Abstract
Inhibitory avoidance (IA) training in rodents initiates a molecular cascade within hippocampal neurons. This cascade contributes to the transition of short- to long-term memory (i.e., consolidation). Here, a differential equation-based model was developed to describe a positive feedback loop within this molecular cascade. The feedback loop begins with an IA-induced release of brain-derived neurotrophic factor (BDNF), which in turn leads to rapid phosphorylation of the cAMP response element-binding protein (pCREB), and a subsequent increase in the level of the β isoform of the CCAAT/enhancer binding protein (C/EBPβ). Increased levels of C/EBPβ lead to increased bdnf expression. Simulations predicted that an empirically observed delay in the BDNF-pCREB-C/EBPβ feedback loop has a profound effect on the dynamics of consolidation. The model also predicted that at least two independent self-sustaining signaling pathways downstream from the BDNF-pCREB-C/EBPβ feedback loop contribute to consolidation. Currently, the nature of these downstream pathways is unknown.
The transformation of short-term memory into long-term memory (LTM) is known as consolidation and can require days to complete. During consolidation, memories pass through several distinct phases. The molecular processes that underlie these distinct phases of consolidation are not well understood.
Inhibitory avoidance (IA) conditioning is an attractive system to examine mechanisms of consolidation (Taubenfeld et al. 2001a,b; Bekinschtein et al. 2010; Radulovic and Tronson 2010). IA is usually acquired in a single training trial, thus providing a well-defined time point for the beginning of consolidation. The present study focuses on the IA protocol described by Bambah-Mukku et al. (2014). Briefly, the IA training and testing chamber is divided into a safe compartment and a shock compartment. The animal is placed in the safe compartment and allowed to spontaneously enter the shock compartment (i.e., step-through), where it is subjected to a foot shock. During subsequent testing, memory of this aversive stimulus is measured by an increased latency for spontaneous entry into the shock compartment. Several brain structures are involved in step-through IA (Baldi and Bucherelli 2015; Izquierdo et al. 2016), including the hippocampus (Kornisiuk et al. 2011; Khakpai et al. 2016).
The consolidation of IA memory is believed to begin with the extracellular release of BDNF, which then acts through the tropomyosin-related kinase B (TrkB) receptor on the plasma membrane to initiate activation of kinases, other intracellular signaling events, and gene expression (Minichiello 2009; Panja and Bramham 2014). Recently, Bambah-Mukku et al. (2014) delineated a BDNF-CREB-C/EBPβ-positive feedback loop in rat hippocampus that contributes to consolidation. In the positive feedback loop, BDNF is released during training, leading to a rapid increase in CREB phosphorylation and a subsequent increase in c/ebpβ expression. The resultant increase in C/EBPβ contributes to a delayed increase in bdnf expression (Chen et al. 2003; Martinowich et al. 2003; Guan et al. 2009; Bambah-Mukku et al. 2014). Anti-BDNF antibody, given before IA training, blocks the increase in pCREB levels at 30 min and 20 h post-training (Chen et al. 2012), demonstrating that increased BDNF levels, in turn, can increase CREB phosphorylation. These reciprocal interactions complete the positive feedback loop. Previous studies found that BDNF activates multiple feedback processes that regulate cellular activities (Cheng et al. 2003; Vaynman et al. 2003; Maharana et al. 2013; Zhang et al. 2014; Tuvikene et al. 2016). The mechanism by which BDNF selectively phosphorylates CREB to activate the BDNF-CREB-C/EBPβ feedback loop after IA training is unclear. However, the existence of this feedback loop and its importance for consolidation of IA memory has been verified by experiments in Bambah-Mukku et al. (2014), as follows: (1) Hippocampal injection of an anti-BDNF antibody inhibits the phosphorylation of CREB by IA; (2) Knockdown of c/ebpβ expression by an antisense oligonucleotide blocks induction of bdnf exon IV mRNA by IA; (3) Knockdown of C/EBPβ expression also impairs IA memory consolidation, and this impairment is rescued by hippocampal injection of BDNF; and (4) Hippocampal injection of an anti-BDNF antibody before training blocks the up-regulation of bdnf exon IV expression post-training. The feedback loop appears to be terminated by increased binding of transcriptional repressors to the bdnf promoter ∼48 h after IA conditioning (Bambah-Mukku et al. 2014).
Here, a mathematical model of the major components of the BDNF feedback loop was developed. Simulations examined the extent to which the loop contributes to the dynamic properties of sensitivity of memory (quantified as a synaptic weight increase) to protein synthesis inhibition and to other disruptions, such as are often studied empirically (Santini et al. 2014), during memory formation and persistence. Ordinary differential equations were used to describe the dynamics of signaling pathways that regulate BDNF, CREB, and C/EBPβ. To fit empirical observations, two independent self-sustaining signaling pathways, regulated by pCREB and C/EBPβ, were hypothesized to contribute to LTM formation and persistence. Simulations suggested that the delayed initiation of the BDNF-positive feedback loop may play a substantial role in the dynamics of time windows for disruption of memory by protein synthesis inhibition and other treatments. The dynamics of the putative biochemical elements that determine the delays in initiation, and in termination, of the BDNF feedback loop were also delineated.
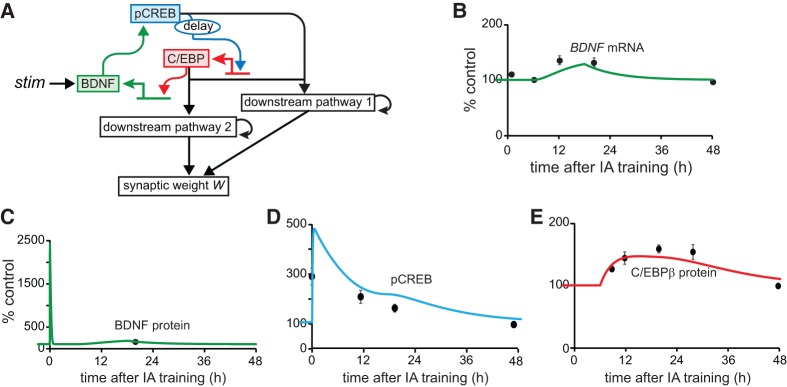
The molecular network that we model is illustrated in Figure 1A. This network embodies and simplifies the essential elements of the above BDNF feedback loop and as such it is a first step in formulating a quantitative model of mechanisms involved in IA conditioning. More detailed future models will need to include other BNDF feedback loops (Vaynman et al. 2003; Maharana et al. 2013; Zhang et al. 2014; Tuvikene et al. 2016), BDNF-dependent activation of translation via activation of phosphatidylinositol-3-kinase and mammalian target of rapamycin (mTOR) kinase, as well as BDNF's contribution to dendritic protein synthesis and the latter's contribution to LTP/LTD (Hou and Klann 2004; Jain and Bhalla 2009).
Figure 1.
Simulating signaling pathways essential to consolidation of IA LTM. (A) Schematic model of the positive BDNF feedback loop that contributes to memory consolidation. The structure of this molecular pathway is based on Bambah-Mukku et al. (2014). Details and equations are given in the main text. Empirical results (black circles) indicate that BDNF protein (C) and pCREB (D) increase for at least 20 h after training but BDNF mRNA (B) and C/EBPβ protein (E) do not significantly increase until 6 h after training (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014), with all species returning to basal levels 48 h after training. Simulated time courses are qualitatively similar to the empirical data.
Standard values for all model parameters are given in Supplemental Table S1. The differential equations of the model are also collected therein. However, the elements of these equations are also set forth here. The synthesis and degradation of BDNF is described by the following differential equation:
where stim represents a stimulus that simulates the initial activation of the BDNF pathway. A single strong stimulus was simulated, such as occurs in one-trial IA training. Stim is 0 except for an increase to 30 µM/sec, for 1 min, to simulate training. BDNF is released by the initial stimulus and, subsequently, produced by the BDNF feedback loop. In simulations, [BDNF] increases abruptly and transiently after stimulus; this increase does not represent new BDNF synthesis, but rather activity-induced vesicular release of pre-existing BDNF (Slipczuk et al. 2009). Bdnf represents bdnf mRNA, with the dynamics described below. ANI (anisomycin, a commonly used protein synthesis inhibitor) represents the effect of protein synthesis inhibition. ANI remains at 0 in the absence of inhibition and increases to 0.8 for 6 h to simulate inhibition. A similar strength of inhibition of the protein synthesis rate (∼80%, for ∼6 h) is commonly reported empirically (Milekic et al. 2006; Bambah-Mukku et al. 2014).
Previous empirical studies indicate that total CREB remains at its basal level for 20 h after IA training in rat hippocampus (Taubenfeld et al. 2001a; Chen et al. 2012; Bambah-Mukku et al. 2014). Therefore, the model keeps total [CREB], denoted by CREBtotal, fixed. The differential equation describing CREB activation by phosphorylation and the conservation relation for fixed total CREB are therefore as follows, with [CREB] denoting non-phosphorylated CREB:
This minimal model does not explicitly include biochemical processes that mediate CREB phosphorylation in response to BDNF elevation. Empirically, these processes are not well characterized. BDNF acts through TrkB receptors to activate MAP kinase signaling cascades, including ERK1/2 isoforms of MAPK, and this MAPK activation is important for induction of bdnf exon IV (Tuvikene et al. 2016). These authors also found that the AP-1 transcription factor is necessary for induction of other exons of BDNF, but not exon IV. However, earlier work demonstrated that CREB activation is necessary for BDNF induction (Tao et al. 1998) and Bambah-Mukku et al. (2014) show that active CREB specifically induces exon IV expression, with exon IV being the most abundant transcript in the hippocampus. Thus, this model assigns pCREB an essential role. MAPK does not appear to phosphorylate CREB directly. Instead, MAPK phosphorylates and activates kinases such as ribosomal S6 kinase or mitogen- and stress-activated kinase, which in turn can phosphorylate CREB (De Cesare et al. 1998; Arthur et al. 2004). The activation of these downstream kinases in response to BDNF has not yet been well characterized.
Subsequently, synthesis of the transcription factor C/EBPβ is up-regulated by active CREB. Empirically, CREB and C/EBPβ are essential for memory consolidation after IA training. pCREB significantly increases 30 min after IA training (Bambah-Mukku et al. 2014), whereas both c/ebpβ mRNA and C/EBPβ protein do not increase until more than 6 h after training (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014; C Alberini, unpublished observations). Experiments have not yet determined the extent to which the increase of the C/EBPβ protein after IA training is a consequence of increased pCREB. However, the expression of c/ebpβ is regulated by a cAMP response element after contextual learning (Impey et al. 1998), and thus presumably by pCREB. Therefore, the model assumes that an increase in pCREB leads to increased expression of C/EBPβ after stimulus. The mechanism underlying the delay in increase in C/EBPβ mRNA and protein is not known. The model assumes that phosphorylated CREB binds to the promoter of C/EBPβ immediately after training. However, the effect of CREB to activate transcription is blocked by repressors that are also bound to the promoter of C/EBPβ (e.g., Sin3a discussed in Bambah-Mukku et al. 2014), until the repressors are removed ∼6 h after training. In the model, the effect of pCREB on C/EBPβ expression is simply reflected by a function Fact_cebp (see Equation 5). To simulate the observed delay in increase of the C/EBPβ protein (Taubenfeld et al. 2001a), Fact_cebp remains at a constant basal synthesis rate kf_cebp_basal for 6 h after stimulus, after which time it is regulated by pCREB. A 6 h delay is selected, because C/EBPβ is still near the basal level at 6 h after training (Taubenfeld et al. 2001a). In the model, starting at 6 h, Fact_cebp is regulated by pCREB so that the C/EBPβ protein (variable [CEBP], Equation 4) starts to increase. By 9 h post-stimulus, [CEBP] is elevated to a level close to empirical observation (Fig. 1E) consistent with data (Taubenfeld et al. 2001a).
It is hypothesized that the ∼6 h delay in c/ebpβ induction is due to the time taken for repressors to unbind from the c/ebpβ promoter after a stimulus. Because the delay is only applied once, with the regulation of C/EBPβ by pCREB beginning at 6 h post-stimulus instead of at 0 h, the differential equation describing the dynamics of C/EBPβ synthesis and degradation is an ordinary differential equation, which is as follows:
with the delay incorporated in the definition of Fact_cebp,
Here CREB and C/EBPβ activate transcription as dimers (homodimers or heterodimers) (Wu et al. 1998; Parkin et al. 2002). Therefore, for simplicity, Hill functions with coefficients of 2 were used to describe the effect of pCREB on c/ebpβ expression and the effect of C/EBPβ on bdnf expression. In Equation 4, CEBPmax denotes the maximum amount of C/EBPβ that can be produced within 48 h, and was used to keep the simulation results within a biologically reasonable concentration boundary.
The transcription of bdnf is regulated by two activators, pCREB and C/EBPβ, and three repressors, Sin3a, MeCP2, and HDAC2 (Bambah-Mukku et al. 2014). The mechanisms underlying the changes in binding of Sin3a, MeCP2, and HDAC2 to the bdnf exon IV promoter are unclear. The dynamics of Sin3a, MeCP2, and HDAC2 protein levels after training are also not well characterized. Therefore, differential equations were not developed to simulate dynamics of binding of these repressors. Instead, a step function is used to approximate the termination of bdnf transcription (Equation 7). The activation of bdnf transcription by C/EBPβ is reflected by Fact_bdnf in Equation 7. The increase in C/EBP activity increases Fact_bdnf and thus induces the expression of bdnf until 18 h post-stimulus, at which time it is assumed that a complex involving the above repressors begins to interfere with the binding of C/EBPβ to bdnf so that bdnf returns to its basal level ∼48 h after training (Bambah-Mukku et al. 2014), terminating the feedback loop. Thus, 18 h after stimulus Fact_bdnf decreases to a constant basal synthesis rate kf_bdnf_basal to reflect this repression. The resulting differential equation describing the synthesis and degradation of bdnf mRNA is
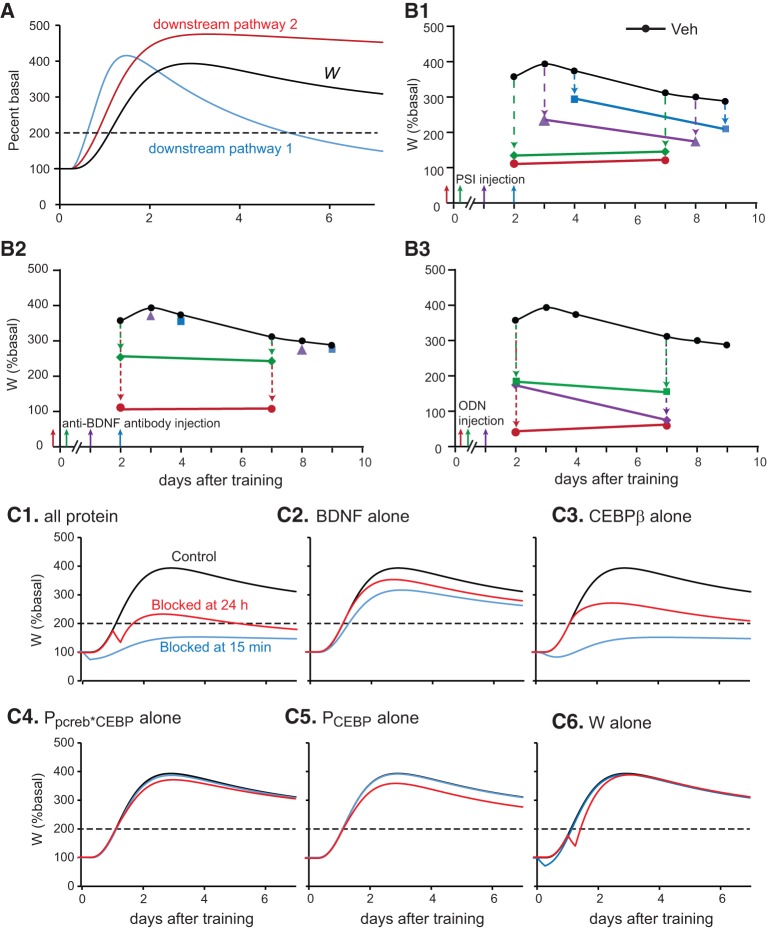
The feedback loop is terminated by 48 h after IA training. BDNF, C/EBPβ, and pCREB all return to basal levels. However, IA memory persists for at least 9 d (Bambah-Mukku et al. 2014). This finding indicates that one or more downstream pathways induced by the BDNF-positive feedback loop are self-sustaining for at least 9 d, remaining activated even after the BDNF loop is turned off at ∼48 h post-IA training. Injection of a protein synthesis inhibitor or anti-BDNF antibody 24 h after IA training does not impair memory at day 3, but impairs memory at day 8 (Bambah-Mukku et al. 2014). If one pathway was responsible for memory both at days 3 and 8, it would be difficult to explain why protein synthesis inhibition at 24 h, lasting ∼6 h, fails to impair this pathway, and memory, at day 3, but does impair memory at day 8. Hence, it is more likely that at least two independent downstream pathways are self-sustaining for some time, enhancing transcription or translation required to sustain synaptic potentiation for 9 d or longer. The model assumes that one such downstream pathway is activated by both pCREB and C/EBPβ immediately after IA training and that the activity of this pathway persists for ∼2 d (Downstream Pathway 1 in Fig. 1A; the activity of this pathway is denoted by PpCREB*CEBP in Equation 8). Another such pathway is activated by C/EBPβ alone (Downstream Pathway 2 in Fig. 1A; the activity of this pathway is denoted by PCEBP in Equation 9). Because of the delayed activation of C/EBPβ expression, the second pathway is slowly activated hours after IA training. The activity of this pathway lasts for at least 9 d. Thus these pathways prolong the effects of the BDNF feedback loop, so that the length of time for which the synaptic weight stays elevated and IA memory persists is up to at least 9 d, in agreement with empirical data (Bambah-Mukku et al. 2014). The differential equation describing the activation of the first downstream pathway by pCREB and C/EBPβ is
and that describing the activation of the second pathway by C/EBPβ is
Both pathways contribute to the elevation of synaptic weight, denoted as W. The differential equation describing the dynamics of W is therefore
To selectively couple the increased gene expression driven by the BDNF-positive feedback loop to increases in the weight of specific synapses that were activated during IA training, as opposed to a general up-scaling of all synaptic weights, it is also necessary to postulate a process similar to synaptic tagging, which data suggest allowing only recently activated synapses to capture plasticity-related proteins synthesized in response to increased activity (Redondo and Morris 2011). Because the present study is interested in modeling transcriptional regulation and associated processes, the dynamics of synaptic tagging are not included in the model.
For this model, the topology of the BDNF feedback loop as diagrammed in Figure 1A is based on substantial empirical data (Bambah-Mukku et al. 2014). However, extant empirical data do not suffice to provide strong constraints on model parameter values. In accordance with data, the model is constrained only to qualitatively simulate the characteristic dynamics of BDNF, bdnf, pCREB, and C/EBPβ after IA training, as well as the effects of protein synthesis inhibitors and oligodeoxynucleotides (ODNs) on IA memory.
The dynamic elements to be simulated include the following:
BDNF increases due to release immediately after training (Slipczuk et al. 2009), but BDNF returns to basal level within 1 h (Finsterwald et al. 2015). BDNF increases again ∼20 h after training (Bambah-Mukku et al. 2014).
pCREB increases immediately after training and remains elevated for at least 20 h (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014).
bdnf and C/EBPβ do not increase until 6 h after training, and then remain elevated for at least 20 h, returning to basal levels ∼48 h after training (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014).
LTM is assumed to be formed (consolidated) if W remains at least 100% higher than its unstimulated (basal) value for more than 2 d. LTM is further considered to be persistent if W remains at least 100% higher than its unstimulated value for more than 7 d.
Parameter sensitivity analysis was performed to test the robustness of the model to parameter variations (Supplemental Fig. S2). Each individual parameter was varied within the range of ±90% of its standard value. For each parameter, 60 evenly spaced values were simulated. Forty-eight hours of time courses of BDNF, bdnf, pCREB, and C/EBPβ after IA training were simulated to investigate how parameter variations affect the qualitative match between simulation and empirical data (Supplemental Fig. S2). Synaptic weight W was also simulated, for 9 d post-training, to quantify sensitivity.
Fourth-order Runge-Kutta integration was used for integration of differential equations, with a time step of 3 sec. The model was programmed in XPP-Auto version 6.1 (www.math.pitt.edu/~bard/xpp/xpp.html). The XPP-Auto code will be provided as requested. The model will also be submitted to the ModelDB database (McDougal et al. 2015).
In Bambah-Mukku et al. (2014), bdnf exon IV mRNA was measured at 40 min and at 6, 12, 20, and 48 h after IA training with quantitative real-time PCR. A delayed increase occurred 12 and 20 h after training, with a return to basal level at 48 h. C/EBPβ protein was significantly increased at 12 h after IA training, BDNF was significantly increased at 20 h, and pCREB remained elevated for 20 h after IA training, returning to basal level at 48 h. BDNF appears to be rapidly released during IA training or neuronal activity (Kojima et al. 2001; Slipczuk et al. 2009). Therefore, to simulate IA training, an immediate brief increase (for 1 min) in the parameter stim generated an initial large, rapid rise in [BDNF]. Simulated time courses of bdnf mRNA, BDNF, pCREB, and C/EBPβ qualitatively agreed with the empirical observations discussed above (Fig. 1B–E), although only a single empirical data point is available for BDNF.
Dynamics of BDNF, pCREB, and C/EBPβ are illustrated in Figure 1B–E. The initial increase in BDNF is due to BDNF release induced by stimulus, and results in rapid CREB phosphorylation. A delayed increase in C/EBPβ expression is induced by this first phase of CREB phosphorylation. This increase in C/EBPβ leads, in turn, to a surge of bdnf mRNA. By 12 h after stimulus, this increase in bdnf mRNA results in increased levels of BDNF, a second phase of increased phosphorylation of CREB, and thus a second phase of increased synthesis of C/EBPβ.
Three examples of parameter sensitivity analyses are illustrated in Supplemental Figure S2. These three examples were selected to illustrate the range of responses (from highly sensitive, column A of Supplemental Fig. S2, to insensitive, column C). For column A, when compared with the control simulation with standard values of parameters (black curves), altering Ka_bdnf by a moderate degree (18%) could substantially change the time courses of pCREB and C/EBPβ post-stimulus. For column B, similar alterations of kminb had modest effects, and for column C, altering kbasalp_creb did not have significant effects. More generally, these sensitivity analyses suggested that parameters directly related to the interactions among bdnf mRNA, BDNF, pCREB, and C/EBPβ (e.g., Ka_bdnf in Supplemental Fig. S2A, Ktrans, Kp_BDNF) have the greatest impact on the dynamics of feedback loops after stimuli and that moderate variations in these parameters could significantly enhance the response of W to stimulus. Thus, this class of parameters might suggest pharmaceutical targets for enhancing memory. The model proved to be relatively insensitive to changes in the remaining parameters, indicating that the qualitative match between simulation and empirical data was generally robust to moderate parameter variations.
With standard parameter values, the increase in synaptic weight (variable W, Equation 10) from days 2 to 9 after IA training was >200% above the basal value of W (Fig. 2A). The inhibition of this increase by several treatments shown by Bambah-Mukku et al. (2014) to impair IA memory was simulated (Fig. 2B). For these simulations, from the day 2 to 9 time points, if W was increased by <100% relative to its basal level, either the formation of memory (from 2 to 4 d) or its persistence (from 7 to 9 d) was considered to be impaired (that is, impairment corresponds to an approximately twofold decrement in the normal increase of W). The corresponding empirical measure of memory impairment is a significant decrease in the average time taken to enter a compartment 2–9 d after a shock has been applied (Bambah-Mukku et al. 2014).
Figure 2.
Simulations of the temporal profile of the protein synthesis, BDNF, and c/ebpβ expression. (A) Simulated time courses of synaptic weight W and of the activities of downstream pathways 1 and 2. LTM is considered to be persistent if W remains at least twofold higher than its basal value (dashed line) for 7 d. Increases in W were impaired by anisomycin (PSI) (B1), anti-BDNF antibody (B2), and ODN (B3). (C) Simulation of effects on W as a result of blocking the synthesis of various components in the model at 15 min (blue curves) or 24 h (red curves) after stimulus. (C1) Blocking synthesis of all proteins. (C2) Blocking the synthesis of only BDNF. (C3) Blocking only the synthesis of C/EBPβ. (C4) Blocking only the synthesis of PpCREB*CEBP. (C5) Blocking only the synthesis of Pcebp. (C6) Blocking only the synthesis of W itself.
Injection of anisomycin in the rat dorsal hippocampus blocks >80% of protein synthesis for ∼6 h (Milekic et al. 2006; Bambah-Mukku et al. 2014). To simulate this effect (Fig. 2B1), the synthesis rates of BDNF and C/EBPβ were reduced by 80% for 6 h. Protein synthesis inhibition was initiated at different times after the initial stimulus. Figure 2B1 illustrates that if inhibition was simulated at 15 min prior to stimulus (shown by red arrow and circles), or immediately after (green arrow and diamonds), and lasting for ∼6 h, W was increased by <100% relative to its basal level at days 2 and 7. This simulation shows impairment of both memory formation (2 d) and memory persistence (7 d). However, if inhibition was simulated at 24 h after stimulus in Figure 2B1 (purple arrow and triangles), W increased by more than 100% relative to its basal level at day 3, but failed to do so at day 8. This simulation shows impairment of only memory persistence, at 8 d. If inhibition occurred 48 h after stimulus (blue arrow and squares), W was increased by more than 100% relative to its basal level at days 4 and 9, which indicates that memory formation and persistence were intact. These results are qualitatively consistent with data (Bambah-Mukku et al. 2014).
In the model, BDNF initiates the positive feedback loop by phosphorylating CREB. No empirical data delineate the strength and duration of the reduction in CREB phosphorylation due to an anti-BDNF antibody. Hence, the effect of anti-BDNF antibody was assumed to have similar strength and duration as protein synthesis inhibition. The phosphorylation rate of CREB was reduced by 80% for 6 h. If the simulated effect of anti-BDNF antibody began 15 min prior to the IA stimulus (red arrow and circles in Fig. 2B2), W was increased by <100% relative to its basal level at days 2 and 7, consistent with impaired memory formation. However, if anti-BDNF antibody was added immediately after (within in 15 min) (green arrow and diamonds in Fig. 2B2) or 24 h after stimulation (purple arrow and triangles), W was increased by more than 100% relative to its basal level from 2 to 9 d. These treatments failed to impair memory formation and persistence. Similarly, memory at days 4 and 9 was unimpaired if anti-BDNF antibody was added 48 h after stimulation (blue arrow and squares). Most of these simulation results are consistent with empirical findings, except that, empirically, anti-BDNF antibody injected immediately after or 24 h after training does impair IA memory at days 8 and 9 in Bambah-Mukku et al. (2014). This discrepancy may be due to an incomplete description in the model of the actions of BDNF. In the model, anti-BDNF antibody reduces W by blocking the phosphorylation of CREB only. Empirically, BDNF is likely to affect a number of additional biochemical targets, such as substrates of ERK MAPK or of mTOR, that are important for IA memory.
Injection of c/ebpβ antisense ODN in rat hippocampus 24 h after training impairs memory 2 d post-training (Taubenfeld et al. 2001a). In contrast, anisomycin injected 24 h after training fails to impair memory at day 3. This discrepancy indicates that at 24 h post-training, anti-c/ebp ODN may more efficiently reduce the expression of C/EBPβ than does anisomycin at the given dose. With the model, if the simulated expression rate of C/EBPβ was reduced by 95% for 24 h, starting either 5 h (red arrow and circles in Fig. 2B3) or 24 h (purple arrow and diamonds) after stimulus, W was increased by <100% relative to its basal level at days 2 and 7, thus memory was considered impaired. Similarly, to simulate the effect of anti-bdnf ODN, the expression rate of BDNF was reduced by 95% for 24 h. In a separate simulation, if anti-bdnf ODN was added 6 h after stimulus (green arrow and squares in Fig. 2B3), W was also increased by <100% relative to its basal level at days 2 and 7. These results are consistent with empirical findings of memory impairment (Bambah-Mukku et al. 2014). Hence, the model can simulate a variety of effects of protein synthesis inhibition and ODN injection on memory formation and persistence.
In the above simulations, protein synthesis inhibition was implemented by reducing the synthesis of BDNF, C/EBPβ PpCREB*CEBP, PCEBP, and W by 80% for 6 h (i.e., increasing ANI in Equations 1, 4, and 8–10 to 0.8 for 6 h). To investigate the individual contribution of each of these components to the stimulus-induced increase in W, simulations were repeated but with inhibition restricted to individual components (i.e., by selectively increasing ANI in the corresponding differential equation). Simulation of protein synthesis inhibition beginning 15 min after stimulation primarily reduced W through its effects on BDNF and C/EBPβ (blue curves in Fig. 2C2, C3). Selective inhibition of PpCREB*CEBP, PCEBP or W had only small effects (blue curves in Fig. 2C4–C6), most likely due to the relatively long half-lives of these components. For inhibition of protein synthesis beginning 24 h after stimulation, reduction of W was still predominately due to BDNF and C/EBPβ (red curves in Fig. 2C1–C6). Note that an accelerated decrease in W was observed, lasting for ∼6 h, in each case with protein synthesis inhibition of W (Fig. 2C1, C6). This effect occurs because W itself depends directly on protein synthesis (Equation 10).
To further develop a temporal profile of sensitivity of memory to protein synthesis inhibition or ODN injection as a function of the time at which application initiates, the simulations were repeated with the time that inhibition began varying from 0 to 50 h post-stimulus. The degree to which W at day 2 or day 7 after stimulus is reduced by treatments was used to represent the degree of sensitivity of memory to protein synthesis inhibition. Specifically, the threshold value of synaptic weight required to maintain memory at either day 2 or day 7 was denoted by Wreference. As above, this threshold was taken to be fixed at a 100% increase of W above its basal value. Impairment at either day 2 or day 7 corresponds to a decrement in W below Wreference. For this model, then, the sensitivity of memory to protein synthesis inhibition or ODN treatment increases with this impairment. Thus the following equation was used to calculate sensitivity at either day 2 or day 7 post-stimulus:
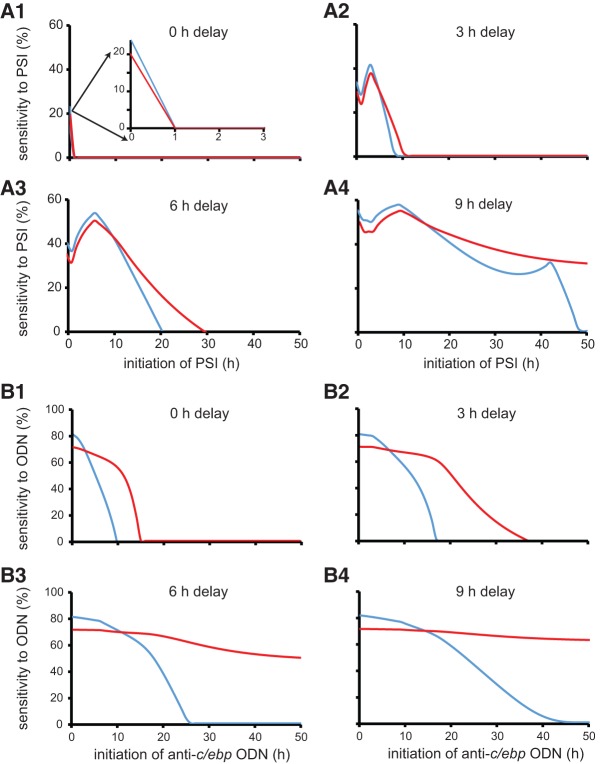
For either day, plots of the variation of this sensitivity versus the time at which protein synthesis inhibition or ODN treatment began were generated. Both impairment and sensitivity were considered to be zero if the right-hand side of Equation 11 was negative. The resulting plot of sensitivity to protein synthesis inhibition versus the time at which inhibition began is shown in Figure 3A3. The sensitivity of memory formation (at 2 d, blue curve in Fig. 3A3), or persistence (at 7 d, red curve in Fig. 3A3) to inhibition displayed a non-monotonic change with the time at which inhibition began. The peaks of sensitivity curves occurred when inhibition initiated at 6 h after stimulus, at the time that C/EBPβ expression was induced and positive feedback was initiated.
Figure 3.
The relationship of the sensitivities of memory formation (2 d) and persistence (7 d) to protein synthesis inhibition (PSI) (A) or anti-c/ebp oligodeoxynucleotide (ODN) (B) versus the time at which inhibition or ODN application began. With increasing delay in the initiation of C/EBPβ expression, from 0 to 9 h (A1–A4), the difference between the sensitivity of memory formation (blue curves) and memory persistence (red curves) to inhibition progressively increases. Similarly, with increasing delay in the initiation of C/EBPβ expression, the difference between the sensitivity of memory formation (blue curves) and memory persistence (red curves) to anti-c/ebp ODN progressively increases (B1–B4).
To investigate the effect on sensitivity of varying the 6 h delay between stimulus-induced pCREB elevation and C/EBPβ expression, these simulations were repeated, with the delay (Equation 5) varying from 0 to 9 h. When no delay was present (i.e., C/EBPβ expression increased immediately after stimulus), the sensitivity of memory formation at day 2 (blue curve in Fig. 3A1), or of persistence at day 7 (red curve in Fig. 3A1) to protein synthesis inhibition displayed a monotonic dependence on the time at which inhibition began. Sensitivity dropped to zero within 1 h after stimulus. When a 3 h delay was present, the sensitivity of memory formation (blue curve in Fig. 3A2), or persistence (red curve in Fig. 3A2) to protein synthesis inhibition displayed a nonmonotonic dependence on the time of inhibition. The sensitivity of memory at day 2 dropped to zero within 10 h after stimulus, whereas it took longer for the sensitivity of memory persistence to drop to zero. With increasing delay in the initiation of C/EBPβ expression and the feedback loop, the sensitivity to protein synthesis inhibition also increased, and displayed a strong nonmonotonic dependence on the time of initiation of inhibition. The difference in the time required for the sensitivity of memory at day 2, versus that at day 7, to drop to zero also increased (Fig. 3A2–A4). When a 9 h delay was present, the sensitivity of memory at day 2 (blue curve in Fig. 3A4), and at day 7 (red curve in Fig. 3A4) to protein synthesis inhibition remained elevated for as long as 48 h after stimulation. This prolonged elevation in sensitivity with the 9 h delay is due to a slower stimulus-induced increase in W with this long delay. For this case, W could be reduced to below Wreference by protein synthesis inhibition initiated as late as 48 h post-stimulus.
When a 6 h or less delay in the initiation of the feedback was present, all the sensitivities dropped to zero by 48 h (Fig. 3A). These results indicated that time windows of sensitivity were dependent on the initiation and termination of the BDNF feedback loop, which is consistent with the previous results that BDNF and C/EBPβ were the main contributors to the effects of protein synthesis inhibition. Note in the case for the 9 h delay in the initiation of the feedback, the sensitivity of memory at day 2 (48 h) had a delayed peak if inhibition of protein synthesis was initiated around 42 h after stimulus (blue curve in Fig. 3A4). This peak occurs because the variable W itself depends directly on protein synthesis (Equation 10).
The corresponding plots of sensitivity to anti-c/ebp ODN versus the time at which ODN treatment began are shown in Figure 3B1–4. Similar to the effects of protein synthesis inhibition, the sensitivity to anti-c/ebp ODN increased, at both days 2 and 7 when the delay in the initiation of C/EBPβ expression and positive feedback was increased. However, unlike the sensitivity to protein synthesis inhibition, the sensitivity of memory at day 2 (blue curves in Fig. 3B1–4) or at day 7 (red curves in Fig. 3B1–4) to anti-c/ebp ODN never appeared to display a nonmonotonic dependence on the time of ODN application. This lack of nonmonotonic dependence may be due to the long duration of the ODN effect (24 h), such that even a prolonged delay (9 h) in the initiation of the feedback will not affect the efficacy of ODN in suppressing the expression of c/ebpβ; whereas for the case of protein synthesis inhibition that lasts ∼6 h, the model might be insensitive to inhibition if the inhibition occurs before the BDNF feedback loop is initiated.
In the current study, we found that for successful simulation of the empirical observations, it was necessary to assume that at least two independent self-sustaining signaling pathways, downstream from pCREB and C/EBPβ, play substantial roles in LTM formation and persistence. These two independent pathways might occur in different synaptic sites of the same neurons, or different neurons in the circuit responsible for memory formation and persistence. This assumption needs to be validated empirically.
Empirical studies support the existence of time windows of sensitivity of consolidation to protein synthesis inhibition, or to inhibition of kinases or of BDNF signaling, during ∼24 h post-training, for some common learning protocols, although the parameters of these windows vary between studies (Grecksch and Matthies 1980; Bourtchouladze et al. 1998; Tiunova et al. 1998; Igaz et al. 2002; Bekinschtein et al. 2007; Slipczuk et al. 2009). Here, it is hypothesized that the observed delay of ∼6 h between IA training and C/EBPβ induction (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014) may help to shape these time windows. This delay leads to a nonmonotonic dependence of sensitivity to protein synthesis inhibition on the time of inhibitor application and implies that memory formation may be less sensitive to inhibition begun at early times before up-regulation of C/EBPβ synthesis occurs. Indeed, in Figure 3A3, with a 6 h delay, there is a peak, or time window, of sensitivity of both 2 and 7 d memory to protein synthesis inhibition, centered around 6 h post-stimulus (the 2 d window is narrower). Although not clearly evident in Figure 3, inhibition that started immediately before training and lasting ∼6 h blocks memory formation in simulations, as it does empirically (Bambah-Mukku et al. 2014).
The simulation results illustrate that the windows of sensitivity of memory formation and persistence to protein synthesis inhibition, as well as to ODN application, move to earlier times rapidly as the delay in initiation decreases (Fig. 3). Therefore, if in a given system memory consolidation depends on a positive feedback loop with a little delay of activation, then there may be a significant early window of decreased sensitivity to protein synthesis inhibition or to ODN administration (Fig. 3A1, B1). An advance in initiation of a positive feedback loop, and therefore of maximal sensitivity to inhibition, might result from a stronger stimulus intensity.
The transcription of bdnf is regulated by three repressors, Sin3a, MeCP2, and HDAC2 (Bambah-Mukku et al. 2014). In the model, a delay of 6 h was used in the onset of pCREB's regulation of the expression of c/ebpβ to take into account the observation that c/ebpβ mRNA remained at basal level for at least 6 h after IA training despite pCREB itself being significantly increased 30 min after training (Taubenfeld et al. 2001a; Bambah-Mukku et al. 2014). What might be the biochemical mechanism underlying this ∼6 h delay? Considering that increased binding of repressors to the promoter of bdnf at 48 h may act as a brake to terminate the BDNF feedback loop (Bambah-Mukku et al. 2014), it is hypothesized that increased binding of Sin3a, or other repressors, to the c/ebpβ promoter during the first few hours after training acts as another brake to delay the initiation of the positive feedback loop. The time required to reduce binding of these repressors and allow c/ebpβ induction could generate the observed delay of ∼6 h between IA training and C/EBPβ increase. However, no empirical studies have yet been performed to investigate this hypothesis. Further empirical studies are needed to examine protein levels of MeCP2, HDAC2, and Sin3a and changes in binding to the bdnf promoter during IA, and the extent to which any changes are dependent on BDNF, CREB, or neuronal activity.
The simulated time courses of BDNF mRNA and protein, pCREB, and C/EBPβ are qualitatively similar to empirical time courses following IA training. However, the empirical data are sparse and additional data will be necessary before it is possible to make more thorough quantitative comparisons between data and the model. Data also indicate that activity of the NMDA receptor and of CaM kinase II are necessary for bdnf induction (Vaynman et al. 2003) and that, reciprocally, elevated BDNF leads to induction of CaM kinase II expression (Chen et al. 2012). However, data have not determined whether they are actual components of the BDNF-positive feedback loop or, instead, permissive factors required for operation of the loop.
In this model, the effect of protein synthesis inhibition lasts ∼6 h. However, inhibition in vivo might last longer, depending upon dosage, brain region, or other parameters. It will be important to extend the model to simulate the effects of varying protein synthesis inhibition duration on memory formation and persistence.
Rapamycin added prior to, but not immediately after, IA training blocks memory at 2 and 7 d post-training (Bambah-Mukku et al. 2014). Hence rapamycin-sensitive protein synthesis appears to be necessary for the early phase of memory induction via a C/EBPβ-independent pathway, which is not simulated in the current model. In addition, the model is simplified by focusing on the C/EBPβ expression induced by BDNF up to 48 h after training. Additional biochemical cascades activated by BDNF and contributing to memory consolidation are reviewed in Panja and Bramham (2014). In view of these additional cascades, as well as other processes (e.g., dendritic spine remodeling, Bosch et al. 2014), it seems unlikely the operation of the BDNF feedback loop modeled here is itself sufficient for consolidation of IA memory or induction of genes such as c/ebpβ. Experiments, however, do suggest that the feedback loop is necessary (Bambah-Mukku et al. 2014).
Models have suggested that positive feedback loops might sustain bistable switches important for memory persistence and consolidation (Bhalla and Iyengar 1999; Miller et al. 2005; Pettigrew et al. 2005; Shema et al. 2007; Liu et al. 2008; Ogasawara and Kawato 2009, 2010; Zhang et al. 2010; Aslam and Shouval 2012; Smolen et al. 2012). In the model presented here, three positive feedback loops were implemented, two of which are hypothetical signaling pathways downstream from pCREB and C/EBP. The role of these hypothetical pathways is limited. They contribute to LTM formation and persistence only by prolonging the effects of the BDNF feedback loop on synaptic weight. For standard parameter values, these two pathways were not bistable because their activities eventually returned to basal levels ∼7 d after training. If parameters were adjusted to generate bistability in these pathways, then W would have lower and upper stable steady states, and the temporary application of inhibitors would only have two lasting outcomes: able to or not able to reduce W from an upper (strong) steady state to a lower steady state. The third positive feedback loop is the BDNF feedback loop. Empirical data suggest that the repressors Sin3a, MeCP2, and HDAC2 are involved in terminating operation of this feedback loop (Bambah-Mukku et al. 2014). To identify the role of this repression in the model, the response to a stimulus was simulated with Fact_bdnf in Equation 7 dependent on C/EBP throughout the simulation:
The simulation results indicated that, in the absence of repressors, the BDNF-positive feedback loop does, with standard parameter values, sustain a bistable switch. One minute of stimulation with an amplitude no lower than 20 µM/sec switched pCREB1 and C/EBP from basal levels to an upper steady state (Supplemental Fig. S1). An amplitude of 20 µM/sec, near the switch threshold, generated slow switching dynamics, yielding a state transition after ∼8 d, with C/EBP transiting to the upper state (orange time course). These results suggest an additional mechanistic possibility for generating bistability in biochemical cascades that sustain memory, which may be terminated by transcription or translation repressors.
Supplementary Material
Acknowledgments
This study was supported by NIH grants MH065635 (to C.M.A.) and NS073974 (to J.H.B.).
Footnotes
[Supplemental material is available for this article.]
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.042044.116.
References
- Arthur JS, Fong AL, Dwyer JM, Davare M, Reese E, Obrietan K, Impey S. 2004. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J Neurosci 5: 4324–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam N, Shouval HZ. 2012. Regulation of cytoplasmic polyadenylation can generate a bistable switch. BMC Syst Biol 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi E, Bucherelli C. 2015. Brain sites involved in fear memory reconsolidation and extinction of rodents. Neurosci Biobehav Rev 53: 160–190. [DOI] [PubMed] [Google Scholar]
- Bambah-Mukku D, Travaglia A, Chen DY, Pollonini G, Alberini CM. 2014. A positive autoregulatory BDNF feedback loop via C/EBPβ mediates hippocampal memory consolidation. J Neurosci 34: 12547–12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P, Cammarota M, Igaz LM, Bevilaqua LR, Izquierdo I, Medina JH. 2007. Persistence of long-term memory storage requires a late protein synthesis- and BDNF-dependent phase in the hippocampus. Neuron 53: 261–277. [DOI] [PubMed] [Google Scholar]
- Bekinschtein P, Katche C, Slipczuk L, Gonzalez C, Dorman G, Cammarota M, Izquierdo I, Medina JH. 2010. Persistence of long-term memory storage: new insights into its molecular signatures in the hippocampus and related structures. Neurotox Res 18: 377–385. [DOI] [PubMed] [Google Scholar]
- Bhalla US, Iyengar R. 1999. Emergent properties of networks of biological signaling pathways. Science 283: 381–387. [DOI] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. 2014. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82: 444–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchouladze R, Abel T, Berman N, Gordon R, Lapidus K, Kandel ER. 1998. Different training procedures recruit either one or two critical periods for contextual memory consolidation, each of which requires protein synthesis and PKA. Learn Mem 5: 365–374. [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302: 885–889. [DOI] [PubMed] [Google Scholar]
- Chen DY, Bambah-Mukku D, Pollonini G, Alberini CM. 2012. Glucocorticoid receptors recruit the CaMKIIα-BDNF-CREB pathways to mediate memory consolidation. Nat Neurosci 15: 1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wang S, Cai J, Rao MS, Mattson MP. 2003. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev Biol 258: 319–333. [DOI] [PubMed] [Google Scholar]
- De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P. 1998. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci 95: 12202–12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterwald C, Steinmetz AB, Travaglia A, Alberini CM. 2015. From memory impairment to posttraumatic stress disorder-like phenotypes: the critical role of an unpredictable second traumatic experience. J Neurosci 35: 15903–15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grecksch G, Matthies H. 1980. Two sensitive periods for the amnesic effect of anisomycin. Pharmacol Biochem Behav 12: 663–665. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. 2009. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Klann E. 2004. Activation of the phosphoinositide-3-kinase – Akt – mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci 23: 6352–6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Vianna MR, Medina JH, Izquierdo I. 2002. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. J Neurosci 22: 6781–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR. 1998. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat Neurosci 1: 595–601. [DOI] [PubMed] [Google Scholar]
- Izquierdo I, Furini CR, Myskiw JC. 2016. Fear memory. Physiol Rev 96: 695–750. [DOI] [PubMed] [Google Scholar]
- Jain P, Bhalla US. 2009. Signaling logic of activity-triggered dendritic protein synthesis: an mTOR gate but not a feedback switch. PLoS Comput Biol 5: e1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakpai F, Nasehi M, Zarrindast MR. 2016. The role of NMDA receptors of the medial septum and dorsal hippocampus on memory acquisition. Pharmacol Biochem Behav 143: 18–25. [DOI] [PubMed] [Google Scholar]
- Kojima M, Takei N, Numakawa T, Ishikawa Y, Suzuki S, Matsumoto T, Katoh-Semba R, Nawa H, Hatanaka H. 2001. Biological characterization and optical imaging of brain derived neurotrophic factor-green fluorescent protein suggest an activity-dependent local release of brain-derived neurotrophic factor in neurites of cultured hippocampal neurons. J Neurosci Res 64: 1–10. [DOI] [PubMed] [Google Scholar]
- Kornisiuk E, Snitcofsky M, Blanco C, Harvey AL, Stone TW, Jerusalinsky D. 2011. Memory impairment in rats by hippocampal administration of serine protease subtilisin. Behav Brain Res 219: 63–67. [DOI] [PubMed] [Google Scholar]
- Liu RY, Fioravante D, Shah S, Byrne JH. 2008. cAMP response element-binding protein 1 feedback loop is necessary for consolidation of long-term synaptic facilitation in Aplysia. J Neurosci 28: 1970–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharana C, Sharma KP, Sharma SK. 2013. Feedback mechanism in depolarization-induced sustained activation of extracellular signal-regulated kinase in the hippocampus. Sci Rep 3: 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–893. [DOI] [PubMed] [Google Scholar]
- McDougal RA, Morse TM, Hines ML, Shepherd GM. 2015. ModelView for ModelDB: online presentation of model structure. Neuroinformatics 13: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milekic MH, Brown SD, Castellini C, Alberini CM. 2006. Persistent disruption of an established morphine conditioned place preference. J Neurosci 26: 3010–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P, Zhabotinsky AM, Lisman JE, Wang XJ. 2005. The stability of a stochastic CaMKII switch: dependence on the number of enzyme molecules and protein turnover. PLoS Biol 3: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minichiello L. 2009. TrkB signaling pathways in LTP and learning. Nat Rev Neurosci 10: 850–860. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Kawato M. 2009. Bistable switches for synaptic plasticity. Sci Signal 2: pe7. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Kawato M. 2010. The protein kinase Mζ network as a bistable switch to store neuronal memory. BMC Syst Biol 4: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panja D, Bramham CR. 2014. BDNF mechanisms in late LTP formation: synthesis and breakdown. Neuropharmacology 76: 664–676. [DOI] [PubMed] [Google Scholar]
- Parkin SE, Baer M, Copeland TD, Schwartz RC, Johnson PF. 2002. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBP (Ig/EBP). J Biol Chem 277: 23563–23572. [DOI] [PubMed] [Google Scholar]
- Pettigrew DB, Smolen P, Baxter DA, Byrne JH. 2005. Dynamic properties of regulatory motifs associated with induction of three temporal domains of memory in Aplysia. J Comput Neurosci 18: 163–181. [DOI] [PubMed] [Google Scholar]
- Radulovic J, Tronson NC. 2010. Molecular specificity of multiple hippocampal processes governing fear extinction. Rev Neurosci 21: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo RL, Morris RG. 2011. Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci 12: 17–30. [DOI] [PubMed] [Google Scholar]
- Santini E, Huynh TN, Klann E. 2014. Mechanisms of translation control underlying long-lasting synaptic plasticity and the consolidation of long-term memory. Prog Mol Biol Transl Sci 122: 131–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema R, Sacktor TC, Dudai Y. 2007. Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKMζ. Science 317: 951–953. [DOI] [PubMed] [Google Scholar]
- Slipczuk L, Bekinschtein P, Katche C, Cammarota M, Izquierdo I, Medina JH. 2009. BDNF activates mTOR to regulate GluR1 expression required for memory formation. PLoS One 4: e6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen P, Baxter DA, Byrne JH. 2012. Molecular constraints on synaptic tagging and maintenance of long-term potentiation: a predictive model. PLoS Comput Biol 8: e1002620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. 1998. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20: 709–726. [DOI] [PubMed] [Google Scholar]
- Taubenfeld SM, Milekic MH, Monti B, Alberini CM. 2001a. The consolidation of new but not reactivated memory requires hippocampal C/EBPβ. Nat Neurosci 4: 813–818. [DOI] [PubMed] [Google Scholar]
- Taubenfeld SM, Wiig KA, Monti B, Dolan B, Pollonini G, Alberini CM. 2001b. Fornix-dependent induction of hippocampal CCAAT enhancer-binding protein β and δ co-localizes with phosphorylated cAMP response element-binding protein and accompanies long-term memory consolidation. J Neurosci 21: 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiunova AA, Anokhin KV, Rose SP. 1998. Two critical periods of protein and glycoprotein synthesis in memory consolidation for visual categorization learning in chicks. Learn Mem 4: 401–410. [DOI] [PubMed] [Google Scholar]
- Tuvikene J, Pruunsild P, Orav E, Esvald EE, Timmusk T. 2016. AP-1 transcription factors mediate BDNF positive feedback loop in cortical neurons. J Neurosci 36: 1290–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaynman S, Ying Z, Gomez-Pinilla F. 2003. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic plasticity. Neuroscience 122: 647–657. [DOI] [PubMed] [Google Scholar]
- Wu X, Spiro C, Owen WG, McMurray CT. 1998. cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J Biol Chem 273: 20820–20827. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Smolen P, Baxter DA, Byrne JH. 2010. The sensitivity of memory consolidation and reconsolidation to inhibitors of protein synthesis and kinases: computational analysis. Learn Mem 17: 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zeng L, Yu T, Xu Y, Pu S, Du D, Jian W. 2014. Positive feedback loop of autocrine BDNF from microglia causes prolonged microglia activation. Cell Physiol Biochem 34: 715–723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.