Abstract
Extracellular-superoxide dismutase (genetic name SOD3) is a secreted anti-oxidative enzyme, and its presence in vascular walls may play an important role in protecting the vascular system against oxidative stress. Oxidative stress has been implicated in the pathogenesis of diabetic retinopathy; therefore, increases in extracellular-superoxide dismutase have been suggested to inhibit the progression of diabetic retinopathy. Incretin-based drugs such as glucagon-like peptide-1 receptor agonists are used in the treatment of type 2 diabetes. Glucagon-like peptide-1 receptor agonists are expected to function as extrapancreatic agents because the glucagon-like peptide-1 receptor is expressed not only in pancreatic tissues, but also in many other tissue types. We herein demonstrated that exendin-4, a glucagon-like peptide-1 receptor agonist, induced the expression of extracellular-superoxide dismutase in human retinal microvascular endothelial cells through epigenetic regulation. The results of the present study demonstrated that exendin-4 induced the expression of extracellular-superoxide dismutase through histone H3 acetylation at the SOD3 proximal promoter region. Moreover, plasma extracellular-superoxide dismutase concentrations in diabetic patients were elevated by incretin-based therapies. Therefore, incretin-based therapies may exert direct extrapancreatic effects in order to protect blood vessels by enhancing anti-oxidative activity.
Keywords: extracellular-superoxide dismutase, incretin-based therapy, exendin-4, epigenetics, diabetic retinopathy
Introduction
Diabetic retinopathy (DR) is one of main causes of visual disorders and ultimately leads to blindness. The development of DR is characterized by intraretinal microvascular abnormalities, particularly those involving dysfunctions in endothelial cells in the initial stage of DR.(1) Oxidative stress is triggered by the excessive endogenous and exogenous production of reactive oxygen species (ROS) and/or their insufficient removal. Enhanced oxidative stress is considered to be one of the main contributors to the pathogenesis of DR.(2) A previous study reported that the administration of antioxidants prevented the development of DR in rats.(3) Exendin-4 (Ex4), a glucagon-like peptide-1 (GLP-1) receptor agonist, was initially discovered in the saliva of the Gila monster, Heloderma suspectum, and shares 53% sequence homology with GLP-1.(4,5) It was approved as a treatment, called “exenatide”, for type 2 diabetes in Japan in 2010. GLP-1 stimulates the secretion of insulin from pancreatic β-cells, but suppresses that of glucagon from α-cells by binding to the GLP-1 receptors on these cells, thereby lowering blood glucose concentrations. The GLP-1 receptor is expressed not only in the pancreas, but also in the heart, intestines, kidney, brain, and other tissues.(6) Therefore, GLP-1 and its receptor agonists are expected to directly exert extrapancreatic effects.
The antioxidant defense system is a crucial component in the maintenance of redox homeostasis, and its impairment leads to the enhancement of oxidative stress. Superoxide dismutase (SOD) is an antioxidative enzyme that protects cells against oxidative stress, and a SOD deficiency has been shown to increase the risk of developing various diseases, such as type 2 diabetes, atherosclerosis, and asthma.(7–9) Three SOD isozymes have been identified in mammals: copper and zinc-containing SOD (Cu,Zn-SOD), manganese-containing SOD (Mn-SOD), and extracellular-SOD (EC-SOD, genetic name SOD3). EC-SOD is the only isozyme of SOD that is secreted into the extracellular space and is widely distributed in tissues.(10,11) In the vascular system, EC-SOD secreted mainly from fibroblasts and smooth muscle cells slowly diffuses and binds to heparan sulfate proteoglycans on the cell surface, in the basal membranes, and in the extracellular matrix. On the other hand, EC-SOD is very weakly expressed in endothelial cells even though these cells are a component of the blood vessel wall.(12,13) The microenvironment created by neutrophil-endothelial cell interactions plays an important role in the progression of vascular injury because the concentration of superoxide released at this site is sufficient to denaturalize redox homeostasis. The presence of a high level of EC-SOD in vessel walls may play an important protective role against vascular disorders induced by superoxide.(14) Previous studies showed that EC-SOD activity was decreased in type 2 diabetes.(15–17) On the other hand, the enhanced expression of EC-SOD has been shown to mitigate diabetic diseases by attenuating oxidative stress.(18)
Epigenetics is defined as mitotically heritable changes in gene expression that do not change the DNA sequence, and include DNA methylation and histone modifications.(19) The methylation of CpG within a gene promoter, in which a methyl group is added to the 5' carbon of cytosine in CpG, is associated with transcriptional gene silencing.(20) On the other hand, modifications in the N-terminal tail of histone, such as acetylation, methylation, and phosphorylation at lysine, arginine, or serine residues, have been shown to induce or suppress gene expression.(21,22) The onset and development of diseases including diabetes has been suggested to occur through abnormalities in epigenetic regulation, such as the promotion of histone deacetylation.(23) A recent study reported that human EC-SOD expression is regulated by DNA methylation and histone acetylation.(24–26)
As described above, diabetes is closely related to EC-SOD expression and/or activity, and the up-regulated expression of EC-SOD has been suggested to mitigate diabetes. Ex4 has recently been shown to epigenetically regulate gene expression.(27,28) In the present study, we determined whether Ex4 regulates the expression of EC-SOD in human retinal endothelial cells (HRECs) via an epigenetic mechanism. Moreover, changes in plasma EC-SOD were assessed in diabetic patients starting incretin-based therapies.
Materials and Methods
Reagents
5-Azacytidine (Aza) was purchased from Wako Pure Chem. Ind., Ltd. (Osaka, Japan). Trichostatin A (TSA) and valproic acid (VPA) were purchased from Cayman Chemical (Ann Arbor, MI). Ex4 and Exendin-(9-39) (Ex9-39) were purchased from AnaSpec (Fremont, CA) and GenScript (Piscataway, NJ), respectively. McrBC was purchased from New England BioLabs (Beverly, MA). Anti-acetyl histone H3 and H4 rabbit polyclonal antibodies and an anti-actin mouse monoclonal antibody were purchased from Millipore Co. (Billerica, MA). Anti-rabbit and -mouse IgG (whole molecule)-peroxidase antibodies were purchased from Sigma-Aldrich, Inc. (St. Louis, MO).
Cell culture
Human retinal microvascular endothelial cells (HRECs) and CSC complete recombinant medium were purchased from DS Pharma Biomedical Co. (Osaka, Japan). HRECs were cultured in CSC complete recombinant medium containing 100 units/ml penicillin and 100 µg/ml streptomycin. Human lung adenocarcinoma A549 cells and human breast cancer MCF-7 cells were cultured in Dulbecco’s modified eagle medium (DMEM) containing 10% (v/v) fetal calf serum (FCS) and antibiotics. Human leukemic THP-1 cells were cultured in RPMI 1640 medium containing inactivated 10% (v/v) FCS and antibiotics. Cells were maintained at 37°C in a humidified 5% CO2 incubator. The culture medium was replaced every 2 days.
Reverse transcriptional-polymerase chain reaction (RT-PCR) analysis
HRECs and other cells were cultured in 60-mm culture dishes overnight and treated with Ex4, Ex9-39, TSA, VPA, or Aza. After being treated for 24 h, cells were washed with cold phosphate-buffered saline (PBS) and total RNA was extracted from cells with TRIzol reagent (Invitrogen, Carlsbad, CA). The preparation of cDNA was performed using a ReverTra Ace® qPCR RT Kit (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. RT-PCR was performed by the method described in our previous study.(29) The primer sequences used in RT-PCR are presented in Table 1. After amplification, aliquots of the PCR mixtures were separated on 2% (w/v) agarose gels, stained with ethidium bromide, and visualized using FLA5100 (Fuji Film, Tokyo, Japan). Real-time RT-PCR was carried out using the ThunderbirdTM SYBR qPCR Mix (Toyobo) according to the manufacturer’s protocol. EC-SOD primers were used in QuantiTect® Primer Assay Hs_SOD3_2_SG (Qiagen, Chatsworth, CA). The other primer sequences used in real-time RT-PCR were presented in Table 1. mRNA levels were normalized to those of 18S rRNA mRNA in each sample.
Table 1.
Primer sequences used in RT-PCR and real-time RT-PCR
| Primer | Sequences | |
|---|---|---|
| EC-SOD | forward | 5'-AGAAAGCTCTCTTGGAGGAG-3' |
| reverse | 5'-ACCGCGAAGTTGCCGAAGTC-3' | |
| Cu,Zn-SOD | forward | 5'-GCGACGAAGGCCGTGTGCGTG-3' |
| reverse | 5'-TGTGCGGCCAATGATGCAATG-3' | |
| Mn-SOD | forward | 5'-CGACCTGCCCTACGACTACGG-3' |
| reverse | 5'-CAAGCCAACCCCAACCTGAGC-3' | |
| HDAC1 | forward | 5'-CCTGAGGAGAGTGGCGATGA-3' |
| reverse | 5'-GTTTGTCAGAGGAGCAGATCGA-3' | |
| HDAC2 | forward | 5'-GCTCTCAACTGGCGGTTCAG-3' |
| reverse | 5'-AGCCCAATTAACAGCCATATCAG-3' | |
| HDAC3 | forward | 5'-CCCAGACTTCACACTTCATCCA-3' |
| reverse | 5'-GGTCCAGATACTGGCGTGAGTT-3' | |
| HDAC4 | forward | 5'-GACCTGACCGCCATTTGC-3' |
| reverse | 5'-GGGAGAGGATCAAGCTCGTTT-3' | |
| HDAC5 | forward | 5'-CAACGAGTCGGATGGGATGT-3' |
| reverse | 5'-GGGATGCTGTGCAGAGAAGTC-3' | |
| HDAC6 | forward | 5'-TGCCTCTGGGATGACAGCTT-3' |
| reverse | 5'-CCTGGATCAGTTGCTCCTTGA-3' | |
| HDAC7 | forward | 5'-AGCAGCTTTTTGCCTCCTGTT-3' |
| reverse | 5'-TCTTGCGCAGAGGGAAGTG-3' | |
| HDAC8 | forward | 5'-CGGCCAGACCGCAATG-3' |
| reverse | 5'-CACATGCTTCAGATTCCCTTT-3' | |
| HDAC9 | forward | 5'-AGGCTCTCCTGCAGCATTTATT-3' |
| reverse | 5'-AAGGGAACTCCACCAGCTACAA-3' | |
| HDAC10 | forward | 5'-ATGACCCCAGCGTCCTTTACT-3' |
| reverse | 5'-CGCAGGAAAGGCCAGAAG-3' | |
| HDAC11 | forward | 5'-CCCCTTGGTCATGGGATTT-3' |
| reverse | 5'-CATCCACACCAGTGCCTATAGC-3' | |
| 18S rRNA | forward | 5'-CGGCTACCACATCCAAGGAA-3' |
| reverse | 5'-GCTGGAATTACCGCGGCT-3' |
Methylation-specific PCR (MSP) analysis
Genomic DNA was isolated from HRECs and fibroblasts using a Puregene Core kit (Qiagen) according to the manufacturer’s protocol. The bisulfite modification of genomic DNA was carried out using an EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol. An aliquot of bisulfite-treated DNA (500 ng) was subjected to MSP amplification. The primer sequences used in MSP of the SOD3 promoter and coding regions were designed for the sodium bisulfite-modified template using MethPrimer software, and these MSP primer pairs are presented in Table 2. After amplification, aliquots of the PCR mixtures were separated on a 2% (w/v) agarose gel, stained with ethidium bromide, and visualized using FLA5100.
Table 2.
Primer sequences used in MSP on SOD3 promoter and coding regions
| Primer | Sequences | |
|---|---|---|
| −173/−35 − (M) | forward | 5'-TGGAGGCGAAGTAATTTTATAATT-3' |
| reverse | 5'-CCTAAAACCTAAACTATTAACGCGA-3 | |
| −173/−35 − (U) | forward | 5'-GGAGGTGAAGTAATTTTATAATTTGG-3' |
| reverse | 5'-CCTAAAACCTAAACTATTAACACAAA-3' | |
| −452/−207 − (M) | forward | 5'-TATAGTTTTGGAGTAAATGTTACGT-3' |
| reverse | 5'-CTCCCATTTTTAAATTTTCGAA-3' | |
| −452/−207 − (U) | forward | 5'-TAGTTTTGGAGTAAATGTTATGT-3' |
| reverse | 5'-CCTCCCATTTTTAAATTTTCAAA-3' | |
| −1,117/−904 − (M) | forward | 5'-TACGAGGTTTTGTTTATTTTTCGTC-3' |
| reverse | 5'-CAACCTACTTACTAACCTACCCGTC-3' | |
| −1,117/−904 − (U) | forward | 5'-TGAGGTTTTGTTTATTTTTTGTTGT-3' |
| reverse | 5'-CAACCTACTTACTAACCTACCCATC-3' | |
| +4,014/+4,241 − (M) | forward | 5'-TCGAGATATGTACGTTAAGGTTACG-3' |
| reverse | 5'-ACTAAAACTATTCGACTCGATCGAA-3' | |
| +4,014/+4,241 − (U) | forward | 5'-TGAGATATGTATGTTAAGGTTATGG-3' |
| reverse | 5'-ACTAAAACTATTCAACTCAATCAAA-3' |
Preparation of histone and Western blotting
HRECs were lysed in extraction buffer (0.1 M Tris-HCl, pH 7.5, containing 0.15 M NaCl, 1.5 mM MgCl2, 0.65% NP-40, 10 mM NaF, 1 mM Na3VO4, 20 mM β-glycerophosphate, 1 mM DTT, and 1 mM PMSF). After centrifugation at 13,200 × g for 10 s, the pellets were mixed with 0.2 M H2SO4 and this was followed by centrifugation at 13,200 × g for 20 min. The supernatant was mixed with 100% trichloroacetic acid and centrifuged at 13,200 × g for 20 min. The pellets were washed with acetone, centrifuged again at 13,200 × g for 5 min, and then dissolved in 0.45 M Tris-HCl, pH 8.8 containing 2% SDS, 6% 2-mercaptoethanol, and 0.01% bromophenol blue.
The histone extracts were boiled for 5 min and then separated by SDS-PAGE on a 15% (w/v) polyacrylamide gel, followed by electrophoretic transferal onto PVDF membranes. The membranes were then incubated with the respective specific primary antibodies (1:3,000). After the membranes had been washed with PBS containing 0.1% Tween 20, the blots were incubated with the anti-rabbit or -mouse IgG-peroxidase antibody (1:5,000). Bands were detected using SuperSignal West Pico (Thermo Scientific, Rockford, IL), and imaged using LAS-3000 UV mini (Fuji Film).
Chromatin immunoprecipitation (ChIP) assay
After cells (5 × 106 cells) had been treated, the protein–DNA complexes were cross-linked using formaldehyde at room temperature for 5 min. After centrifugation at 1,000 × g for 3 min, the pellets were sequentially washed with PBS and NP-40 buffer (10 mM Tris-HCl, pH 8.0, containing 10 mM NaCl and 0.5% NP-40), dissolved in 100 µl of SDS buffer (50 mM Tris-HCl, pH 8.0, containing 1% SDS and 10 mM EDTA), and added to 400 µl of ChIP dilution buffer (50 mM Tris-HCl, pH 8.0, containing 167 mM NaCl, 1.1% Triton X-100, 0.11% deoxycholic acid, 10 mM NaF, 1 mM Na3VO4, 20 mM β-glycerophosphate, 5 µg/ml leupeptin, 1 mM DTT, and 1 mM PMSF). Genomic DNA was sheared using the ultrasonic homogenizer Vibra-cell VC100 (Sonic & Materials, Danbury, CT) in order to achieve an estimated DNA size range of 150 to 800 bp, and 500 µl of ChIP dilution buffer was added. Sheared genomic DNA was incubated with primary antibodies overnight. The solution was then incubated with Dynabeads Protein G (Invitrogen) for 2 h. After being incubated, beads were sequentially washed with RIPA buffer I (50 mM Tris-HCl, pH 8.0, containing 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% deoxycholic acid, and proteinase inhibitors), RIPA buffer II (50 mM Tris-HCl, pH 8.0, containing 500 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% deoxycholic acid, and proteinase inhibitors), and TE buffer (10 mM Tris-HCl, pH 8.0, containing 1 mM EDTA), and then incubated in ChIP elution buffer (10 mM Tris-HCl, pH 8.0, containing 300 mM NaCl, 5 mM EDTA, and 0.5% SDS) with RNase A at 37°C for 30 min and with proteinase K at 65°C for 2 h. After phenol-chloroform extraction and ethanol precipitation, genomic DNA was eluted in 20 µl of TE buffer. The abundance of SOD3 promoter regions in ChIP precipitates was quantified using a real-time RT-PCR analysis. The primer sequences for EC-SOD were sense 5'-GTG GAG GCG AAG CAA TTC TA-3'; antisense 5'-CTG TTA GCG CGA GTG CAG GA-3' (126 bp). Real-time RT-PCR was performed using the ThunderbirdTM SYBR qPCR Mix (Toyobo) according to the manufacturer’s protocol.
Histone deacetylase (HDAC) activity analysis
HRECs (seeded at 2 × 104 cells/well in 96-well plates) were cultured overnight and then treated with Ex4. After the treatment, HDAC activity was determined using a HDAC Cell-Based Activity Assay Kit (Cayman Chemical) according to the manufacturer’s protocol. Fluorescent intensities (excitation; 365 nm, emission; 410–460 nm) were read using the GloMax®-Multi Detection System (Promega, Madison, WI).
Patients
The study protocol and informed consent documents were reviewed and approved by the Ethics Committees of Gifu University Graduate School of Medicine and Gifu Pharmaceutical University. All study subjects provided written informed consent prior to participation. The protocols were carried out under the provisions of the Declaration of Helsinki. Twelve diabetic patients (6 men and 6 women) were started on incretin-based therapies comprising GLP-1 receptor agonists or dipeptidyl peptidase 4 (DPP4) inhibitors in addition to conventional diabetic medication. Blood samples were obtained before the incretin treatment and after the start of incretin treatment. Patient profiles are shown in Table 3.
Table 3.
Clinical characteristics of the study population
| Number of patients (male/female) | 12 (6/6) |
| Age (min-max) | 64.7 (42–83) |
| Height (m) | 1.58 ± 0.08 |
| Weight (kg) | 60.8 ± 16.9 |
| Body mass index | 24.4 ± 6.8 |
| History of diabetes (years, min-max) | 7.82 (0–28) |
| Presence of neurotic disorders | 3 (25.0%) |
| Presence of diabetic nephropathy (>phase 2) | 3 (25.0%) |
| Presence of diabetic retinopathy | 2 (16.7%) |
| Concomitant insulin use | 8 (66.7%) |
| Incretin-based drugs | |
| Sitagliptin | 3 (25.0%) |
| Vildagliptin | 6 (50.0%) |
| Teneligliptin | 2 (16.7%) |
| Liraglutide | 1 (8.3%) |
Laboratory measurements
Fasting blood samples were obtained. Plasma EC-SOD concentrations were determined by ELISA as described in our previous study.(30) LDL-cholesterol (LDL-C) and triglycerides (TG) were measured using standard clinical laboratory methods.
Data analysis
Data are presented as the mean ± SD from at least three experiments. Data were analyzed by the Mann-Whitney U test. Changes in plasma EC-SOD, LDL-C and TG before and after incretin-based therapies were analyzed by the paired t test. A p value of less than 0.05 was considered significant.
Results
Effects of Ex4 on SOD expression in HRECs
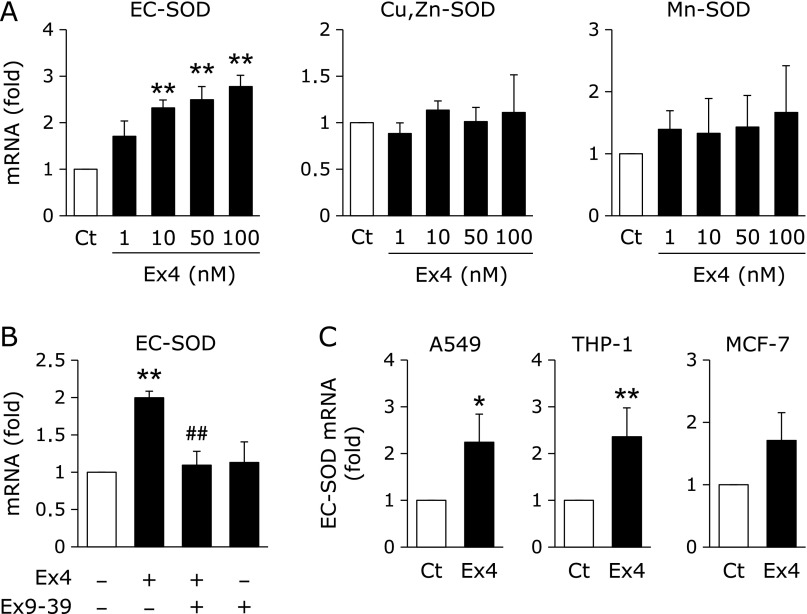
The treatment of HRECs with Ex4 for 24 h significantly induced the expression of EC-SOD, but not Cu,Zn-SOD or Mn-SOD (Fig. 1A). Moreover, the Ex4-induced up-regulated expression of EC-SOD was significantly suppressed by the addition of Ex9-39, an antagonist of the GLP-1 receptor (Fig. 1B), suggesting that Ex4 induced the expression of EC-SOD by binding to the GLP-1 receptor. Ex4 significantly up-regulated the EC-SOD expression in A549 cells and THP-1 cells, and tended to up-regulate that in MCF-7 cells (Fig. 1C).
Fig. 1.
Effects of Ex4 on EC-SOD expression through binding to the GLP-1 receptor. (A) HRECs were treated with the indicated concentrations of Ex4 for 24 h, followed by the measurement of SOD mRNA levels. Data are shown as the mean ± SD (n = 3). **p<0.01 vs vehicle. (B) HRECs were pretreated with (+) or without (–) 200 nM Ex9-39 for 1 h, and then treated with (+) or without (–) 100 nM Ex4 for 24 h. Real-time RT-PCR data were normalized using 18S rRNA levels. Data are shown as the mean ± SD (n = 3). **p<0.01 vs vehicle, ##p<0.01 vs Ex4-treated cells. (C) Cells were treated with or without 100 nM Ex4 for 24 h, followed by the measurement of EC-SOD mRNA level. Data are shown as the mean ± SD (n = 3). *p<0.05, **p<0.01 vs vehicle.
Effects of Ex4 on the DNA-methylated regulation of EC-SOD
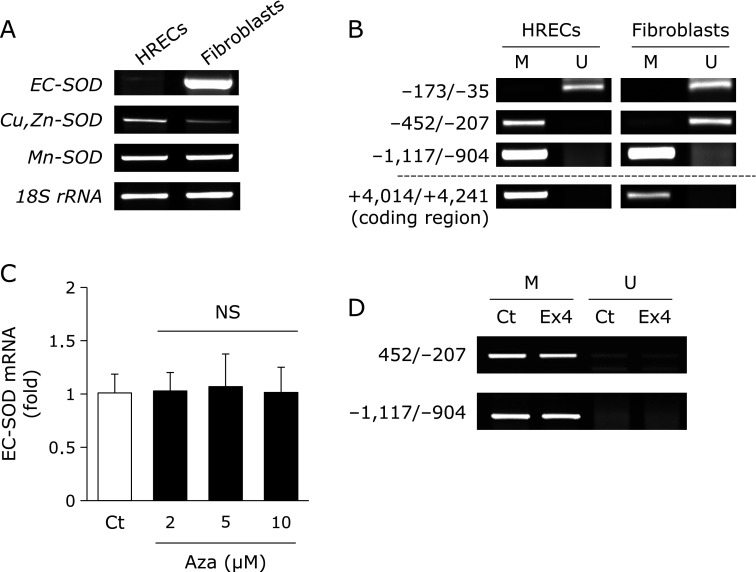
EC-SOD is known to be strongly expressed in fibroblasts.(12,13) The expression of EC-SOD was weaker in HRECs than in fibroblasts (Fig. 2A). We determined whether DNA methylation within the SOD3 promoter and coding regions was involved in the expression of EC-SOD in HRECs and fibroblasts because its expression is known to be regulated epigenetically.(24–26) As shown in Fig. 2B, the MSP analysis revealed that the SOD3 promoter region from −173 to −35 was not methylated, while that from −1,117 to −904 and the coding region from +4,014 to +4,241 were methylated in HRECs and fibroblasts. The region from −452 to −207 of the SOD3 promoter was methylated in HRECs, but not in fibroblasts (Fig. 2B). We investigated the effects of Aza on DNA demethylation within the SOD3 promoter region and the expression of EC-SOD. The expression of EC-SOD in HRECs was not increased by the treatment with Aza (Fig. 2C) despite the regions from −452 to −207 and from −1,117 to −904 in the promoter regions being demethylated by this treatment (data not shown). These results suggested that the expression of EC-SOD in HRECs was not induced by DNA demethylation within the SOD3 promoter region. Additionally, the results of the MSP analysis demonstrated that the treatment with Ex4 did not increase DNA demethylation within the SOD3 promoter region in HRECs (Fig. 2D) because Ex4 potentially regulates DNA demethylation in pancreatic cells and A549 cells.(27,28)
Fig. 2.
Involvement of Ex4 in DNA demethylation within the SOD3 promoter region in HRECs. cDNA or genomic DNA from HRECs and fibroblasts were purified, and followed by RT-PCR (A) or methylation-specific PCR (MSP) (B). MSP was carried out with methylation (M) and unmethylation (U) site primers after these genomic DNA were treated with bisulfite. (C) HRECs were treated with the indicated concentrations of 5-azacytidine (Aza) for 72 h, followed by the measurement of EC-SOD mRNA levels. Real-time RT-PCR data were normalized using 18S rRNA levels. Data are shown as the mean ± SD (n = 3). (D) HRECs were treated with 100 nM Ex4 for 24 h. After the treatment, the genomic DNA of HRECs was purified and treated with bisulfite, and this was followed by MSP amplification.
Involvement of Ex4 in the histone-acetylated regulation of EC-SOD expression
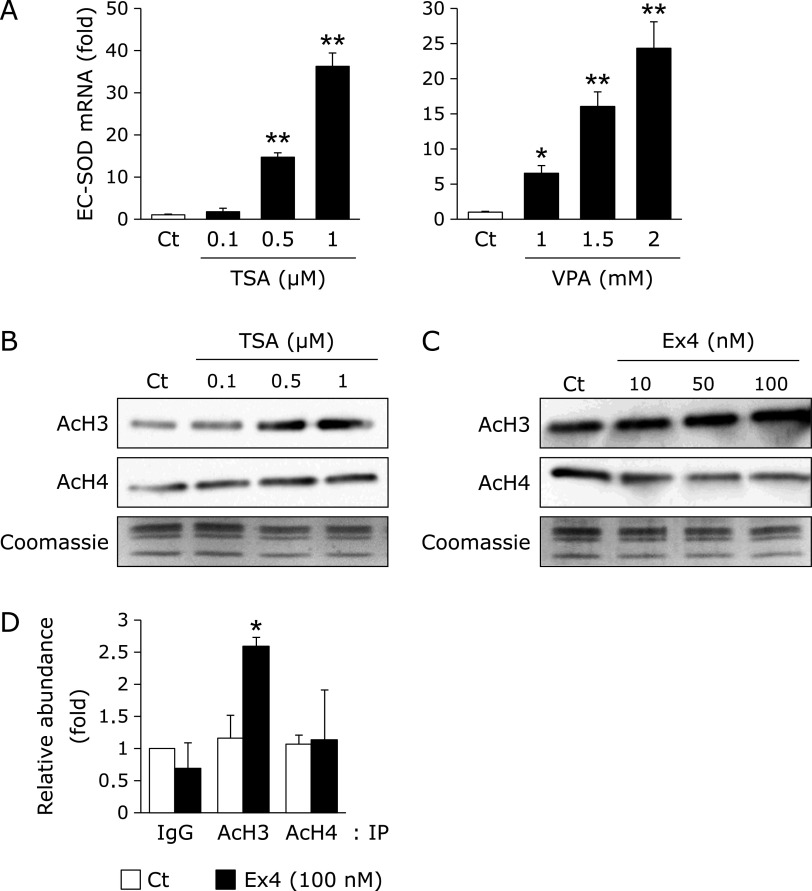
In order to further elucidate the epigenetic regulation of EC-SOD expression, we investigated the effects of TSA and VPA, which are HDAC inhibitors,(31,32) on EC-SOD expression and histone acetylation. The treatment with TSA and VPA for 24 h significantly induced the expression of EC-SOD in a concentration-dependent manner (Fig. 3A), but not that of Cu,Zn-SOD or Mn-SOD (data not shown). Moreover, the treatment with TSA for 24 h increased global histone H3 and H4 acetylation in HRECs (Fig. 3B). These results suggest that the up-regulated expression of EC-SOD mRNA in HRECs is mediated by histone acetylation. A previous study reported that Ex4 potentially regulates histone acetylation in pancreatic cells.(27) The treatment with Ex4 for 24 h increased global histone H3 acetylation in HRECs (Fig. 3C) and significantly enhanced the histone H3 acetylation status at the SOD3 proximal promoter (Fig. 3D). These results suggested that Ex4 induced the expression of EC-SOD through histone H3 acetylation within the SOD3 promoter region.
Fig. 3.
Involvement of Ex4 in histone acetylation within the SOD3 promoter region in HRECs. (A) HRECs were treated with the indicated concentrations of TSA or VPA for 24 h, followed by the measurement of EC-SOD mRNA levels. Real-time RT-PCR data were normalized using 18S rRNA levels. Data are shown as the mean ± SD (n = 3). *p<0.05, **p<0.01 vs vehicle. (B) HRECs were treated with the indicated concentrations of TSA for 24 h. After the treatment, the histones of HRECs were extracted, and this was followed by a Western blotting analysis for acetylated histone H3 (AcH3) and acetylated histone H4 (AcH4). The loading of histones was monitored by Coomassie staining. (C) HRECs were treated with the indicated concentrations of Ex4 for 24 h. After the treatment, the histones of HRECs were extracted, and this was followed by a Western blotting analysis for AcH3 and AcH4. The loading of histones was monitored by Coomassie staining. (D) HRECs were treated with 100 nM Ex4 for 24 h. After the treatment, a ChIP assay was performed as described in the Materials and methods section. Relative binding to the promoter region is expressed as the percentage amount over input (%). *p<0.05 vs vehicle (AcH3).
Effects of Ex4 on HDAC expression and activity
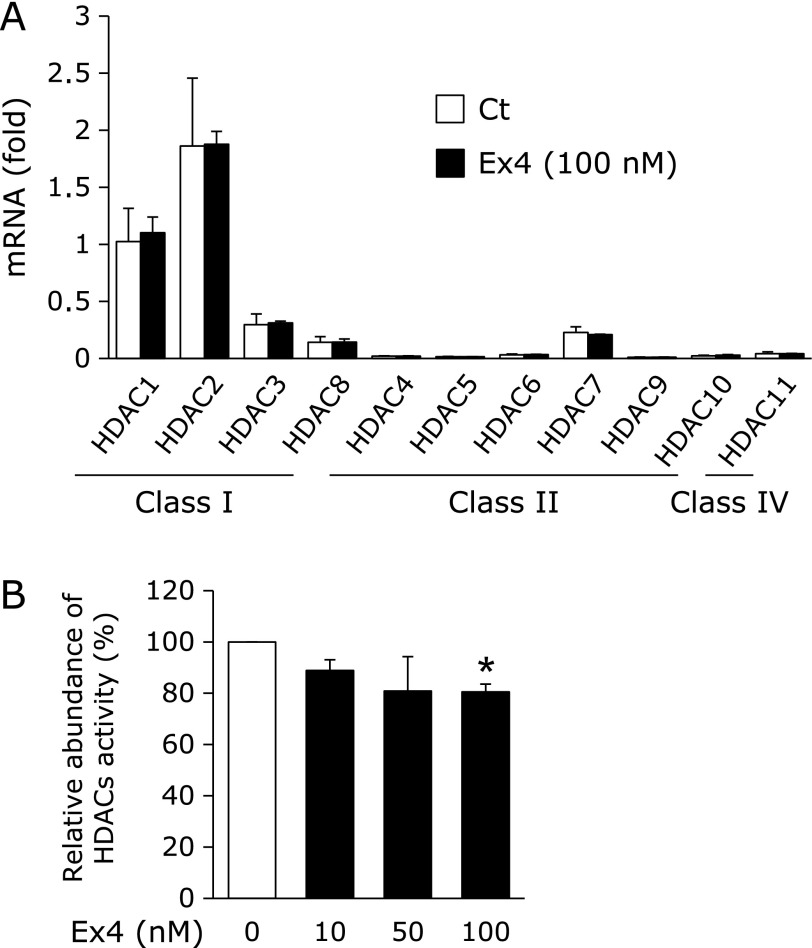
Histone acetylation/deacetylation is regulated by HDACs. The expression of each HDAC subclass was not changed by the treatment with Ex4 (Fig. 4A). On the other hand, the treatment with Ex4 significantly decreased HDAC activity (Fig. 4B), suggesting that the enhancement observed in the histone H3 acetylation status at the SOD3 proximal promoter by Ex4 was mediated by the inactivation of HDACs.
Fig. 4.
Expression and activity of HDACs by the treatment with Ex4. (A) HRECs were treated with 100 nM Ex4 for 24 h. After the treatment, real-time RT-PCR was carried out. Real-time RT-PCR data were normalized using 18S rRNA levels. Data are shown as the mean ± SD (n = 3). (B) HRECs were treated with the indicated concentrations of Ex4. After the treatment, HDAC activities were measured. Data are shown as the mean ± SD (n = 3). *p<0.05 vs vehicle.
Changes in plasma EC-SOD levels by incretin-based therapies
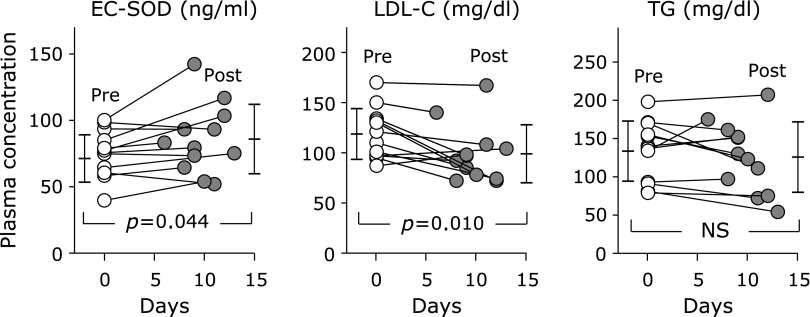
We investigated changes in plasma EC-SOD levels in diabetic patients who started incretin-based therapies. As shown in Fig. 5, the administration of incretin-based drugs to 12 diabetic patients (one patient was treated with a GLP-1 receptor agonist and 11 patients with DPP4 inhibitors) significantly increased plasma EC-SOD concentrations (75.5 ± 17.8 ng/ml to 85.8 ± 26.1 ng/ml, p = 0.044). We also observed a significant decrease in plasma LDL-C levels (118.7 ± 25.3 mg/dl to 98.9 ± 28.9 mg/dl, p = 0.010). On the other hand, no significant changes were noted in plasma TG levels (133.5 ± 39.3 mg/dl to 125.7 ± 46.0 mg/dl).
Fig. 5.
Changes in plasma EC-SOD, LDL-C, and TG levels by incretin-based therapies. Data are presented as the mean ± SD of plasma levels in patients with incretin-based therapies (n = 12) pre-treatment (pre) and post-treatment (post). Significant differences (p<0.05) before and after treatments were analyzed.
Discussion
DR is a major diabetic complication that ultimately leads to vision impairments. Oxidative stress, caused by an imbalance between the production and elimination of ROS, has been shown to result in the progression of DR.(2) The binding of EC-SOD to endothelial cell surfaces is very important for defending these cells against exogenously produced ROS.(10,11,14) However, EC-SOD was very weakly expressed in HRECs (Fig. 2A). Furthermore, previous studies reported that EC-SOD activity was decreased with type 2 diabetes.(15–17) Therefore, the up-regulated expression of EC-SOD in HRECs is considered to be important for suppressing the progression of DR. The expression of EC-SOD was recently reported to be regulated by epigenetic mechanisms.(24–26) The state of DNA methylation within the SOD3 promoter region was investigated in an attempt to elucidate the epigenetic mechanism underlying EC-SOD expression in HRECs. As shown in Fig. 2, CpG sites within the SOD3 promoter region, except the region from −173 to −35, were methylated in HRECs. Previous studies demonstrated that CpG sites within the SOD3 promoter region in A549 cells are strongly methylated and a treatment with Aza increased the expression of EC-SOD.(25,28) However, the treatment with Aza did not up-regulate the expression of EC-SOD in HRECs in the present study (Fig. 2C). We also observed that the treatment of A549 cells with Ex4 induced the demethylation of −149 and −93 CpG sites in the SOD3 promoter region and up-regulated the expression of EC-SOD.(28) These results suggest that the expression of EC-SOD in HRECs does not depend on DNA methylation because SOD3 promoter regions, at least those from −173 to −35, in HRECs are not methylated (Fig. 2B). On the other hand, Ex4 did not induce DNA demethylation within the SOD3 promoter regions from −452 to −207 or from −1,117 to −904 (Fig. 2D).
We subsequently investigated the involvement of histone acetylation in EC-SOD expression in HRECs. Histone acetylation is proceeded by the activation of histone acetyltransferase (HAT) and/or inactivation of HDAC, and enhances gene transcription by neutralizing the positive charge of histone tails.(33–35) We previously reported that the 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced expression of EC-SOD in monocytic THP-1 cells was positively related with the acetylation status of histones H3 and H4.(26) As shown in Fig. 3A and B, the treatment with TSA or VPA significantly increased histone H3 and H4 acetylation and EC-SOD expression. These results suggest that the expression of EC-SOD in HRECs depends on histone acetylation/deacetylation.
Ex4 is a well-known therapeutic agent for diabetes that binds to GLP-1 receptors on pancreatic β-cells.(5) Pinney et al.(27) recently demonstrated that Ex4 regulated pancreatic and duodenal homobox-1 (Pdx1) gene expression through the histone acetylation and demethylation of promoter DNA in the Pdx1 gene in intrauterine growth retardation (IUGR) pancreatic islets. Moreover, the GLP-1 receptor is expressed not only in the pancreas, but also in the heart, intestines, kidney, brain, and other tissues, suggesting that it has the ability to regulate various genes through an epigenetic mechanism. In the present study, Ex4 induced the expression of EC-SOD in HREC, but not Cu,Zn-SOD or Mn-SOD by binding to the GLP-1 receptor (Fig. 1A and B). The up-regulation of EC-SOD mRNA by Ex4 was observed also in other cell lines (Fig. 1C). We determined whether Ex4 induced the acetylation of histones H3 and H4 at the SOD3 promoter region because the expression of EC-SOD in HRECs is up-regulated by HDAC inhibitors (Fig. 3A). The treatment with Ex4 increased the acetylation of global histone H3 and enhanced the histone H3 acetylation status at the SOD3 proximal promoter region (Fig. 3C and D), indicating that Ex4 may expand the chromatin configuration and facilitate the binding of transcription factors, such as Sp1/3 or C/EBPβ, to the SOD3 promoter region. GLP-1 has been shown to decrease HDAC activity in INS-1 cells.(36) The results shown in Fig. 4 revealed that the treatment with Ex4 decreased HDAC activity, but did not change the mRNA level of each HDAC subclass, suggesting that Ex4 induced the expression of EC-SOD by decreasing HDAC activity and the acetylation of histone H3 at the SOD3 promoter region. The GLP-1R is well known to activate adenylate cyclase and convert ATP to cAMP, leading to the activation of second messenger pathways such as cAMP-dependent protein kinase (PKA).(37,38) However, the molecular mechanisms responsible for GLP-1 induces core histone H3 protein modifications through the regulation of HAT and/or HDAC are almost uncertain. Kim et al.(36) proposed a model in which GLP-1 activate mitogen- and stress-activated kinase-1 (MSK-1) via PKA, with MSK-1 as the final mediator responsible for histone H3 modification. Extracellular signal-regulated kinase 1/2 and p38 might involve in the above pathway, because inhibitors of these mitogen-activated protein kinases reversed the inhibitory effect of GLP-1 on HDAC activity.(36)
Protective ability of EC-SOD against oxidative stress has been revealed by the in vivo studies using EC-SOD null mice and in vitro experiments using siRNA and/or miRNA.(39,40) We observed that treatment with siRNA of EC-SOD reduced the viability of cell against oxidative stress induced by 6-hydroxydopamine (unpublished data). The expression of EC-SOD in culture cells lines is known to be regulated by numerous substances such as cytokines.(41,42) For example, tumor necrosis factor α (TNFα), a kind of proinflammatory cytokine that increases with insulin resistance, is known to decrease EC-SOD expression.(43,44) We previously reported that a treatment with pioglitazone, an anti-diabetic agent, increased plasma EC-SOD and adiponectin levels and decreased TNFα levels, indicating that plasma EC-SOD levels negatively correlate with insulin resistance in type 2 diabetic patients.(16) In the present study, plasma EC-SOD concentrations significantly increased in diabetic patients who started incretin-based therapies, and this was accompanied by decreases in LDL-cholesterol concentrations (Fig. 5). Incretin-based therapies are known to be associated with reductions in LDL-cholesterol levels in type 2 diabetic patients, in the uptake of oxLDL by macrophages, and in the suppression of arteriosclerosis.(45–47)
In conclusion, the results of the present study indicate that the Ex4-induced expression of EC-SOD in HRECs is regulated by histone H3 acetylation through reductions in the activities of HDACs. Additionally, incretin-based therapies increased plasma EC-SOD levels, suggesting that the expression of EC-SOD in not only retinal vessels, but also other tissues is increased through epigenetic regulation by incretin-based therapies. Ex4 has been widely approved as a treatment for type 2 diabetes and exerts its pancreatic and extrapancreatic effects by binding to GLP-1 receptors distributed in a number of tissues. We speculated that GLP-1 and GLP-1 receptor agonists may exert direct vasoprotective effects by up-regulating EC-SOD expression and may also function in the maintenance of physiological homeostasis by epigenetically regulating various genes.
Acknowledgments
This study was supported in part by a JSPS KAKENHI Grant (to TA, No. 25460654), and a Grant-in-Aid for Drug Development/Discovery Research from Gifu Pharmaceutical University and Gifu University (to TA).
Abbreviations
- Aza
5-azacytidine
- DPP-4
dipeptidyl peptidase 4
- DR
diabetic retinopathy
- EC-SOD
extracellular-superoxide dismutase
- Ex4
exendin-4
- Ex9-39
exendin-(9-39)
- GLP-1
glucagon-like peptide 1
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HRECs
human retinal endothelial cells
- IUGR
intrauterine growth retardation
- MSP
methylation-specific polymerase chain reaction
- Pdx1
pancreatic and duodenal homobox-1
- ROS
reactive oxygen species
- TPA
12-O-tetradecanoylphorbol-13-acetate
- TSA
trichostatin A
- VPA
valproic acid
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Lu QY, Chen W, Lu L, Zheng Z, Xu X. Involvement of RhoA/ROCK1 signaling pathway in hyperglycemia-induced microvascular endothelial dysfunction in diabetic retinopathy. Int J Clin Exp Pathol. 2014;7:7268–7277. [PMC free article] [PubMed] [Google Scholar]
- 2.Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 3.Kowluru RA, Odenbach S. Effect of long-term administration of alpha-lipoic acid on retinal capillary cell death and the development of retinopathy in diabetic rats. Diabetes. 2004;53:3233–3238. doi: 10.2337/diabetes.53.12.3233. [DOI] [PubMed] [Google Scholar]
- 4.Buse JB, Henry RR, Han J, Kim DD, Fineman MS, Baron AD, ; Exenatide-113 Clinical Study Group Effects of exenatide (exendin-4) on glycemic control over 30 weeks in sulfonylurea-treated patients with type 2 diabetes. Diabetes Care. 2004;27:2628–2635. doi: 10.2337/diacare.27.11.2628. [DOI] [PubMed] [Google Scholar]
- 5.Thorens B, Porret A, Bühler L, Deng SP, Morel P, Widmann C. Cloning and functional expression of the human islet GLP-1 receptor. Demonstration that exendin-4 is an agonist and exendin-(9-39) an antagonist of the receptor. Diabetes. 1993;42:1678–1682. doi: 10.2337/diab.42.11.1678. [DOI] [PubMed] [Google Scholar]
- 6.Ban K, Noyan-Ashraf MH, Hoefer J, Bolz SS, Drucker DJ, Husain M. Cardioprotective and vasodilatory actions of glucagon-like peptide 1 receptor are mediated through both glucagon-like peptide 1 receptor-dependent and -independent pathways. Circulation. 2008;117:2340–2350. doi: 10.1161/CIRCULATIONAHA.107.739938. [DOI] [PubMed] [Google Scholar]
- 7.Cave AC, Brewer AC, Narayanapanicker A, et al. NADPH oxidases in cardiovascular health and disease. Antioxid Redox Signal. 2006;8:691–728. doi: 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- 8.Wright E, Jr, Scism-Bacon JL, Glass LC. Oxidative stress in type 2 diabetes: the role of fasting and postprandial glycaemia. Int J Clin Pract. 2006;60:308–314. doi: 10.1111/j.1368-5031.2006.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vachier I, Chanez P, Le Doucen C, Damon M, Descomps B, Godard P. Enhancement of reactive oxygen species formation in stable and unstable asthmatic patients. Eur Respir J. 1994;7:1585–1592. doi: 10.1183/09031936.94.07091585. [DOI] [PubMed] [Google Scholar]
- 10.Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc Natl Acad Sci USA. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ookawara T, Imazeki N, Matsubara O, et al. Tissue distribution of immunoreactive mouse extracellular superoxide dismutase. Am J Physiol. 1998;275 (3 Pt 1):C840–C847. doi: 10.1152/ajpcell.1998.275.3.C840. [DOI] [PubMed] [Google Scholar]
- 12.Marklund SL. Expression of extracellular superoxide dismutase by human cell lines. Biochem J. 1990;266:213–219. doi: 10.1042/bj2660213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strålin P, Karlsson K, Johansson BO, Marklund SL. The interstitium of the human arterial wall contains very large amounts of extracellular superoxide dismutase. Arterioscler Thromb Vasc Biol. 1995;15:2032–2036. doi: 10.1161/01.atv.15.11.2032. [DOI] [PubMed] [Google Scholar]
- 14.Takatsu H, Tasaki H, Kim HN, et al. Overexpression of EC-SOD suppresses endothelial-cell-mediated LDL oxidation. Biochem Biophys Res Commun. 2001;285:84–91. doi: 10.1006/bbrc.2001.5114. [DOI] [PubMed] [Google Scholar]
- 15.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35:236–256. doi: 10.1016/s0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 16.Adachi T, Inoue M, Hara H, Maehata E, Suzuki S. Relationship of plasma extracellular-superoxide dismutase level with insulin resistance in type 2 diabetic patients. J Endocrinol. 2004;181:413–417. doi: 10.1677/joe.0.1810413. [DOI] [PubMed] [Google Scholar]
- 17.Fujita H, Fujishima H, Chida S, et al. Reduction of renal superoxide dismutase in progressive diabetic nephropathy. J Am Soc Nephrol. 2009;20:1303–1313. doi: 10.1681/ASN.2008080844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Call JA, Chain KH, Martin KS, et al. Enhanced skeletal muscle expression of extracellular superoxide dismutase mitigates streptozotocin-induced diabetic cardiomyopathy by reducing oxidative stress and aberrant cell signaling. Circ Heart Fail. 2015;8:188–197. doi: 10.1161/CIRCHEARTFAILURE.114.001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen LS, Nielsen HM, Hansen LL. Epigenetics and cancer treatment. Eur J Pharmacol. 2009;625:131–142. doi: 10.1016/j.ejphar.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 23.Zhong Q, Kowluru RA. Role of histone acetylation in the development of diabetic retinopathy and the metabolic memory phenomenon. J Cell Biochem. 2010;110:1306–1313. doi: 10.1002/jcb.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zelko IN, Stepp MW, Vorst AL, Folz RJ. Histone acetylation regulates the cell-specific and interferon-γ-inducible expression of extracellular superoxide dismutase in human pulmonary arteries. Am J Respir Cell Mol Biol. 2011;45:953–961. doi: 10.1165/rcmb.2011-0012OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic Biol Med. 2010;48:895–904. doi: 10.1016/j.freeradbiomed.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamiya T, Machiura M, Makino J, Hara H, Hozumi I, Adachi T. Epigenetic regulation of extracellular-superoxide dismutase in human monocytes. Free Radic Biol Med. 2013;61:197–205. doi: 10.1016/j.freeradbiomed.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Pinney SE, Jaeckle Santos LJ, Han Y, Stoffers DA, Simmons RA. Exendin-4 increases histone acetylase activity and reverses epigenetic modifications that silence Pdx1 in the intrauterine growth retarded rat. Diabetologia. 2011;54:2606–2614. doi: 10.1007/s00125-011-2250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuda H, Mizukami K, Hayashi M, Kamiya T, Hara H, Adachi T. Exendin-4 promotes extracellular-superoxide dismutase expression in A549 cells through DNA demethylation. J Clin Biochem Nutr. 2016;58:34–39. doi: 10.3164/jcbn.15-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adachi T, Teramachi M, Yasuda H, Kamiya T, Hara H. Contribution of p38 MAPK, NF-κB and glucocorticoid signaling pathways to ER stress-induced increase in retinal endothelial permeability. Arch Biochem Biophys. 2012;520:30–35. doi: 10.1016/j.abb.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 30.Adachi T, Hara H, Yamada H, et al. Heparin-stimulated expression of extracellular-superoxide dismutase in human fibroblasts. Atherosclerosis. 2001;159:307–312. doi: 10.1016/s0021-9150(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 32.Khan N, Jeffers M, Kumar S, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are hitone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 34.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 35.Gregory PD, Wagner K, Hörz W. Histone acetylation and chromatin remodeling. Exp Cell Res. 2001;265:195–202. doi: 10.1006/excr.2001.5187. [DOI] [PubMed] [Google Scholar]
- 36.Kim SJ, Nian C, Mclntosh CHS. Glucose-dependent insulinotropic polypeptide and glucagon-like peptide-1 modulate β-cell chromatin structure. J Biol Chem. 2009;284:12896–12904. doi: 10.1074/jbc.M809046200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayo KE, Miller LJ, Bataille D, et al. International Union of Pharmacology. XXXV. The glucagon receptor family. Pharmacol Rev. 2003;55:167–194. doi: 10.1124/pr.55.1.6. [DOI] [PubMed] [Google Scholar]
- 38.Quoyer J, Longuet C, Broca C, et al. GLP-1 mediates antiapoptotic effect by phosphorylating Bad through a β-arrestin 1-mediated ERK1/2 activation in pancreatic β-cells. J Biol Chem. 2010;285:1989–2002. doi: 10.1074/jbc.M109.067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon MJ, Jeon YJ, Lee KY, Kim TY. Superoxide dismutase 3 controls adaptive immune responses and contributes to the inhibition of ovalbumin-induced allergic airway inflammation in mice. Antioxid Redox Signal. 2012;17:1376–1392. doi: 10.1089/ars.2012.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YS, Cheon IS, Kim BH, Kwon MJ, Lee HW, Kim TY. Loss of extracellular superoxide dismutase induces severe IL-23-mediated skin inflammation in mice. J Invest Dermatol. 2013;133:732–741. doi: 10.1038/jid.2012.406. [DOI] [PubMed] [Google Scholar]
- 41.Marklund SL. Regulation by cytokines of extracellular superoxide dismutase and other superoxide dismutase isoenzymes in fibroblasts. J Biol Chem. 1992;267:6696–6701. [PubMed] [Google Scholar]
- 42.Strålin P, Marklund SL. Multiple cytokines regulate the expression of extracellular superoxide dismutase in human vascular smooth muscle cells. Atherosclerosis. 2000;151:433–441. doi: 10.1016/s0021-9150(99)00427-x. [DOI] [PubMed] [Google Scholar]
- 43.Adachi T, Toishi T, Wu H, Kamiya T, Hara H. Expression of extracellular superoxide dismutase during adipose differentiation in 3T3-L1 cells. Redox Rep. 2009;14:34–40. doi: 10.1179/135100009X392467. [DOI] [PubMed] [Google Scholar]
- 44.Adachi T, Toishi T, Takashima E, Hara H. Infliximab neutralizes the suppressive effect of TNF-alpha on expression of extracellular-superoxide dismutase in vitro. Biol Pharm Bull. 2006;29:2095–2098. doi: 10.1248/bpb.29.2095. [DOI] [PubMed] [Google Scholar]
- 45.Sun F, Wu S, Wang J, et al. Effect of glucagon-like peptide-1 receptor agonists on lipid profiles among type 2 diabetes: a systematic review and network meta-analysis. Clin Ther. 2015;37:225–241. doi: 10.1016/j.clinthera.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Parlevliet ET, Geerling JJ, et al. Exendin-4 decreases liver inflammation and atherosclerosis development simultaneously by reducing macrophage infiltration. Br J Pharmacol. 2014;171:723–734. doi: 10.1111/bph.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terasaki M, Nagashima M, Nohtomi K, et al. Preventive effect of dipeptidyl peptidase-4 inhibitor on atherosclerosis is mainly attributable to incretin’s actions in nondiabetic and diabetic apolipoprotein E-null mice. PLoS One. 2013;8:e70933. doi: 10.1371/journal.pone.0070933. [DOI] [PMC free article] [PubMed] [Google Scholar]