpeters, but are phase delayed in their time of arrival. In this way, they synchronize their periodicity while assuming unique phase relationships. Period synchrony (also called frequency entrainment) does not require oscillators to peak together. Instead, synchronized oscillators can establish unique, and stable, phase relationships with other oscillators in the population (phase synchrony or phase locking). In nature, noise (internal and external to the oscillators) introduces a small, bounded variation in the phase differences. Many studies of mechanical, electrical, chemical and biological oscillators have focused on mechanisms that can produce period synchrony and conditions that can alter phase synchrony (Kurths, 2003; Strogatz, 2003).
What is circadian synchrony?
Daily changes at both cellular and systemic levels arise from biological oscillators that keep near 24-hour rhythms and entrain to the 24-h cues associated with day and night. These self-sustained circadian rhythms are intrinsic to individual cells. The period of the individual cells depends predominantly on their genetics and light-dark history, and less so on the ambient temperature (i.e. their period is temperature compensated) or other environmental inputs. These cells must synchronize to each other and the environment to coordinate daily rhythms including feeding-fasting, waking-sleeping, hormone levels, metabolism and gene expression. Circadian synchrony describes when cells (or organisms) express the same, near 24-h period (Bloch, Herzog, Levine, & Schwartz, 2013). Much like the synchronized marching of a band of musicians, circadian clocks are often comprised of populations of cells that share the same daily period, but with some cells leading (by up to 12 hours) other cells. Critically, oscillators may share the same period and a constant phase relationship for one of three reasons: 1) they communicate with each other, 2) they both receive the same synchronizing signal from other cells or the environment or 3) coincidence. By measuring circadian synchrony following a perturbation, we can distinguish whether cells are entraining each other, to their environment, or simply express the same near 24-h period by chance.
Synchrony among circadian cells has been described in single celled organisms like cyanobacteria and dinoflagellates and metazoans including plants, fungi, flies and rodents. In a few cases, there is evidence that the synchrony arises primarily due to environmental inputs (e.g. cyanobacteria, dinoflagellates and plants) while cells in other systems appear to have evolved the ability to synchronize to each other (e.g. fungi, flies and rodents).
To illustrate how to measure and use synchrony in a circadian system, this chapter will focus on the mammalian suprachiasmatic nucleus (SCN). The SCN of mice and humans contains approximately 20,000 cells with many of them functioning as individual self-sustained circadian oscillators. SCN cells receive information about local day-night changes indirectly from other cell types. For example, the cells of the SCN normally entrain to input from the retina and other brain areas so that their peak metabolism and electrical activity occur during the day. For the SCN to function as a circadian pacemaker, individual SCN cells must synchronize to each other to coordinately drive rhythms in neural activity and transmitter release. Strikingly, the degree of phase synchrony among SCN cells can change with conditions. During short winter days, for example, SCN cells tend to peak together whereas, they spread out their times of peak activity during the long days of summer.
Goals of this review
This chapter aims to review how to measure synchrony between circadian cells with a focus on analyzing single-cell SCN slice bioluminescence recordings. Briefly, we summarize methods for discriminating circadian rhythms from single cells. We then discuss the strengths and weaknesses of independent methods that quantify period and phase synchrony among a population of oscillating cells. Finally we provide examples of how perturbations affect cell-cell synchrony in the SCN.
Monitoring SCN rhythms with cellular resolution
To study synchrony among SCN cells, researchers have used a variety of direct and indirect indicators of circadian physiology. The best methods share the following features: 1) relatively non-invasive monitoring of single-cell physiology, 2) high frequency sampling for more than four days, 3) sensitive enough to detect circadian rhythms above background, 4) a dynamic range that allows recording of daily, biological changes without saturating and 5) can be combined with genetic or pharmacological perturbations. To date, circadian synchrony has been assessed based on daily rhythms in cytosolic calcium, gene expression, firing rate and cAMP activity (Table 1). Figure 1 illustrates a representative, long-term recording of PERIOD2 (PER2) protein levels from SCN neurons using the PER2-luciferase (PER2::LUC) knockin reporter.
Table 1.
Methods that have been used to monitor circadian rhythms with cellular resolution.
| Daily rhythms in: | Method | Examples | Sampling period | References |
|---|---|---|---|---|
| Intracellular calcium | Fluorescent calcium-sensitive reporter | Yellow Cameleon 2.1 or 3.6 or 6.0, or GCaMP3-WPRE | 0.5 sec every 60 minutes | (Brancaccio, Maywood, Chesham, Loudon, & Hastings, 2013; Ikeda & Ikeda, 2014; Ikeda et al., 2003; Enoki, Kuroda, et al., 2012; Enoki, Ono, Hasan, Honma, & Honma, 2012; Irwin & Allen, 2013) |
| Gene expression | Bioluminescent (luciferase) or fluorescent (e.g. destabilized GFP) reporter of transcription or translation | Per1::Luc, Per1:GFP, Per1-Venus, PER2::LUC, Per2-DsRED or Bmal1::Luc | Integrated over 15–60 minutes | (Day & Schaufele, 2008; Hastings, Reddy, McMahon, & Maywood, 2005; Herzog et al., 2004; Welsh et al., 2005; Welsh & Kay, 2005; Welsh & Noguchi, 2012; Yoo et al., 2004; Cheng et al., 2009; Kuhlman, Quintero, & McMahon, 2000; Yamaguchi et al., 2003; Yamazaki et al., 2000) |
| Firing rate | Multielectrode array | MEA 60 or MED 64 | Sampled every 50 μsec to report spikes per second | (Herzog, 2007; Honma et al., 2012) |
| cAMP activity | Bioluminescent (luciferase) reporter of CREB activity or fluorescent (e.g. destabilized GFP) reporter cAMP levels | CRE::Luc, ELISA kit or ICUE2 | Integrated over 60 minutes | (Brancaccio, Maywood, Chesham, Loudon, & Hastings, 2013; An, Irwin, Allen, Tsai, & Herzog, 2011; O’Neill, Maywood, Chesham, Takahashi, & Hastings, 2008) |
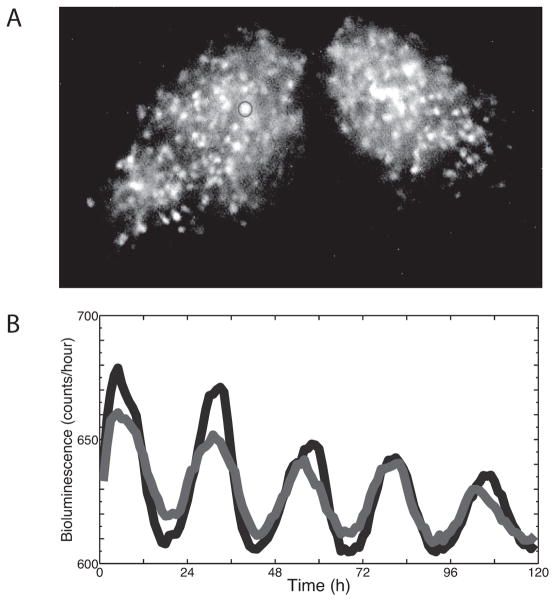
Figure 1.
Recording circadian rhythms in gene expression from a SCN slice culture. (A) An image of a SCN carrying the PER2::Luc reporter construct with two representative cells encircled with Regions Of Interest (ROI). With single-cell resolution, at least 100 ROIs can be identified from each half of the bilateral SCN. (B) The bioluminescence over time of the two cells from Figure 1A. Note that one cell (blue line) consistently peaked a few hours before the other cell (red line).
Isolating data from single cells
Once data have been collected from a population of cells, we discriminate rhythms expressed by single cells. For example, extracellular spikes originating from individual neurons can be separated from the activities of other neurons based on their shape (e.g. spike height, polarity and duration) and confirmation that they occur at intervals greater than 1 ms (i.e. do not violate the absolute refractory period of neuronal firing). Once separated, these spikes can be counted to produce firing rate (in spikes/second) as a function of recording time.
In imaging experiments (e.g. calcium or gene expression), the experimenter should be able to track light intensity from each cell by defining a region of interest over each cell. We use ImageJ software (Rasband, 1997–2014) to track circadian rhythms from cells in movies of bioluminescence reporters. To date, no one has reported an algorithm that allows for automated tracking of circadian rhythms from single cells that can dim below detection, move and overlap in space. Therefore, we provide step-by-step instructions for setting up and analyzing cellular bioluminescence from SCN slices at the end of this chapter.
Resolving the contributions of single cells allows us to resolve the contributions of each cell to the amplitude and phase of the population rhythm. The approach, however, requires a user’s time and subjective decision about which cells can be tracked over many frames in a movie. Conversely, analyses can be performed on every pixel in the image (Evans, Leise, Castanon-Cervantes, & Davidson, 2011, 2013; Foley et al., 2011; Myung et al., 2012; Pauls et al., 2014). This has the advantage of being relatively quick and automated and possible when the recorded light is too dim to visualize individual cells. It has the disadvantage that it does not track the activity of individual cells so that measures of circadian similarity (e.g. phase or period synchrony) reflect regions that contain unknown numbers of cells.
Defining a rhythm
Once we have a physiological measure from a cell over multiple days, we can assess its rhythmicity. Rhythmicity cannot be determined with methods that evaluate simply whether the data vary over time (e.g. One-way ANOVA). Instead, rhythmic data typically fulfill the requirement of being fit with a periodic (e.g. cosine) function for the duration of the recording. For each cell, we estimate its amplitude and period and its phase relative to other cells. Waveform (e.g. a cosine versus a square wave or saw-tooth function) and stability of amplitude (e.g. damping) and period (e.g. cycle-to-cycle phase variation) can dramatically influence the estimates of period, amplitude and phase. We therefore prefer to present the data in formats that allow easy visual inspection for rhythmicity and results from multiple, independent algorithms for estimating rhythmicity.
Plotting rhythmic data for visual inspection
To illustrate how firing rate, gene expression or other physiological measures vary on a daily basis, experimenters will superimpose the traces of many cells using multiple colors (Fig. 2). This format allows us to inspect rapidly the amplitude and phase of the cells within the population without paying too much attention to data from each cell. To easily visualize the phase relationships between cells, we present the data from each cell as a row in a raster plot. Raster plots can be generated in ImageJ by importing the inverted normalized values for each cell as a text image.
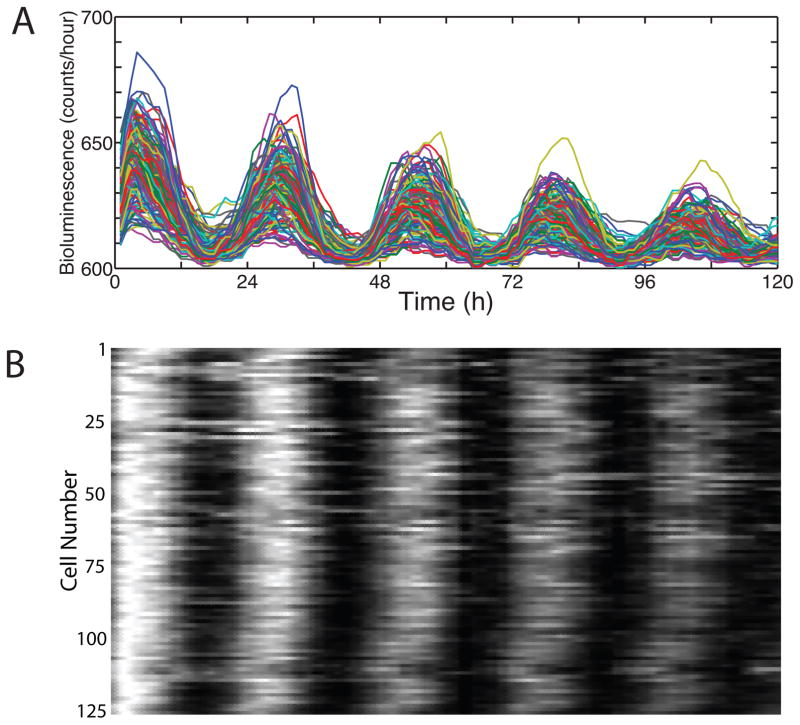
Figure 2.
Methods to visual phase and period synchrony in circadian cells. A) Superimposed traces from 125 cells, normalized to intensity values between 0.7 and 0.9. B). A raster plot of the same data. This sort of synchrony can arise from cell-cell communication within the culture, an external drive on the culture, or, although unlikely, coincidence.
We inspect the data for trends and outliers. Some methods of period estimation are particularly sensitive to gradual increases or decreases in the baseline, amplitude or period. Removing trends or outliers that are not of interest can then be applied during the analysis of the data. For example, when recording bioluminescence from dim cells, single cosmic radiation events can cause the apparent gene expression to increase more than 1000-fold for a single time point. We replace such values with the average of the two adjacent time points. Methods for detrending and removing outliers in circadian data have been nicely reviewed elsewhere (Welsh & Kay, 2005; Welsh & Noguchi, 2012; Zielinski, Moore, Troup, Halliday, & Millar, 2014).
Period Synchrony: Methods to extract and compare periods between cells
Once we have inspected the data in the time series and raster plots, we estimate the period of each cell. Most methods assume that the period and amplitude are stationary. If there were perturbations that could cause a phase shift or period change (e.g. addition of a drug) during the recording, we estimate separately the period from the data before and after the perturbation. Then, we seek to confirm any period estimates using independent methods. It is important to realize that each method has its limitations. Any report of biological periodicity approximates period and amplitude and should, ideally, include an estimate of confidence or variability. We use the period and phase estimation from rhythmic cells to characterize the synchrony in the network. Here we briefly review how three independent methods can be implemented (Table 2). Other excellent reviews compare these methods and detail others (Zielinski et al., 2014; Refinetti, Lissen, & Halberg, 2007; Refinetti, 2004; Welsh, Imaizumi, & Kay, 2005; Parati, 2004; Sokolove & Bushell, 1978; Moore, Zielinski, & Millar, 2014; Shono et al., 2000; Levine, Funes, Dowse, & Hall, 2002; Diez-Noguera, 2013; Dowse, 2009).
Table 2.
Comparison of available methods for period and phase estimation. Each method provides an independent assessment of rhythmicity with different algorithms and assumptions. Reporting similar periods and phases obtained with multiple methods gives strength to the assessment of rhythmicity and synchrony.
| Characteristics extracted
|
|||||
|---|---|---|---|---|---|
| Period | Amplitude | Phase | P value | Notes | |
| Chi-squared Periodogram | Yes | Circadian amp | No | 95% CI | Weakened by cycle-to-cycle period or amplitude variations. Typically requires at least 5 days of data for reasonable period estimates. |
| FFT-NLLS | Yes | Power or Rel-amp | Yes | 95% CI | Weakened by cycle-to-cycle period or amplitude variations. Typically requires at least 5 days of data for reasonable period estimates. Can be computationally slow. |
| Cross-over | Yes | Peak-to-trough | Yes | Not yet implemented | Does not assume period and amplitude stationarity. Provides cycle-to-cycle estimates of period and phase. Fails if noisy data produce excessive daily crossings. |
| MESA | Yes | Spectral density | No | Signal-to-Noise ratio | Weakened by cycle-to-cycle period or amplitude variations. Typically requires at least 5 days of data for reasonable period estimates. |
| Autocorrelation | Yes | Power | No | 95% CI | Weakened by cycle-to-cycle period or amplitude variations. Typically requires at least 5 days of data for reasonable period estimates. |
| Wavelet | Yes | Power | Yes | Not yet implemented | Provides continuous estimates of period and phase. |
Chi-squared Periodogram
Perhaps the most popular method for estimating circadian period is the Chi-squared Periodogram as developed by Sokolove and Bushell (Sokolove & Bushell, 1978). The method estimates the power at all periods in the data based on a simple, fast algorithm. At its core, the method cuts the data at different period lengths (e.g. between 18 and 32 hours) and quantifies the variance in the data explained by each period. It has no assumptions about waveform and does not fit the data with a function. The resulting plot of power as a function of test periods indicates which periods exceed a user-defined confidence interval (e.g. 99%). Typically, researchers report the period with the maximal power, termed the dominant circadian period. The magnitude of this peak above the confidence interval is termed circadian amplitude and provides an estimate of the strength of rhythmicity at the dominant period. Circadian amplitude increases with the amplitude of the recorded rhythm (e.g. the difference from peak to trough gene expression). Circadian amplitude, however, diminishes if the period or amplitude varies from cycle-to-cycle (e.g. with reduced period precision or amplitude damping). Many software packages support Chi-squared Periodogram analysis (Fig. 3a) including Clocklab and Lumicycle Analysis (Actimetrics Inc., Evanston, IL), BioDare (www.biodare.ed.ac.uk)(Zielinski et al., 2014), El Temps (www.el-temps.com), Circadian Rhythm Lab (www.circadian.org/periodogram.html), or Hutlab (hutlab.nl/).
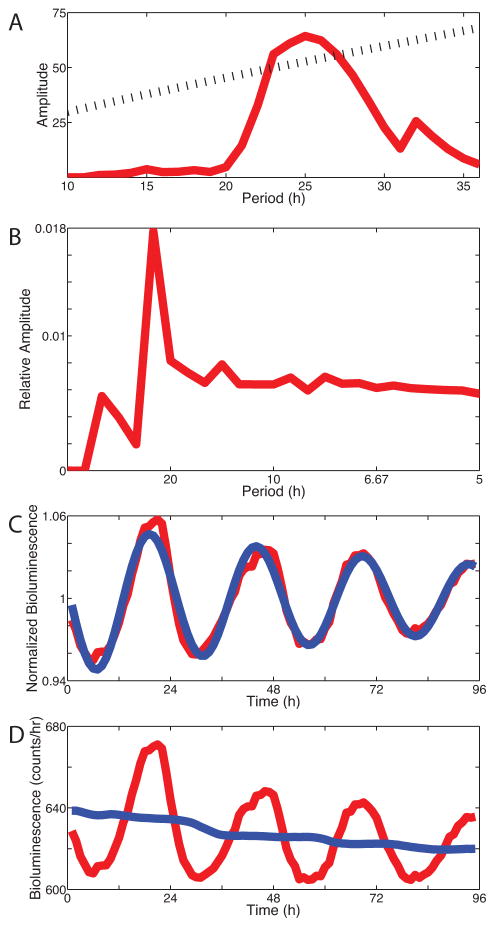
Figure 3.
Four independent methods to compare period, phase and amplitude of circadian cells. (A) The Chi-squared periodogram estimated the dominant period at 25.0 h, B) FFT-NLLS estimated the period at 23.8 h, (C) Cosinor analysis estimates the period at 24.8 h and (C) Cross-over analysis found the average period was 24.5 h based on cycle-by-cycle period measurement.
Fast Fourier Transform (FFT)
FFT is an independent method based on fitting the data with several cosine functions that differ in their periods, amplitudes and phase relationships (Johnson & Frasier, 1985; Moore et al., 2014). The dominant periodicity in the data is fit by the cosine function with the highest amplitude. We use FFT-NLLS(Straume, Frasier-Cadoret, Johnson, & Lakowicz, 1991) to estimate the dominant circadian period, its amplitude and phase, with estimates of their significance based on Monte Carlo resampling of the randomized raw data (Fig. 3b). FFT-NLLS, a variation of cosinor analysis(Halberg, 1969) (Fig 3c), is reported to be less sensitive to missing or noisy data and computationally faster than Chi-squared periodogram. We implement FFT-NLLS in Matlab; it is also available through BioDare (Zielinski et al., 2014)(www.biodare.ed.ac.uk).
Cross-over analysis
We also use “cross-over analysis” to estimate the period, peak-to-trough amplitude and phase from cycle-to-cycle because, unlike Chi-squared periodogram and FFT-NLLS, it does not assume these parameters are constant in the biological data. This approach calculates two running averages from the raw data, with a 3-h and a 24-h window, respectively. The 24-h smooth removes non-circadian trends and provides a baseline, while the 3-h smooth reveals the circadian oscillations around this baseline. We calculate the period from the daily intersections of these two lines (Fig. 3d). The method is based on a simplified Poincaré-section based recurrence time analysis(Kurths, 2003). To date, no statistical tests have been integrated into this method and it can be sensitive to missing or noisy data, but it is fast, intuitive, and provides details about how the rhythm changes from day to day.
Other methods of period estimation
The above list emphasizes some of the most common methods used to estimate circadian periodicity. Other methods including Maximum Entropy Spectral Analysis (MESA) and autocorrelation (Levine et al., 2002) have been used, especially by researchers studying fly circadian biology. In addition, wavelet-based methods are gaining popularity in the field (Leise, 2013; Leise, Indic, Paul, & Schwartz, 2013; Bours, Muthuraman, Bouwmeester, & van der Krol, 2012; Harang, Bonnet, & Petzold, 2012; Leise & Harrington, 2011; Meeker et al., 2011; Price, Baggs, Curtis, Fitzgerald, & Hogenesch, 2008; Chan, Wu, Lam, Poon, & Poon, 2000) and are reviewed elsewhere in this volume (Leise et al. Chapter). The field will benefit from a comprehensive toolbox that allows easy comparisons of periodicity estimated by multiple, independent methods. Furthermore, we lack careful validation of all of these methods using realistic, synthetic data (e.g. with known period and noise characteristics that resemble data we collect (Deckard, Anafi, Hogenesch, Haase, & Harer, 2013; Refinetti, 1993). Although we must always balance sampling rate and sampling duration against experimental realities of time, resources and money consumed, we have arrived at a point where we have multiple, independent methods that, when they converge on a period estimate, allow us to assess whether circadian cells are synchronized.
Do cells share the same period?
To test whether cells are likely to have synchronized their circadian periods, we next examine the distribution of dominant periods in the population (Fig. 4a). We use the standard deviation of the distribution as an indicator of the degree of period synchrony. Statistical tests comparing the variation between populations (e.g. Levene’s or Brown-Forsythe’s) can provide additional evidence for changes in the degree of period synchrony.
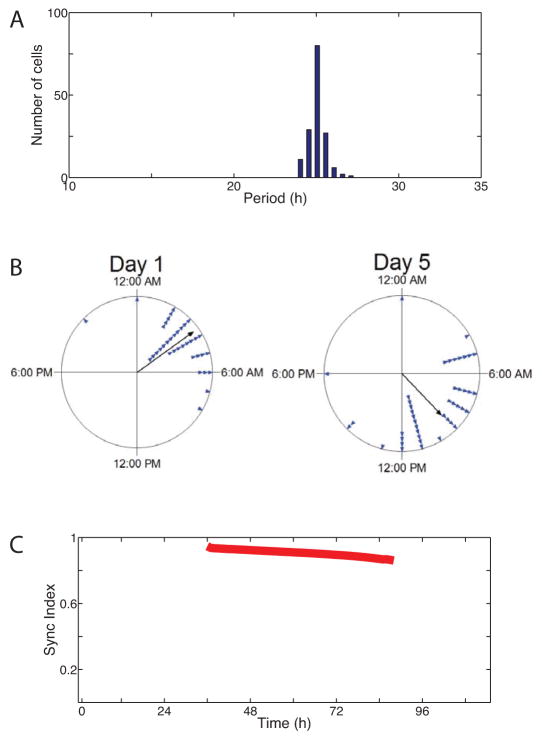
Figure 4.
Period and phase synchrony estimation in a population of cells. (A) The period distribution of the population of cells within the cultured SCN from Fig 1A. Note that the narrow distribution indicates the cells were period synchronized. (B) The times of peak Per2 expression for each cell (triangles) on days 1 and 5 of a recording presented as Rayleigh plots. Note that phases remained significantly clustered during the recording. (C) The phase synchrony (termed Sync Index, Kuramoto Order Parameter or Raleigh Statistic, R) of all cells (n=125) indicates that synchrony decreased modestly over the recording.
Phase Synchrony: Methods to extract and compare phase relationships between cells
Just as a raster plot (Fig. 2b) provides a convenient method to visualize whether the phase relationships between cells remain stable during a recording, Rayleigh plots provide a statistical test of phase clustering among cells (Fig. 4b). The first step is to identify a marker that reliably indicates the phase of the oscillating cell. For example, the time of daily peak of PER2 expression is often used as a reliable phase marker of SCN cells. For the most accurate estimation of clock phase within each cell, conventional wisdom holds that you should use the phase marker with the least variability from cycle to cycle (Herzog, Aton, Numano, Sakaki, & Tei, 2004; Liu & Reppert, 2000).
Next, we apply the Rayleigh test (Batschelet & Batschelet, 1981), from circular statistics to quantify the degree of synchrony among cells. This test is appropriate because phase (e.g. the time of day when a cell fires maximally) cannot be outside a specific range (e.g. 0–24 hours or 0–360 degrees). For this reason, it is not possible to test whether cells are synchronized using linear statistics like a One-Way ANOVA. The resulting Rayleigh Statistic, R (also termed the “Sync Index”), quantifies the degree of clustering among cells, ranging from 0 (uniformly distributed phases across the day) to 1.0 (all cells peak at the same time of day). The resulting p-value indicates the likelihood that the observed clustering occurred by chance. We implement the Rayleigh Test with Oriana Software (Kovach Computing Services, Pentraeth, UK).
It is important to understand that Sync Index is an imperfect measure of synchrony. A population of synchronized oscillators can have a low or high R value. For example, cells that communicate to have identical periods and phases will have an R equal to 1 while cells that coordinate to have identical periods, but that peak in anti-phase, will have an R equal to 0. In addition, phase definition inconsistencies can induce a biased (often increased) R(Kralemann, Cimponeriu, Rosenblum, Pikovsky, & Mrowka, 2008). Therefore, it can be helpful to monitor how cellular periods and their sync index change over time or as a result of a perturbation.
Perturbations reveal synchronization mechanisms
To test whether synchrony changes during a recording, we can apply the Rayleigh Test for each cycle of data. Alternatively, we can use methods like wavelet to extract the phase of each cell at all times in the recording and calculate the Sync Index over time (Fig. 4c). The Sync Index provides a convenient metric to monitor when treatments perturb phase synchrony among cells.
This approach has been applied to test, for example, whether the neuropeptide, vasoactive intestinal polypeptide (VIP), is required for circadian synchrony. Normally, the Sync Index of dispersed SCN cells cultured on multi-electrode arrays approaches 0.4 based on firing rate recordings. When the gene for Vip or its receptor, Vipr2, were deleted, the sync index dropped to 0.05 (Aton, Colwell, Harmar, Waschek, & Herzog, 2005). Similarly, pharmacological disruption of cell-cell communication with drugs like tetrodotoxin or pertussis toxin can reduce the sync index of PER2::Luc rhythms in a SCN slice from nearly 0.7 to 0.1(Aton, Huettner, Straume, & Herzog, 2006; Webb, Taylor, Thoroughman, Doyle III, & Herzog, 2012). Importantly, the reduction in phase synchrony was accompanied by a broadened distribution of periods expressed by the cells, demonstrating that these genetic and pharmacological interventions disrupted the mechanisms underlying synchronization.
Methods awaiting application in circadian biology
Over the past 20 years, fields such as non-linear physics have been inspired by biological systems to develop methods for measuring synchrony. Here, we highlight a few that could be applied in circadian biology. The Perturbation-free Method evaluates statistically significant levels of synchrony based on a twin-surrogate analysis of recurrence plots(Thiel, Romano, Kurths, Rolfs, & Kliegl, 2006). Synchrony induced by direct connections (versus indirect connections) between oscillators can be distinguished using Granger Causality(Nawrath et al., 2010). Synchrony measures have been developed for systems like the bilateral SCN where groups of oscillators interact(Kiss, Quigg, Chun, Kori, & Hudson, 2008). When the network topology is known, cell-specific measures of Sync Index have been proposed to characterize spatially-organized, partially synchronized states(Kuramoto & Battogtokh, 2002). The application of these methods awaits circadian data with more samples per day and for more days.
Step-by-step instructions for measuring synchrony in SCN slice
Bioluminescence recordings using a charge-coupled device (CCD) camera
To record from a cultured SCN slice containing a bioluminescence reporter (e.g. PER2::LUC), we use spatial (2 x 2 pixels) and temporal binning (e.g. 1-h integration) to reliably collect enough light to track over 100 cells distributed throughout the unilateral SCN.
-
We record for at least 4 days.
Note: Culturing the SCN slice for at least 2 days before recording allows the slice to flatten and minimizes SCN cell movement during the recording.
Image processing
Using an image processing program (e.g. ImageJ), we maximize the signal to noise ratio from glowing cells in three standard steps:
Adjacent Frame Minimization: We compare each pixel’s intensity in adjacent frames and assign the lower value to that pixel. This effectively removes noise due to dark current in the CCD camera and absorbed cosmic radiation.
Thresholding: If cosmic radiation persists after adjacent frame minimization, we can set the max threshold of the movie at the value of the cosmic radiation. This takes advantage of the brightness of cosmic spots in comparison to the rest of the movie.
Post-collection temporal binning: We can bin frames in segments up to 4 hours, this makes individual cells brighter and also cuts down on the length of the movie – facilitating tracking.
Single cell tracking
We identify single cells that persist for the duration of the movie by eye.
We encircle each cell with a region of interest (ROIs).
-
We quantify the integrated intensity within each ROI for the duration of the movie.
Note: Some cells cannot be tracked by this method. If a cell disappears for more than a two hours, we do not include it in the final analysis. This can be particularly problematic, for example, when measuring dim reporters where cells have expression near the detection limits of available cameras (e.g. PER2::Luc in the ventral SCN or Bmal1:Luc in astrocytes).
Data presentation
Raster Plot: Raster plots use inverted normalized data. First, we normalize the data from each individual ROI, with the highest and lowest bioluminescence values in the recording becoming 1 and 0, respectively. These data are inverted and saved as a text file.
We import the normalized bioluminescence over time into ImageJ,
-
We generate a raster plot using the text image function as a raster plot.
Note: When normalizing data, it becomes critical that cosmic radiation has been removed. Looking at the individual raw data traces can help identify any spurious peaks.
Rayleigh Plot: For any 24 hours of data, we measure and plot the times of peak of bioluminescence from each cell in degrees (0–360).
Acknowledgments
The authors thank members of the St. Louis Clocksclub for valuable discussions, Drs. Stephanie Taylor, Kirsten Meeker and Linda Petzold for help implementing code in Matlab and Drs. John Hogenesch and Andrew Millar for generously developing and maintaining analytic tools for the field. This work is dedicated to Elise Herzog and was supported in part by NIGM grants 96873 and 104991.
References
- An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011;105(5):2289–2296. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci US A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschelet E, Batschelet E. Circular statistics in biology. Academic Press; London: 1981. p. 371. [Google Scholar]
- Bloch G, Herzog ED, Levine JD, Schwartz WJ. Socially synchronized circadian oscillators. Proceedings of the Royal Society B: Biological Sciences. 2013;280(1765):20130035. doi: 10.1098/rspb.2013.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bours R, Muthuraman M, Bouwmeester H, van der Krol A. OSCILLATOR: A system for analysis of diurnal leaf growth using infrared photography combined with wavelet transformation. Plant Methods. 2012;8(1):29. doi: 10.1186/1746-4811-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78(4):714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FH, Wu BM, Lam FK, Poon PW, Poon AM. Multiscale characterization of chronobiological signals based on the discrete wavelet transform. IEEE Trans Biomed Eng. 2000;47(1):88–95. doi: 10.1109/10.817623. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, Obrietan K. Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009;18(16):3110–3124. doi: 10.1093/hmg/ddp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RN, Schaufele F. Fluorescent protein tools for studying protein dynamics in living cells: a review. J Biomed Opt. 2008;13(3):031202. doi: 10.1117/1.2939093. [DOI] [PubMed] [Google Scholar]
- Deckard A, Anafi RC, Hogenesch JB, Haase SB, Harer J. Design and analysis of large-scale biological rhythm studies: a comparison of algorithms for detecting periodic signals in biological data. Bioinformatics. 2013:btt541. doi: 10.1093/bioinformatics/btt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Noguera A. Methods for serial analysis of long time series in the study of biological rhythms. J Circadian Rhythms. 2013;11(1):7. doi: 10.1186/1740-3391-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowse HB. Analyses for physiological and behavioral rhythmicity. Methods in enzymology. 2009;454:141–174. doi: 10.1016/S0076-6879(08)03806-8. [DOI] [PubMed] [Google Scholar]
- Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S, Honma K. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A. 2012;109(52):21498–21503. doi: 10.1073/pnas.1214415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki R, Ono D, Hasan MT, Honma S, Honma KI. Single-cell resolution fluorescence imaging of circadian rhythms detected with a Nipkow spinning disk confocal system. Journal of neuroscience methods. 2012;207(1):72–79. doi: 10.1016/j.jneumeth.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011;6(1):e15869. doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80(4):973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NC, Tong TY, Foley D, LeSauter J, Welsh DK, Silver R. Characterization of orderly spatiotemporal patterns of clock gene activation in mammalian suprachiasmatic nucleus. European Journal of Neuroscience. 2011;33(10):1851–1865. doi: 10.1111/j.1460-9568.2011.07682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halberg F. Frequency spectra and cosinor for evaluating circadian rhythms in rodent data and in man during Gemini and Vostok flights. Life sciences and space research. 1969;8:188–214. [PubMed] [Google Scholar]
- Harang R, Bonnet G, Petzold LR. WAVOS: a MATLAB toolkit for wavelet analysis and visualization of oscillatory systems. BMC Res Notes. 2012;5:163. doi: 10.1186/1756-0500-5-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, McMahon DG, Maywood ES. Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods in enzymology. 2005;393:579–592. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]
- Herzog ED. Neurons and networks in daily rhythms. Nature Reviews Neuroscience. 2007;8(10):790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms. 2004;19(1):35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- Honma S, Ono D, Suzuki Y, Inagaki N, Yoshikawa T, Nakamura W, Honma K. Suprachiasmatic nucleus: cellular clocks and networks. Progress in brain research. 2011;199:129–141. doi: 10.1016/B978-0-444-59427-3.00029-0. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ikeda M. Bmal1 Is an Essential Regulator for Circadian Cytosolic Ca2+ Rhythms in Suprachiasmatic Nucleus Neurons. The Journal of Neuroscience. 2014;34(36):12029–12038. doi: 10.1523/JNEUROSCI.5158-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. Circadian Dynamics of Cytosolic and Nuclear Ca2+ in Single Suprachiasmatic Nucleus Neurons. Neuron. 2003;38(2):253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- Irwin RP, Allen CN. Simultaneous electrophysiological recording and calcium imaging of suprachiasmatic nucleus neurons. J Vis Exp. 2013;(82):50794. doi: 10.3791/50794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Frasier SG. Nonlinear least squares analysis. Methods Enzymology. 1985;117:301–342. [Google Scholar]
- Kiss IZ, Quigg M, Chun SHC, Kori H, Hudson JL. Characterization of synchronization in interacting groups of oscillators: application to seizures. Biophysical journal. 2008;94(3):1121–1130. doi: 10.1529/biophysj.107.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralemann B, Cimponeriu L, Rosenblum M, Pikovsky A, Mrowka R. Phase dynamics of coupled oscillators reconstructed from data. Physical Review E. 2008;77(6):066205. doi: 10.1103/PhysRevE.77.066205. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport. 2000;11(7):1479–1482. [PubMed] [Google Scholar]
- Kuramoto Y, Battogtokh D. Coexistence of Coherence and Incoherence in Nonlocally Coupled Phase Oscillators. NONLINEAR PHENOMENA IN COMPLEX SYSTEMS. 2002;5(4):380–385. [Google Scholar]
- Kurths J. Synchronization: A universal concept in nonlinear sciences. Vol. 12. Cambridge University Press; 2003. [Google Scholar]
- Leise TL. Wavelet analysis of circadian and ultradian behavioral rhythms. J Circadian Rhythms. 2013;11(1):5. doi: 10.1186/1740-3391-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leise TL, Harrington ME. Wavelet-based time series analysis of circadian rhythms. Journal of biological rhythms. 2011;26(5):454–463. doi: 10.1177/0748730411416330. [DOI] [PubMed] [Google Scholar]
- Leise TL, Indic P, Paul MJ, Schwartz WJ. Wavelet meets actogram. Journal of biological rhythms. 2013;28(1):62–68. doi: 10.1177/0748730412468693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25(1):123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Meeker K, Harang R, Webb AB, Welsh DK, Doyle FJ, 3rd, Bonnet G, Herzog ED, Petzold LR. Wavelet measurement suggests cause of period instability in mammalian circadian neurons. J Biol Rhythms. 2011;26(4):353–362. doi: 10.1177/0748730411409863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A, Zielinski T, Millar AJ. Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. Methods Mol Biol. 2014;1158:13–44. doi: 10.1007/978-1-4939-0700-7_2. [DOI] [PubMed] [Google Scholar]
- Myung J, Hong S, Hatanaka F, Nakajima Y, De Schutter E, Takumi T. Period coding of Bmal1 oscillators in the suprachiasmatic nucleus. The Journal of Neuroscience. 2012;32(26):8900–8918. doi: 10.1523/JNEUROSCI.5586-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath J, Romano MC, Thiel M, Kiss IZ, Wickramasinghe M, Timmer J, Kurths J, Schelter B. Distinguishing direct from indirect interactions in oscillatory networks with multiple time scales. Physical review letters. 2010;104(3):038701. doi: 10.1103/PhysRevLett.104.038701. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320(5878):949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parati G. Assessing circadian blood pressure and heart rate changes: advantages and limitations of different methods of mathematical modelling. Journal of hypertension. 2004;22(11):2061–2064. doi: 10.1097/00004872-200411000-00005. [DOI] [PubMed] [Google Scholar]
- Pauls S, Foley N, Foley D, LeSauter J, Hastings M, Maywood E, Silver R. Differential contributions of intra-cellular and inter-cellular mechanisms to the spatial and temporal architecture of the suprachiasmatic nucleus circadian circuitry in wild-type, cryptochrome-null and vasoactive intestinal peptide receptor 2-null mutant mice. European Journal of Neuroscience. 2014 doi: 10.1111/ejn.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TS, Baggs JE, Curtis AM, Fitzgerald GA, Hogenesch JB. WAVECLOCK: wavelet analysis of circadian oscillation. Bioinformatics. 2008;24(23):2794–2795. doi: 10.1093/bioinformatics/btn521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. Bethesda, Maryland, USA: U.S. National Institutes of Health; 1997–2014. http://imagej.nih.gov/ij/) [Google Scholar]
- Refinetti R. Laboratory instrumentation and computing: comparison of six methods for the determination of the period of circadian rhythms. Physiology & behavior. 1993;54(5):869–875. doi: 10.1016/0031-9384(93)90294-p. [DOI] [PubMed] [Google Scholar]
- Refinetti R. Non-stationary time series and the robustness of circadian rhythms. Journal of theoretical biology. 2004;227(4):571–581. doi: 10.1016/j.jtbi.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Refinetti R, Lissen GC, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res. 2007;38(4):275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shono M, Shono H, Ito Y, Muro M, Maeda Y, Sugimori H. A new periodogram using one-way analysis of variance for circadian rhythms. Psychiatry Clin Neurosci. 2000;54(3):307–308. doi: 10.1046/j.1440-1819.2000.00690.x. [DOI] [PubMed] [Google Scholar]
- Sokolove PG, Bushell WN. The chi square periodogram: its utility for analysis of circadian rhythms. Journal of Theoretical Biology. 1978;72(1):131–160. doi: 10.1016/0022-5193(78)90022-x. [DOI] [PubMed] [Google Scholar]
- Straume M, Frasier-Cadoret SG, Johnson ML. Topics in fluorescence spectroscopy. Springer US; 2002. Least-squares analysis of fluorescence data; pp. 177–240. [Google Scholar]
- Strogatz S. Sync: The emerging science of spontaneous order. Hyperion; 2003. [Google Scholar]
- Thiel M, Romano MC, Kurths J, Rolfs M, Kliegl R. Twin surrogates to test for complex synchronisation. EPL (Europhysics Letters) 2006;75(4):535. [Google Scholar]
- Webb AB, Taylor SR, Thoroughman KA, Doyle FJ, III, Herzog ED. Weakly circadian cells improve resynchrony. PLoS computational biology. 2012;8(11):e1002787. doi: 10.1371/journal.pcbi.1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods in enzymology. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Kay SA. Bioluminescence imaging in living organisms. Current opinion in biotechnology. 2005;16(1):73–78. doi: 10.1016/j.copbio.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harbor Protocols. 2012;2012(8) doi: 10.1101/pdb.top070607. pdb-top070607. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302(5649):1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi RI, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, … Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ. Strengths and limitations of period estimation methods for circadian data. PLoS One. 2014;9(5):e96462. doi: 10.1371/journal.pone.0096462. [DOI] [PMC free article] [PubMed] [Google Scholar]