Simonini et al. present an alternative auxin-sensing mechanism in which the auxin response factor ARF3/ETTIN controls gene expression through interactions with process-specific transcription factors.
Keywords: Arabidopsis, auxin signaling, IAA, plant development, transcription factor complex, ETTIN
Abstract
Tissue patterning in multicellular organisms is the output of precise spatio–temporal regulation of gene expression coupled with changes in hormone dynamics. In plants, the hormone auxin regulates growth and development at every stage of a plant's life cycle. Auxin signaling occurs through binding of the auxin molecule to a TIR1/AFB F-box ubiquitin ligase, allowing interaction with Aux/IAA transcriptional repressor proteins. These are subsequently ubiquitinated and degraded via the 26S proteasome, leading to derepression of auxin response factors (ARFs). How auxin is able to elicit such a diverse range of developmental responses through a single signaling module has not yet been resolved. Here we present an alternative auxin-sensing mechanism in which the ARF ARF3/ETTIN controls gene expression through interactions with process-specific transcription factors. This noncanonical hormone-sensing mechanism exhibits strong preference for the naturally occurring auxin indole 3-acetic acid (IAA) and is important for coordinating growth and patterning in diverse developmental contexts such as gynoecium morphogenesis, lateral root emergence, ovule development, and primary branch formation. Disrupting this IAA-sensing ability induces morphological aberrations with consequences for plant fitness. Therefore, our findings introduce a novel transcription factor-based mechanism of hormone perception in plants.
Precise orchestration of organ patterning and polarity establishment during plant growth depends on close integration of environmental, hormonal, and cellular responses. A crucial factor facilitating such integration is the phytohormone auxin, which coordinates growth and development at every stage of a plant's life cycle (for review, see Benjamins and Scheres 2008; Vanneste and Friml 2009). Canonical auxin signaling occurs through binding of the auxin molecule to F-box proteins of the TIR1/AFB class, which forms part of the SCFTIR1/AFB complex (Dharmasiri et al. 2005; Kepinski and Leyser 2005). This allows interaction with the transcriptional repressor proteins of the Aux/IAA family, which, in the absence of auxin, repress auxin response factor (ARF) proteins, preventing them from regulating their targets. The interaction between Aux/IAAs and ARFs occurs through the PB1 (Phox/Bem1p) domain, which is present in the C-terminal part of most ARFs (for review, see Guilfoyle 2015; Korasick et al. 2015; Salehin et al. 2015). Different combinations of the 23 ARFs, 29 Aux/IAAs, and six TIR1/AFB auxin receptors in Arabidopsis have been proposed to contribute to the complexity of auxin responses during plant development (Calderón Villalobos et al. 2012).
The ETTIN (ETT)/ARF3 gene encodes an atypical ARF lacking the PB1 domain (Guilfoyle 2015), and it is therefore unclear how or whether ETT functions within the canonical auxin signaling machinery during plant development. Mutations in the ETT gene lead to severe polarity defects in the female reproductive organ, the gynoecium, with overproliferation of apical tissue and reduced ovary development (Fig. 1A,B; Supplemental Fig. 1A,B; Sessions et al. 1997). In addition to its role in the gynoecium, ETT functions during lateral root (LR) formation and ovule integument development, in the establishment of leaf polarity, and in the stem–pedicel fusion process (Garcia et al. 2006; Marin et al. 2010; Kelley et al. 2012; Zhao et al. 2013). It has been suggested that ETT has a role in the interpretation of auxin levels (Nemhauser et al. 2000; Pekker et al. 2005). However, a mechanistic model for how ETT may respond to changes in auxin concentrations has not yet been proposed.
Figure 1.
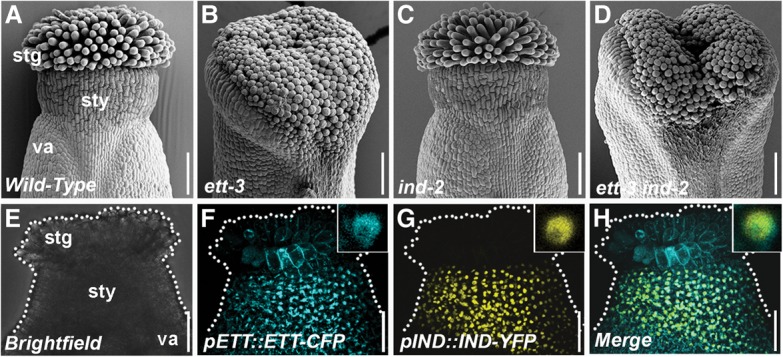
ETT and IND genetically interact to regulate gynoecium development. (A–D) Scanning electron micrographs (SEMs) of apices from stage 12 gynoecia of Col-0 (A), ett-3 (B), ind-2 (C), and ett-3 ind-2 (D). (E–H) Confocal images of stage 11 gynoecium (E) from double-transgenic lines expressing pETT::ETT-CFP (F) and pIND::IND-YFP (G) and their colocalization in the nucleus (H). Insets show close-ups of nuclei. (stg) Stigma; (sty) style; (va) valve. Bars, 100 µm.
INDEHISCENT (IND) is a basic helix–loop–helix (bHLH) transcription factor (TF) required for formation of the Arabidopsis valve margins, which comprise the tissue that allows the fruit to open upon seed dispersal (Liljegren et al. 2004). In addition, IND controls polarity at the apex of the gynoecium together with the bHLH protein SPATULA (SPT) (Girin et al. 2011; Moubayidin and Østergaard 2014), and, in both processes, IND mediates its function at least in part by controlling auxin distribution (Sorefan et al. 2009; Girin et al. 2011; Moubayidin and Østergaard 2014). IND coordinates directional auxin flux by direct repression of the PINOID (PID) gene (Sorefan et al. 2009). PID is a serine–threonine kinase that mediates polar auxin transport and is fundamental for proper symmetry establishment (Benjamins et al. 2001; Friml et al. 2004; Moubayidin and Østergaard 2014).
Here we show that ETT and IND interact to form a complex that mediates gynoecium patterning and that auxin affects the activity of this complex in the regulation of gene expression. Our data demonstrate the existence of a novel auxin-sensing mechanism comprised of ETT and IND that exhibits a strong preference for the naturally occurring auxin indole 3-acetic acid (IAA) over synthetic auxins and is fundamental for coordinating growth and patterning during carpel development. Moreover, our findings demonstrate that the IAA-sensing function is an intrinsic property of ETT, and the disruption of this IAA-sensing ability triggers morphological aberration in other developmental contexts in addition to gynoecium patterning such as LR emergence, ovule development, and primary branch formation. Thus, our findings introduce a novel TF-based mechanism of hormone signaling in plants.
Results
ETT and IND control polarity at the gynoecium apex by direct transcriptional regulation of the serine–threonine kinase PID
Understanding how auxin can coordinate growth with symmetry establishment and tissue patterning is essential to obtain an integrated comprehension of plant development. The Arabidopsis gynoecium is particularly well suited for such purposes due to its division into highly distinct tissues with different symmetries (for review, see Reyes-Olalde et al. 2013; Liu and Franks 2015). Both ETT and IND control polarity in the gynoecium through precise coordination of auxin distribution (Nemhauser et al. 2000; Girin et al. 2011). We therefore tested whether genetic interactions between them contribute to gynoecial polarity establishment. Although ind-2 mutant gynoecia have no obvious radiality defects (Fig. 1A,C; Supplemental Fig. 1A,C,E,G), the radial style deformation of the ett-3 single mutant is exacerbated in the ett-3 ind-2 double mutant, which fails to achieve complete closure at the apex (Fig. 1A–D; Supplemental Fig. 1A–H), suggesting overlapping roles of these regulators. Analyses of the pETT::ETT-CFP and pIND::IND-YFP reporter lines revealed that ETT and IND expression patterns overlap in the apical region and that the fusion proteins colocalize in the nuclei in the cells of this domain (Fig. 1E–H; Supplemental Fig. 1I–K). We demonstrated previously that IND promotes radial symmetry at the gynoecium apex via interaction with another bHLH protein, SPT (Moubayidin and Østergaard 2014). The data presented here suggest that this may occur via cooperation with ETT. Indeed, a genetic interaction between SPT and ETT was reported previously (Heisler et al. 2001).
The controlled expression of PID at the gynoecium apex is fundamental to ensure proper symmetry establishment (Moubayidin and Østergaard 2014). Given that IND is a direct regulator of PID gene expression (Sorefan et al. 2009), we therefore tested whether ETT contributes to this regulation. The PID promoter contains a number of potential auxin response elements (AuxREs) with the consensus sequence TGTCNN (or NNGACA on the opposite strand). Phylogenetic shadowing using mVISTA (Mayor et al. 2000) demonstrated that two AuxREs in positions −429 and −447 compared with the start codon are highly conserved among PID genes in the Brassicaceae family and therefore are strong candidates for being recognition sites for ARF proteins (Fig. 2A,B). Chromatin immunoprecipitation (ChIP) using the pETT::ETT-GFP (ett-3) line revealed a particularly strong interaction with a region in the PID promoter containing these two elements (Fig. 2B; Supplemental Fig. 2A,B). Moreover, a yeast one-hybrid (Y1H) assay demonstrated that ETT specifically interacts with these sequences, while this interaction is disrupted when they are mutated to non-AuxREs (Supplemental Fig. 2C).
Figure 2.
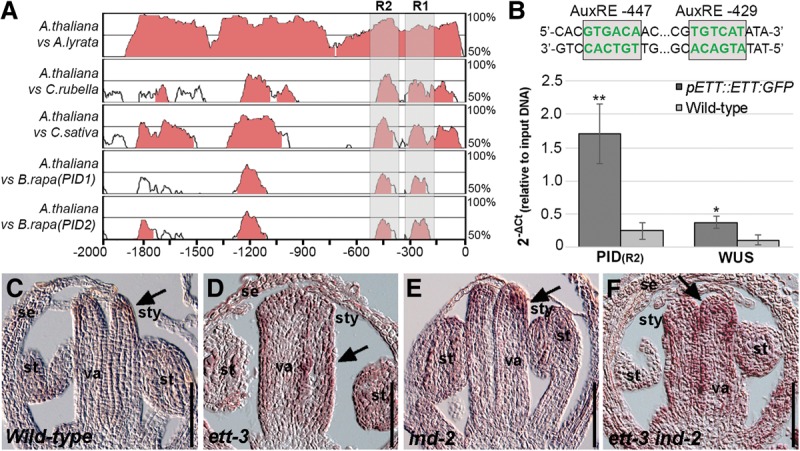
ETT and IND function together to regulate target genes during gynoecium development. (A) Phylogenetic shadowing using mVISTA of a 2-kb genomic region upstream of the translational start site of the PID gene with pairwise alignments of Arabidopsis thaliana with Arabidopsis lyrata, Capsella rubella, Camelina sativa, and Brassica rapa. Regions 1 and 2 (R1 and R2) are indicated by shaded areas. (B) ChIP with the pETT::ETT-GFP line showing enrichment of a fragment containing the conserved AuxRE sites at −429 and −447. The WUS promoter was used as a positive control. Error bars show standard deviation. (*) P < 0.01; (**) P < 0.001. (C–F) In situ hybridization of PID mRNA at the apex of stage 8 gynoecia from Col-0 (C), ett-3 (D), ind-2 (E), and ett-3 ind-2 (F). (se) Sepal; (st) stamen; (stg) stigma; (sty) style; (va) valve. Bars, 50 μm.
To test whether the interaction of ETT and IND with the PID promoter is of relevance for PID gene expression, we performed in situ hybridization with a probe against the PID mRNA on longitudinal sections of developing wild-type gynoecia. In agreement with previous data (Girin et al. 2011), we detected very weak PID expression at the apex of the gynoecium (Fig. 2C). This expression pattern was not significantly changed in the ind-2 mutant but was markedly expanded in the ett-3 mutant gynoecium (Fig. 2D,E). Moreover, in the ett-3 ind-2 double mutant, strong ectopic and precocious PID expression was observed, extending the wild-type PID expression domain from the apex of the gynoecium into the carpel body (Fig. 2C,F). Analyses of the pPID::PID-GUS reporter line in an ett-3 ind-2 background corroborated the in situ hybridization observations (Supplemental Fig. 2D–G) and together suggest that IND and ETT repress PID expression during early stages of gynoecium growth to ensure proper carpel development.
ETT and IND proteins interact
Given the genetic interaction between ETT and IND, their overlapping expression pattern, and the fact that both proteins together directly regulate PID expression, we investigated whether ETT and IND proteins interact. To this end, we used bimolecular fluorescence complementation (BiFC) (Fig. 3A–C), yeast two-hybrid (Y2H) assays (Fig. 3D), fluorescence resonance energy transfer/fluorescence lifetime imaging (FRET/FLIM) (Fig. 3E–I), and in vivo FRET (Supplemental Fig. 3A). All of the techniques revealed strong interactions between the two proteins, suggesting that ETT and IND most likely heterodimerize to regulate expression of their target genes.
Figure 3.
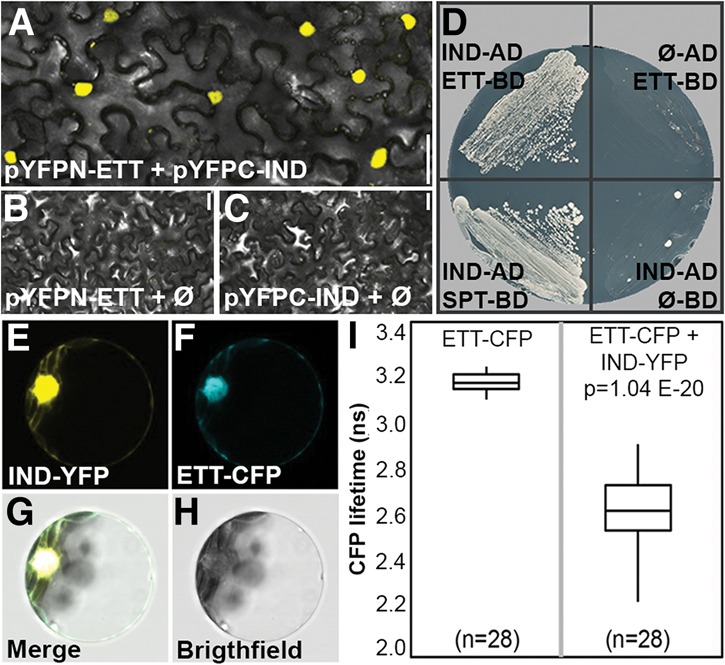
ETT and IND proteins interact. (A–C) BiFC with pYFPN-ETT and pYFPC-IND (A) and respective negative controls pYFPN-ETT + pYPFC empty (B) and pYFPN empty + pYFPC-IND (C). (D, left) Y2H assay showing positive interaction between IND-AD and ETT-BD and between IND-AD and SPT-BD. (Right) Negative controls (IND-AD with empty BD vector and ETT-BD with empty AD vector) are shown. (E–H) FRET/FLIM analysis in Arabidopsis protoplast of IND-YFP (E) and ETT-CFP (F) and the merged signal in the nucleus (G–H). (I) Quantification of CFP lifetime with ETT-CFP and empty YFP vector (left) and ETT-CFP with IND-YFP (right). Bars: A–C, 50 µm.
A deletion analysis demonstrated that the C-terminal ETTIN-specific (ES) domain of 215 amino acids is sufficient for interaction with IND (Supplemental Fig. 3B). IND has been divided previously into three domains, including the bHLH domain in the C-terminal half and a central HEC domain with homology with the closest homologs of IND, the HECATE proteins (Girin et al. 2011; Gremski et al. 2007). The N-terminal part of IND, comprising 56 amino acids, has no sequence homology with any other protein in Arabidopsis and was therefore named the IND-specific (IS) domain. In contrast to ETT, all IND domains appear necessary to maintain the interaction with ETT (Supplemental Fig. 3B). Moreover, the interaction with IND was specific to ETT, since no interaction could be detected between IND and the ETT homolog ARF4 (Supplemental Fig. 3B). These results further support a synergistic role for ETT and IND in the control of gynoecium development.
ETT and IND interaction is IAA-sensitive
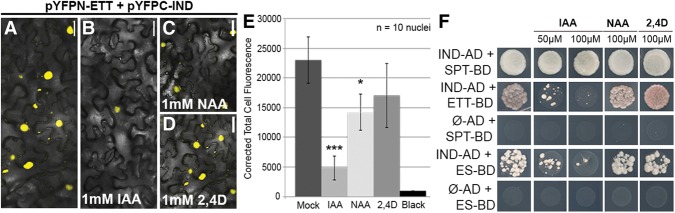
Members of the ARF family are capable of forming complexes with components of the auxin signaling pathway, including other ARFs and Aux/IAA repressor proteins (Vernoux et al. 2011; Boer et al. 2014; Korasick et al. 2014). Since ETT does not contain a C-terminal PB1 domain and does not appear to interact with Aux/IAA proteins (Piya et al. 2014), it is possible that ETT is regulated post-transcriptionally by auxin via a different pathway. To test this, we added the naturally occurring auxin IAA to the in vivo interaction assays. Interestingly, we found that the YFP signal with ETT and IND in the BiFC assay decreased in the presence of IAA (applied in lanolin) (Fig. 4A,B,E; Supplemental Fig. 3C), suggesting that IAA affected the ETT–IND dimerization. Performing the Y2H assay in the presence of IAA showed, in agreement with the BiFC data, that the ETT–IND interaction was sensitive and suppressed at IAA concentrations similar to those used previously for testing the activity of TIR1 and AFBs in yeast (Fig. 4F; Calderón Villalobos et al. 2012). In contrast, the previously established interaction between IND and SPT was not sensitive to IAA (Fig. 4F). These data suggest that the dimerization between ETT and IND is modulated by IAA.
Figure 4.
ETT and IND protein interact in an IAA-sensitive manner. (A–D) BiFC with pYFPN-ETT and pYFPC-IND in the presence of IAA (B), NAA (C), and 2,4-D (D). All hormonal treatments were with 1 mM in lanolin. (E) Fluorescence quantification (CTCF) of split YFP signal between ETT and IND without treatment and with IAA, NAA, and 2,4-D. (*) P < 0.01; (***) P < 0.0001. Error bars show the standard deviation. (F) Y2H assays with increasing concentration of IAA, NAA, and 2,4D. Bars: A–D, 50 µm.
The effect on the ETT–IND complex was most pronounced with IAA, whereas the synthetic auxin NAA had a weaker effect in the BiFC assay. In yeast, neither NAA nor 2,4-D showed any effect (Fig. 4A–F). This IAA preference differs distinctly from the established SCFTIR1/AFB-mediated auxin signaling pathway that is strongly activated also by these synthetic analogs (Dharmasiri et al. 2005; Kepinski and Leyser 2005).
A potential biological consequence of the IAA-sensing mechanism emerged when we performed ChIP on ETT-GFP inflorescences following exogenous IAA treatment. While regions upstream in the PID promoter exhibited decreased association with ETT upon IAA treatment, a striking increase in the enrichment of a region close to the transcriptional start site of PID was detected (region 1) (Supplemental Fig. 4A,B). Although region 1 contains potential AuxREs, our phylogenetic shadowing analysis shows that none of these are conserved in PID genes of the other Brassicaceae species in the analysis. Moreover, the interaction between ETT and region 2 (containing the two conserved AuxREs) is not disrupted by the IAA treatment in both the ChIP and Y1H assays (Supplemental Fig. 4A,C). One way to explain this could be that the increased association of ETT with region 1 in the presence of IAA may be indirect and possibly occurs via another protein bound to this region and facilitated by the modified ETT–IND interaction (see the model in Supplemental Fig. 4B). The potential importance of sequences within region 1 for PID regulation is reflected in its high overall level of conservation in the phylogenetic shadowing analysis (Fig. 2A).
Additional support of IAA affecting IND activity emerged when we carried out a microarray experiment using a 35S::IND-GR line treated with or without dexamethasone (DEX) and with or without IAA. In this analysis, 1734 genes significantly changed expression (>1.5 fold; P < 0.05) following induction of IND. However, out of these, 796 genes required the presence of IAA for IND to have an effect (Supplemental Fig. 5; Supplemental Table 1).
IAA perception is fundamental for proper gynoecium morphogenesis
To assess the biological and functional relevance of the IAA-sensing ability in planta, we aimed to identify point mutations in IND and the ETT-ES domain that permitted the formation of the ETT–IND dimer while rendering it insensitive to IAA.
A screening for random point mutations introduced in the IND ORF resulted in the isolation of an aspartate-to-glycine substitution at position 30 (D30G) (Fig. 5A), which caused the ETT–IND interaction to become IAA-insensitive (Fig. 5B). Expressing this INDD30G variant under the IND promoter (pIND::INDD30G) in the strong ind-2 mutant background led to a striking overproliferation of stigmatic tissue accompanied by reduced pollen tube density and decreased fertility, which was not seen when ind-2 was transformed with a wild-type pIND::IND construct or in the untransformed ind-2 mutant (Fig. 5C–E; Supplemental Fig. 6A–F). These results strongly suggest that the IAA sensitivity of the ETT–IND complex is important for proper gynoecium development and plant reproduction.
Figure 5.
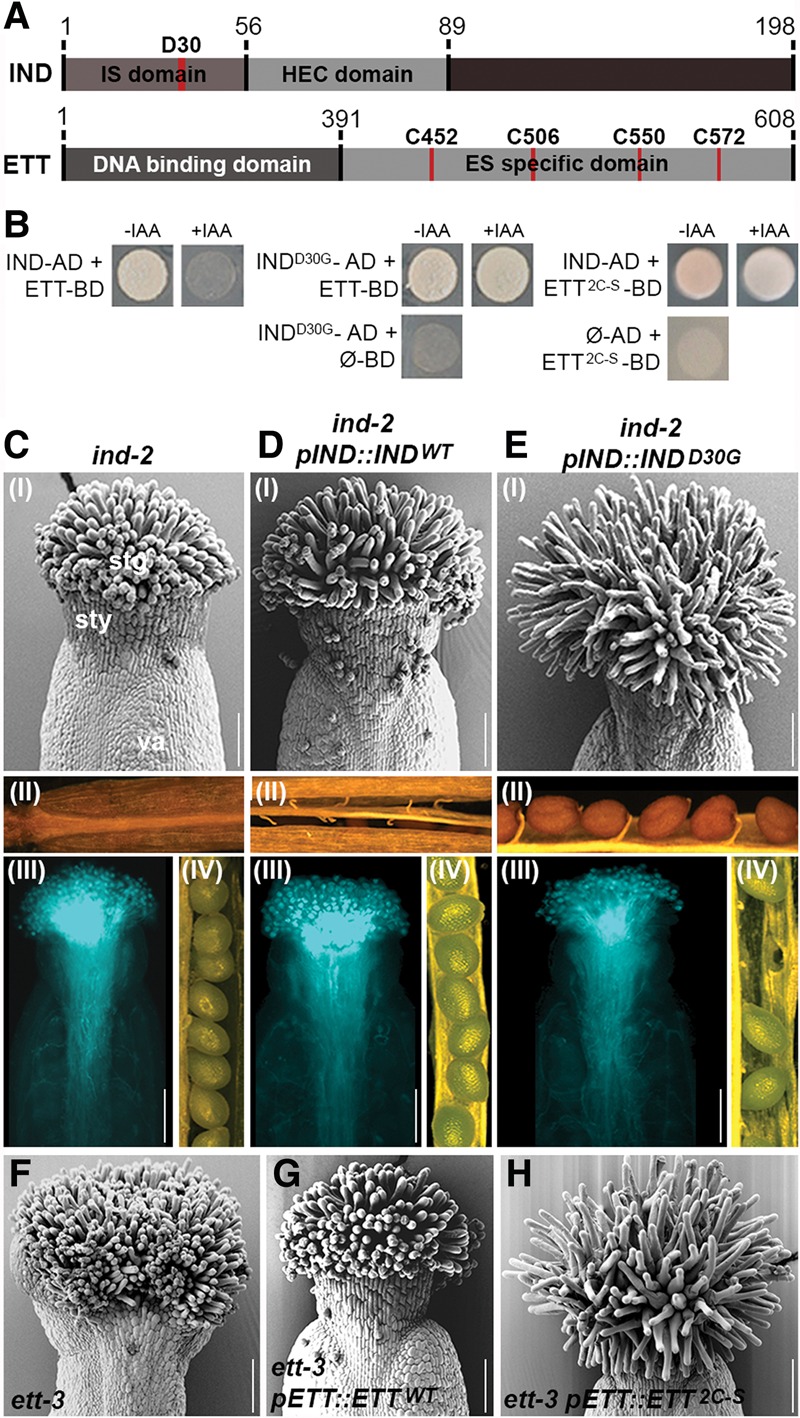
Auxin sensitivity is required for proper gynoecium morphogenesis. (A) Schematic representation of protein domains of IND and ETT. Numbers indicate amino acid positions. (B) Y2H assays of IND/INDD30G versions with ETT-BD and ETT/ETT2C-S versions with IND-AD and their sensitivity to IAA. Controls of INDD30G, ETT2C-S, and empty plasmids are shown below. (C–E) Phenotypic analyses of ind-2 (C), pIND::INDWT in ind-2 (D), and pIND::INDD30G in ind-2 (E) with SEM images of stage 13 gynoecia (panel I), dehiscence (panel II), pollen tube growth (panel III), and a seed set (panel IV). (F–H) SEM images of stage 13 gynoecia from ett-3 (F), pETT::ETTWT in ett-3 (G), and pETT::ETT2C-S in ett-3 (H). (stg) Stigma; (sty) style; (va) valve. Bars, 100 μm.
In contrast to the gynoecium defect, expressing pIND::INDD30G in the ind-2 mutant background was perfectly able to rescue its indehiscent phenotype (Fig. 5E; Supplemental Fig. 6E). This observation shows that the INDD30G protein is functional and that the IAA sensitivity of the IND and ETT interaction is not required for specification of the valve margin. This is in agreement with the lack of ETT expression in this tissue (Supplemental Fig. 1J).
Since both ETT and IND are TFs, we tested whether the IAA insensitivity of the ETT–INDD30G complex affected the regulation of downstream target genes. In situ hybridization showed that PID expression was reduced in gynoecia of the pIND::INDD30G line as compared with wild type (Supplemental Fig. 6G,H), thus suggesting that IAA regulates the activity of the ETT–IND complex toward its downstream targets. In the case of PID expression, it is conceivable that ETT–IND acts as a repressor in the absence of IAA, while accumulation of IAA leads to an ETT–IND-dependent induction of PID expression. Additional evidence of an ETT–IND-mediated effect on auxin dynamics in the gynoecium comes from a persisting DR5::GFP signal at the late developmental stages in the pIND::INDD30G line (Supplemental Fig. 6I). This emphasizes a defect in auxin dynamics when disrupting the IAA-sensitive ETT–IND activity.
As opposed to the random mutagenesis approach with IND, we took a site-directed approach with ETT due to the presence of four cysteine residues in the ES domain. Cysteines have been shown previously to modulate transactivational activity of TFs (Klatt et al. 1999; Pineda-Molina et al. 2001; Dietz 2008; Li et al. 2009) and could therefore also play a role in the IAA-sensitive response uncovered here. We substituted each cysteine for serine and tested them individually in Y2H assays with wild-type IND. No single substitution interfered with the ETT–IND interaction but also did not render the interaction with IND IAA-insensitive (Supplemental Fig. 7A,B). In contrast, combining C-to-S mutations in positions 452 and 506 led to growth of yeast also in the presence of IAA (Fig. 5A,B). Remarkably, expressing this pETT::ETT-C452S-C506S IAA-insensitive variant (referred to here as pETT::ETT2C-S) in the ett-3 mutant background led to the same overproliferated stigmatic tissue phenotype observed in the pIND::INDD30G line (Fig. 5F–H), while other defects of the ett-3 gynoecium were rescued (Supplemental Fig. 7C–H). These data therefore strongly support the hypothesis that gynoecium development requires the ETT–IND-mediated IAA-sensing pathway.
ETT can interact with proteins belonging to different TF families in an IAA-sensitive fashion
Abnormal carpel development is the most obvious and severe defect in ett mutants; however, ETT has been reported to function in a diverse range of developmental contexts in addition to the gynoecium; namely, LR initiation, ovule integument development, and stem–pedicel fusion (Marin et al. 2010; Kelley et al. 2012; Zhao et al. 2013). Analyses of the pETT(8Kb)::GUS reporter line (Ng et al. 2009) and the pETT::ETT-GFP complementation line confirmed the presence of ETT expression in domains relevant to these three processes. ETT expression was detected (1) in the pericycle (the tissue that will give rise to LRs) and during all stages of LR formation (Fig. 6C; Supplemental Fig. 8A,B); (2) during ovule development, coinciding with the region from which the integuments will later emerge (Fig. 6E; Supplemental Fig. 9A–F); and (3) at the abaxial side of the new developing primary branches, marking the region that corresponds to the point of conjunction between the stem and the branch (Fig. 6K; Supplemental Fig. 10A–C).
Figure 6.
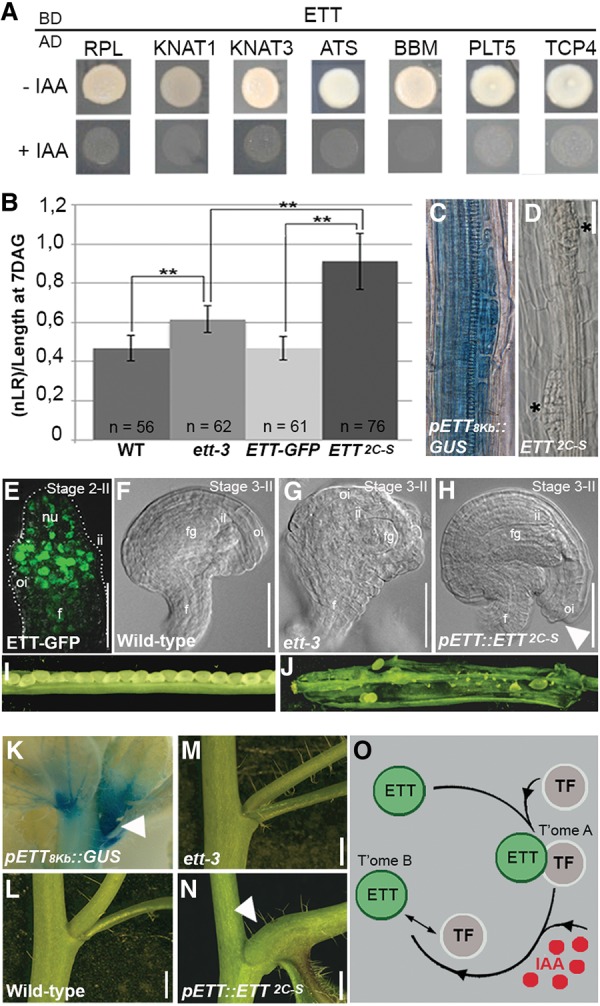
Auxin sensitivity is required for proper organ morphogenesis. (A) Y2H assay showing IAA sensitivity of interactions of ETT-BD and TFs (fused to the AD domain) identified in the REGIA (Regulatory Gene Initiative in Arabidopsis) library. Assays were carried out in the absence of IAA (−IAA; top) and presence of IAA (+IAA; bottom). (B–D) Chart summarizing the comparison of LR numbers among the genotypes (normalized against root length), showing a statistically higher number of LR in the pETT::ETT2C-S line (shown in B). GUS staining of the pETT(8Kb)::GUS line in the root (C) and LR defects in pETT::ETT2C-S (D), with two adjacent LRs developing (indicated by asterisks). (E) pETT::ETT-GFP expression profile in ovule primordia at stage 2-II; the signal is localized predominantly in the region from which the integuments will develop. (F–H) Clearing of wild-type (F), ett-3 (G), and pETT::ETT2C-S (H) ovules showing abnormal overgrowth of the outer integument in pETT::ETT2C-S. (I,J) Stage 17 fruits from wild type (I) and pETT::ETT2C-S (J), with one valve removed to expose the defect in fertility in the pETT::ETT2C-S line. (K–N) GUS staining of the pETT(8Kb)::GUS line during lateral branch formation (K) and close-up views of the primary branch–stem internode in wild type (L), ett-3 (M), and pETT::ETT2C-S (N). (O) Schematic model illustrating how the IAA-induced change in the ETT/TF complex dimerization state determines the regulation of downstream targets. As shown for PID in this study, transcriptomes A and B share genes regulated differently by the repressor/activator states. (nu) Nucellus; (ii) inner integument; (oi) outer integument; (fg) female gametophyte; (f) funiculus. Bars: C–H, 20 µm; L–M, 5 mm.
In contrast, IND function appears to be very specific to gynoecium and fruit development (Liljegren et al. 2004; Girin et al. 2011). It is therefore possible that ETT constitutes the principal prerequisite for the IAA-sensing ability and that it interacts with different protein partners in other processes. To test this hypothesis, we performed a Y2H screening of ETT against the REGIA (Regulatory Gene Initiative in Arabidopsis) library of TFs (Paz-Ares et al. 2002). This analysis led to the identification of several putative ETT interactors (Fig. 6A; Supplemental Fig. 11) belonging to different TF families, including HOMEOBOX (REPLUMLESS [RPL], KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 [KNAT1], and KNAT3), AP2 domain (BABYBOOM [BBM]), and TCP (TEOSINTE BRANCHED, CYCLOIDEA AND PCF4 [TCP4] and TCP18). Since ETT already has been reported to interact with ABERRANT TESTA SHAPE (ATS; also known as KANADI4) during ovule development (Kelley et al. 2012), ATS was included in this test. Furthermore, due to its high sequence homology with BBM, we also included PLETHORA5 (PLT5). Both ATS and PLT5 were found to interact with ETT in this assay (Fig. 6A). Similar to the interaction with IND, the interaction between ETT and these TFs was sensitive to the presence of IAA (Fig. 6A), thus suggesting that the IAA-sensitive activity of ETT involves tissue- and process-specific TF partners. Interestingly, the factors on this list function in the same processes as ETT, including the development of roots (KNAT3, BBM, and PLT5) (Serikawa et al. 1997; Galinha et al. 2007; Truernit and Haseloff 2007; Hofhuis et al. 2013), ovules (ATS) (Kelley et al. 2012), stem–pedicel fusion (TCP18) (Aguilar-Martínez et al. 2007), and carpels (RPL and KNAT1) (Roeder et al. 2003; Alonso-Cantabrana et al. 2007). Remarkably, RPL was shown recently to directly bind a fragment of the PID promoter located ∼1.5 kb upstream of the PID start codon (Bencivenga et al. 2016). This region is enriched in the ETT-GFP ChIP assay (region 4 in Supplemental Fig. 2A,B) despite the lack of conserved AuxREs (Fig. 2A). As proposed above, it is thus possible that ETT associates with DNA in this region indirectly via IAA-controlled protein–protein interactions (see the model in Supplemental Fig. 4B).
We next wondered whether the IAA-sensing activity of ETT is a generally conserved mechanism for modulating organ and tissue morphogenesis in response to high IAA concentrations. Indeed, LR initiation, ovule integument development, and primary branch fusion are strictly connected to and dependent on auxin accumulation (Benková et al. 2003; Gallavotti 2013). Therefore, we performed a morphological analysis and compared wild-type, ett-3, pETT::ETT-GFP, and pETT::ETT2C-S genotypes with respect to these three different developmental contexts.
In accordance with the ETT expression pattern, the ett-3 mutant displays defects in all the three tissues: a significantly higher number of developing LRs (Fig. 6B; Supplemental Fig. 8C–F), aberrant ovule integument morphology (Fig. 6F,G; Supplemental Fig. 9K–N; Kelley et al. 2012), and a mild deformity at the stem–primary branch fusion (Fig. 6L,M; Supplemental Fig. 10D,E,H,I; Zhao et al. 2013). Notably, the pETT::ETT-GFP construct was able to complement all of these defects (Fig. 6B; Supplemental Figs. 8E,F, 9O,R, 10F,J). In contrast, analyses of the pETT::ETT2C-S line revealed morphological aberrations in all three developmental contexts that could not be explained by complementation of the ett-3 phenotypes.
First, during the LR emergence process, we detected a significant overproduction of LRs along the root length and in proximity to the root tip (Fig. 6B,D; Supplemental Fig. 8C–F). Second, during ovule development, we observed an extensive overgrowth of the outer integument that brings it particularly close to the funiculus (Fig. 6H; Supplemental Fig. 9S–V), thus placing the micropile aperture, the point of entrance of the pollen tube, in an unfavorable position for fertilization. As a consequence, pETT::ETT2C-S plants displayed very high ovule abortion frequency and a drastically reduced seed set (Fig. 6I,J). Last, in pETT::ETT2C-S plants, abnormal development at the point of conjunction between the stem and the primary branch was observed (Fig. 6N), with the latter developing more horizontally in comparison with wild type (Supplemental Fig. 10D–K). These results demonstrate that ETT-mediated IAA perception plays multiple diverse roles throughout plant development.
Discussion
A common theme in plant hormone signaling has emerged involving ubiquitination and degradation of transcriptional repressors (for review, see Santner and Estelle 2009). For auxin, this mechanism involves repression of ARFs at low auxin concentrations through interaction between PB1 protein domains of ARFs and Aux/IAAs. Derepression of ARFs at high auxin levels occurs when the Aux/IAA repressor proteins are degraded (for review, see Guilfoyle 2015). ETT belongs to the ARF family and has been associated previously with auxin-related processes during gynoecium development (Nemhauser et al. 2000); however, given that ETT lacks the PB1 domain, it seems unlikely that ETT would function via the canonical auxin signaling pathway. Indeed, the data presented here demonstrate the existence of an alternative mechanism through which IAA regulates ETT activity. This mechanism is fundamentally different from the canonical auxin signaling pathway, as it involves an IAA-mediated direct effect on TF activity without the involvement of a ubiquitination and degradation step. Both the ETT–TF and TIR1/AFB pathways are short, allowing for rapid transcriptional responses to auxin. However, since the Aux/IAAs must be resynthesized to reset repression, the ETT–TF pathway may have an advantage in the speed with which it can return to a nonauxin state, depending on the reversibility of the IAA effect. This may be particularly important during processes of developmental switching such as in the patterning of the gynoecium apex or the initiation of LRs.
Our data suggest that IAA affects the activity of ETT toward downstream targets by directly interfering with ETT–TF interactions (Fig. 6O). Specific ETT–TF complexes may therefore control two separate transcriptomes dependent on the absence/presence of IAA (Fig. 6O, transcriptomes A and B, respectively). The same genes may be present in both transcriptomes but simply be controlled differently under the two conditions. One such example is PID: At low IAA levels, an ETT–IND complex represses PID expression, whereas high IAA levels change the manner in which ETT interacts with the PID promoter, most likely via IAA-induced changes in protein interactions correlating with induction of PID expression (Supplemental Fig. 4A,B). The IAA-dependent modulation of ETT protein interactions implicates that, in a precise spatio–temporal window that coincides with ETT expression, a specific set of ETT partners must be available to achieve a tissue-specific transcriptional response. The identification here of a number of proteins from diverse TF families that interact with ETT in an IAA-sensitive manner supports this hypothesis.
The genetic results as well as the protein–protein interaction data indicate that IAA directly affects the ETT–TF dimer, opening the possibility that the IAA perception occurs via direct contact between the IAA molecule and the ETT protein. If so, this mechanism may be reminiscent of the way by which the thyroid hormone receptor (TR) regulates its targets in animals (for review, see Tsai and O'Malley 1994; Xu et al. 1999). The TR is constitutively bound to the promoter of its target genes. However, in the absence of the hormone ligand, TR functions primarily as a repressor, whereas binding of the ligand changes it into an activator of the same targets (Tsai and O'Malley 1994; Xu et al. 1999). The molecular nature of the interaction between ETT and IAA and the effect it has on the ETT–TF complex will be the focus of future biochemical and structural studies.
In summary, we identified a novel IAA-sensing mechanism in plant development. While adding to the complexity of auxin signaling, these findings will contribute to our understanding of how auxin can regulate such a vast number of biological processes. Above and beyond the specific mechanism by which the ETT–TF sensor complex controls organ morphology, identification of this novel signaling conduit may have far-reaching implications for the existence of similar alternative mechanisms for hormonal regulation of plant growth and development.
Materials and methods
Plant materials and growth conditions
Plants were grown on soil in long-day conditions (16 h light/8 h dark). Mutant line ind-2 is a strong loss-of-function allele in the Col-0 background (Liljegren et al. 2004); ett-3, a mutant allele of medium strength and originally in the Ler ecotype (Sessions et al. 1997), was introgressed in the Col-0 background via five backcrosses; and pETT(8Kb)::GUS was kindly provided by Toshiro Ito (Ng et al. 2009). Selection of transgenic lines was carried out by plating seeds on 0.8% MS supplemented with the appropriate antibiotic; the plates were subsequently stratified for 4 d in the dark at 4°C and then moved to long-day conditions (16 h light/8 h dark).
Scanning electron micrographs (SEMs), sectioning, and GUS staining
SEMs and sectioning were performed as described previously (Moubayidin and Østergaard 2014). GUS staining was performed as described previously (Liljegren et al. 2004). SEM images were used as the source for stigmatic papilla length quantification.
ChIP
ChIP was performed in triplicate according to Schiessl et al. (2014) using pETT::ETT-GFP in the ett-3 line, and wild-type plants were used as a negative control. Data were analyzed as in Schiessl et al. (2014). A WUS promoter fragment was used as a positive control as described previously (Liu et al. 2014). IAA treatment was performed by spraying pETT::ETT-GFP and wild-type plants with a solution containing 0.1 mM IAA, 10 µM NPA, and 0.03% silwet and collecting material 6 h after treatment. Primers sequences are in Supplemental Table 2.
Y1H assay, Y2H assay, and REGIA Y2H library screening
The Y1H assay was performed in strains Y187 and PJ64a following the protocol described in Roccaro et al. (2005). PID promoter fragments were cloned in the pHISi vector (Clontech). Interaction was tested on selective yeast synthetic dropout (YSD) medium lacking Leu (L) and His (H) supplemented with different concentrations (20, 40, and 60 mM) of 3-aminotriazole (3-AT). IAA (Sigma) was dissolved in ethanol and added at the desired concentration directly to the cooling medium.
The Y2H assays were performed at 28°C in the yeast strain AH109 (Clontech) using the cotransformation technique (Egea-Cortines et al. 1999). Coding sequences were cloned into pGAD424 and pGBKT9 vectors (Clontech) using the SmaI and PstI sites or cloned into the Gateway vector GAL4 system (pGADT7 and pGBKT7; Clontech) passing through pDONR207 (Life Technologies). Strength of interaction was tested on selective YSD lacking Leu (L), Trp (W), adenine (A), and His (H) supplemented with different concentrations (1, 2.5, and 5 mM) of 3-AT. IAA, NAA, and 2,4D (Sigma) were dissolved in ethanol and added at the desired concentrations directly to the cooling medium.
Screening of the REGIA library was performed with the mating technique; the ETT coding sequence was cloned in the pDEST32 plasmid (Invitrogen) passing through the pDONR207 (Invitrogen) and transformed in the PJ69a strain. The REGIA library was in the PJ69alfa mating type. After mating, cells were resuspended in sterile water and plated on YSD − W-L-H-A supplemented with different concentration of 3-AT. Plasmids were extracted from positive clones and sequenced.
BiFC
ORFs of full-length IND and ETT were cloned in the pYFPN43 and pYFPC43 (http://www.ibmcp.upv.es/ferrandolabvectors.php) passing through pDONR207 (Life Technologies). The pEAQ-HT vector (Sainsbury et al. 2009), which expresses the silencing suppressor p19 of tomato bushy stunt virus, was added to the vector combination. The positive pYFPN43-SNF + pYFPC43-SNF control was used as described previously (Ferrando et al. 2001). BiFC was performed as described previously (Belda-Palazón et al. 2012).
Exogenous IAA, NAA, and 2,4D applications were performed by mixing lanolin with the hormone as described previously (Reinhardt et al. 2003); a thin layer of lanolin–hormone mixture was applied on the infiltrated area 24 and 48 h after infiltration. Images were taken 72 h after infiltration.
In situ hybridization
The PID digoxygenin-labeled antisense and sense RNA probes were generated by in vitro transcription according to the instructions provided with the DIG RNA labeling kit (SP6/T7; Roche) using cDNA as template. Developing inflorescences were fixed and embedded in Paraplast Plus embedding medium, cut in 8-μm sections, and then hybridized as described previously (Dreni et al. 2007). For in situ of the ind-2 pIND-INDD30G line, a 3× PID antisense probe quantity was used to amplify the signal. Sections were observed using a Leica DM6000 equipped with differential interface contrast (DIC) optics.
FRET/FLIM
FRET/FLIM was performed as described previously (Boer et al. 2014) using transfected Arabidopsis (Columbia wild-type) mesophyll protoplasts (Rademacher et al. 2012). Images of 128 × 128 pixels were acquired with acquisition times of 120 sec and an average count rate of 104 photons per second. Analysis of the data was as described (Rademacher et al. 2012). Several cells (n = 28) were analyzed, and average FLIMs of different combinations were exported for generating a box plot. The statistical significance of differences between samples was determined using a two-tailed Student's t-test.
Phylogenetic footprinting
Phylogenetic shadowing was performed using the mVISTA program (http://genome.lbl.gov/vista/index.shtml). PID genomic sequences from Brassicaceae species were obtained from public databases via BLAST search on http://www.ncbi.nlm.nih.gov using the Arabidopsis PID gene as the query sequence.
Microarray data analyses using GeneSpring
35S::IND:GR seeds were grown for 7 d in 5 mL of 0.5% (w/v) glucose, 0.5× Murashige, and Skoog medium with constant shaking and constant light. Seedlings were treated for 6 h with either DMSO mock or 10 μM IAA ± 10 μM DEX. RNA was extracted from three biological replicates (total of 12 samples). The Robust Multiarray Average (RMA) algorithm was used to normalize the microarray data using Affymetrix expression console. Affymetrix transcriptome analysis console (TAC) software was used for differential gene expression analysis. Genes with a >1.5-fold differential expression and a P-value of <0.05 were selected using one-way ANOVA.
Pollen tube guidance
Pistils were emasculated and pollinated after 24 h with wild-type pollen. After 16–18 h, pistils were carefully isolated from the plants and fixed in a solution of acetic acid and absolute ethanol (1:3), cleared with 8 M NaOH, and labeled with aniline blue (Sigma). Images were taken using a Leica DM6000.
In planta colocalization and in vivo CFP–YFP FRET
pIND (3.2kb)::IND-YFP and pETT(3.7kb)::ETT-CFP constructs were assembled using the Golden Gate method and appropriate plasmids as described previously (Engler et al. 2008, 2014) and introduced in wild-type plants. Samples was imaged with a Zeiss LSM 780 confocal microscope. YFP fluorescence was excited using the 514-nm line from an argon ion laser, and the emitted light was captured between 530 and 579 nm. CFP fluorescence was excited using the 458-nm line from an argon ion laser, and the emitted light was captured between 470 and 535 nm. Confocal Z-section stacks were collected at 1-µm spacing throughout the depth of the tissue. In vivo FRET was performed with a Zeiss LSM 780 confocal microscope following the acceptor photobleaching method reported in Karpova and McNally (2006). Two-hundred nuclei were imaged for the ETT-CFP/IND-YFP-expressing cells (stylar cells at stage 10–11). As a control, 200 nuclei from a tissue expressing ETT:CFP only (valves at stages 4–5) were imaged. The background value was subtracted from all the values.
ETT constructs and marker line
For the pETT::ETT-GFP marker line, the ETT gene (including a 5-kb promoter sequence upstream of the ATG exons and introns) was cloned in the pPZP222 plasmid already harboring the GFP-coding region and the tNOS terminator and transformed in ett-3 (Col-0 background). Thirteen independent T1 lines were analyzed for GFP expression pattern. Quantitative real-time PCR analysis was used to estimate the numbers of transgene copies in these individual lines, similar to the approach reported previously (Bartlett et al. 2008). Line number 1, which contained one copy of the transgene and fully complemented the ett-3 phenotype, was propagated to obtain ett-3 homozygous plants with the homozygous pETT::ETT-GFP construct.
For construction of ETT versions with cysteines mutated to serines, the ETT gene (including a 5-kb promoter sequence upstream of the ATG, exons, and introns) was cloned in the pPZP222 plasmid already harboring a tNOS terminator and transformed in ett-3 (Col-0 background). Cysteine-to-serine mutations were introduced by site-specific mutagenesis. Primer sequences are in Supplemental Table 2.
Error-prone mutagenesis for INDD30G identification
Error-prone PCR mix was composed of G2 polymerase buffer (Promega) and G2 polymerase, 5 mM MgCl2, 0.15 mM MnCl2, and unbalanced dNTP mix (2.5 mM dATP, 2.5 mM dGTP, 10 mM dCTP, 10 mM dTTP). The pool of PCR fragments was incubated with the pGADT7-rec plasmid (Clontech) following the cotransformation protocol described in the Matchmaker library construction and screening user manual (Clontech). Cells were plated on YSD-selective medium lacking Trp (W), Leu (L), Ade (A), and His (H) and supplemented with 100 μM IAA. Plasmids were recovered from those colonies able to grow in the presence of IAA and sequenced (total of 27 colonies). One clone carried five mismatches, including the D30G mutation, and three had a deletion of 12 amino acids, including the D30 residue. The remaining 23 clones contained several errors that affected the correct frame and for this reason were discarded. The IND promoter (3.2 kb upstream of the ATG) plus the IND/INDD30G-coding region were cloned into the pCGN1547 vector already harboring the tNOS terminator. The Agrobacterium tumefaciens strain GV3101 containing the final vector was transformed into ind-2 plants using the floral dip method.
Supplementary Material
Acknowledgments
We thank Norwich Research Park Bioimaging, Grant Calder, Roy Dunford, Caroline Smith, Paul Thomas, and Mark Youles for technical support; Charlie Scutt, Alejandro Ferrando, and George Lomonossoff for plasmids; Toshiro Ito for seeds; Brendan Davies and Barry Causier for the REGIA library; and Mark Buttner, Simona Masiero, Fabio Rossi, Doris Wagner, and Jun Xiao for help and material. We are also grateful to Stefano Bencivenga, Marie Brüser, Friederike Jantzen, Lukasz Langowski, Xinran Li, and Nicola Stacey for discussions and helpful comments on the manuscript. This work was supported by grants BB/M004112/1 and BB/I017232/1 (Crop Improvement Research Club) to L.Ø. from the Biotechnological and Biological Sciences Research Council, and Institute Strategic Programme grant (BB/J004553/1) to the John Innes Centre. S.S., J.D., and L.Ø conceived the experiments. S.S., J.D., L.M., A.F.-R., M.V., and K.S. performed the experiments. S.S., J.D., L.M. P.S., A.F.-R., M.V., K.S., and L.Ø analysed the data. S.S, J.D., and L.Ø. wrote the manuscript, and L.M., P.S., K.S., J.F., and D.W. commented on it. All authors read and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.285361.116.
Freely available online through the Genes & Development Open Access option.
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, Martínez-Laborda A. 2007. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134: 2663–2671. [DOI] [PubMed] [Google Scholar]
- Bartlett JG, Alves SC, Smedley M, Snape JW, Harwood WA. 2008. High-throughput Agrobacterium-mediated barley transformation. Plant Methods 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farrás R, Carrasco P, Ferrando A. 2012. Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Serrano-Mislata A, Bush M, Fox S, Sablowski R. 2016. Control of oriented tissue growth through repression of organ boundary genes promotes stem morphogenesis. Dev Cell 39: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. 2008. Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R. 2001. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128: 4057–4067. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WA, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliviera C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. 2012. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dietz KJ. 2008. Redox signal integration: from stimulus to networks and genes. Physiol Plant 133: 459–468. [DOI] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. 2007. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J 52: 690–699. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H. 1999. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843. [DOI] [PubMed] [Google Scholar]
- Ferrando A, Koncz-Kálmán Z, Farràs R, Tiburcio A, Schell J, Koncz C. 2001. Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plants cells. Nucleic Acids Res 29: 3685–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Gallavotti A. 2013. The role of auxin in shaping shoot architecture. J Exp Bot 64: 2593–2608. [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938. [DOI] [PubMed] [Google Scholar]
- Girin T, Paicu T, Stephenson P, Fuentes S, Körner E, O'Brien M, Sorefan K, Wood TA, Balanzá V, Ferrándiz C, et al. 2011. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23: 3641–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremski K, Ditta G, Yanofsky MF. 2007. The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134: 3593–3601. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ. 2015. The PB1 domain in Auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M, Atkinson A, Bylstra YH, Walsh R, Smyth DR. 2001. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. 2013. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr Biol 23: 956–962. [DOI] [PubMed] [Google Scholar]
- Karpova T, McNally JG. 2006. Detecting protein–protein interactions with CFP–YFP FRET by acceptor photobleaching. Curr Protoc Cytom 135: 12.7.1–12.7.11. [DOI] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. 2012. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Klatt P, Molina EP, De Lacoba MG, Padilla CA, Martinez-Galesteo E, Barcena JA, Lamas S. 1999. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J 13: 1481–1490. [DOI] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Jez JM, Strader LC. 2015. Refining the nuclear auxin response pathway through structural biology. Curr Opin Plant Biol 27: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. 2009. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. 2004. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116: 843–853. [DOI] [PubMed] [Google Scholar]
- Liu Z, Franks RG. 2015. Molecular basis of fruit development. Front Plant Sci 6: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dinh TT, Li D, Shi B, Li Y, Cao X, Guo L, Pan Y, Jiao Y, Chen X. 2014. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J 80: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. 2010. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Østergaard L. 2014. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr Biol 24: 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J, Feldmann LJ, Zambryski PC. 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Ng KH, Yu H, Ito T. 2009. AGAMOUS controls GIANT KILLER, a multifunctional chromatin modifier in reproductive organ patterning and differentiation. PLoS Biol 7: e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Regia Consortium. 2002. REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics 3: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Molina E, Klatt P, Vázquez J, Marina A, García de Lacoba M, Pérez-Sala D, Lamas S. 2001. Glutathionylation of the p50 subunit of NF-κB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry 40: 14134–14142. [DOI] [PubMed] [Google Scholar]
- Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T. 2014. Protein–protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front Plant Sci 5: 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G, et al. 2012. Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Reyes-Olalde JI, Zuñiga-Mayo VM, Chávez Montes RA, Marsch-Martínez N, de Folter S. 2013. Inside the gynoecium: at the carpel margin. Trends Plant Sci 18: 644–655. [DOI] [PubMed] [Google Scholar]
- Roccaro M, Li Y, Masiero S, Saedler H, Sommer H. 2005. ROSINA (RSI), a novel protein with DNA-binding capacity, acts during floral organ development in Antirrhinum majus. Plant J 43: 238–250. [DOI] [PubMed] [Google Scholar]
- Roeder AH, Ferrándiz C, Yanofsky MF. 2003. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 13: 1630–1635. [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693. [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. 2009. Recent advances and emerging trends in plant hormone signaling. Nature 459: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Schiessl K, Muiño JM, Sablowski R. 2014. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc Natl Acad Sci 111: 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serikawa KA, Martinez-Laborda A, Kim HS, Zambryski PC. 1997. Localization of expression of KNAT3, a class 2 knotted1-like gene. Plant J 11: 853–861. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. 1997. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galván-Ampudia CS, Offringa R, Friml J, Yanosfky MF, Østergaard L. 2009. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459: 583–586. [DOI] [PubMed] [Google Scholar]
- Truernit E, Haseloff J. 2007. A role for KNAT class II genes in root development. Plant Signal Behav 2: 10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 63: 451–486. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136: 1005–1016. [DOI] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev 9: 140–147. [DOI] [PubMed] [Google Scholar]
- Zhao H, Liu L, Mo H, Qian L, Cao Y, Cui S, Li X, Ma L. 2013. The ATP-binding cassette transporter ABCB19 regulates postembryonic organ separation in Arabidopsis. PLoS One 8: e60809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.