Abstract
Adult-onset Niemann–Pick disease type C (NPC) is an infrequent presentation of a rare neurovisceral lysosomal lipid storage disorder caused by autosomal recessive mutations in NPC1 (∼95%) or NPC2 (∼5%). Our patient was diagnosed at age 33 when he presented with a 10-yr history of difficulties in judgment, concentration, speech, and coordination. A history of transient neonatal jaundice and splenomegaly with bone marrow biopsy suggesting a lipid storage disorder pointed to NPC; biochemical (“variant” level cholesterol esterification) and ultrastructural studies in adulthood confirmed the diagnosis. Genetic testing revealed two different missense mutations in the NPC1 gene—V950M and N1156S. Symptoms progressed over >20 yr to severe ataxia and spasticity, dementia, and dysphagia with aspiration leading to death. Brain autopsy revealed mild atrophy of the cerebrum and cerebellum. Microscopic examination showed diffuse gray matter deposition of balloon neurons, mild white matter loss, extensive cerebellar Purkinje cell loss with numerous “empty baskets,” and neurofibrillary tangles predominantly in the hippocampal formation and transentorhinal cortex. We performed whole-genome sequencing to examine whether the patient harbored variants outside of the NPC1 locus that could have contributed to his late-onset phenotype. We focused analysis on genetic modifiers in pathways related to lipid metabolism, longevity, and neurodegenerative disease. We identified no rare coding variants in any of the pathways examined nor was the patient enriched for genome-wide association study (GWAS) single-nucleotide polymorphisms (SNPs) associated with longevity or altered lipid metabolism. In light of these findings, this case provides support for the V950M variant being sufficient for adult-onset NPC disease.
Keywords: atrophy/degeneration affecting the cerebrum, central nervous system degeneration, falls, hepatosplenomegaly, neurodegeneration, neuronal loss in central nervous system, progressive encephalopathy, progressive forgetfulness, progressive language deterioration, progressive psychomotor deterioration, slowed slurred speech, social and occupational deterioration
INTRODUCTION
Niemann–Pick represents a group of progressive neurodegenerative diseases characterized by defects in lipid metabolism. Typically, Niemann–Pick disease type A presents in infancy, whereas type B presents in mid-childhood. In contrast, type C disease is marked by greater variability in timing, both of presentation and eventual demise. The progression of neurological abnormalities typically begins with ataxia and frequently includes spasticity and seizures. Dystonia, vertical supranuclear gaze palsy, dementia, and psychiatric manifestations are also common.
At the cellular level, Niemann–Pick disease type C (NPC) is characterized by defects in cholesterol storage and esterification. Consistent with this, NPC1 and NPC2 are believed to function in a coordinated fashion in the endosomal processing of cholesterol, as well as other lipids (Vanier and Millat 2003). Specifically, cells cultured from patients with NPC were found to have a severe deficit in cholesterol esterification, whereas this process remains intact in cells from Niemann–Pick type A and B patients (Pentchev et al. 1985). As such, the diagnosis of NPC traditionally included a cholesterol esterification assay from cultured fibroblasts, as well as filipin staining to demonstrate excess intracellular cholesterol. Recently, the search for blood-based tests for biomarkers has yielded a number of diagnostic metabolites that can be detected sensitively and measured accurately by mass spectrometry (for review, see Vanier et al. 2016). These include the cholesterol oxidation product cholestane-3β,5α,6β-triol, which appears to be elevated because of the combination of increased oxidative stress and deposition of free cholesterol in NPC (Porter et al. 2010), as well as bile acid metabolites 3β-hydroxy-7β-N-acetylglucosaminyl-5-cholenoic acid and N-(3β,5α,6β-trihydroxy-cholan-24-oyl)glycine, which have the potential to be used as part of a newborn screen because of their detectability in dried blood spots (Jiang et al. 2016; Mazzacuva et al. 2016). Mutations in NPC1 account for 95% of patients with NPC, with the remainder coming from NPC2 (Vanier and Millat 2003). Within NPC1 and NPC2, a wide spectrum of DNA sequence variants can cause clinically significant disease; more than 150 known causal mutations have been identified in NPC1 alone (Garver et al. 2010; Adebali et al. 2016).
The adult-onset form of the disease, which is believed to comprise ∼10% of cases, typically presents in the second or third decade with neurological symptoms. Recent predictions of the frequency of disruptive mutations in NPC1 and NPC2 from large exome data sets suggest that the incidence of the more easily missed adult-onset form may be more common than previously thought (Wassif et al. 2016). Given the variable disease presentation and incomplete understanding of the spectrum of variation that can cause NPC, it is important to understand how a patient-specific mutation in NPC1 as well as modifying genetic factors contribute to phenotypic heterogeneity.
Even in highly penetrant Mendelian diseases, variability in phenotype can be a result of both differences at the disease locus and modifiers elsewhere in the genome. Cystic fibrosis (CF) patients with the compound heterozygous CFTR genotype R117H/ΔF508 show a less severe phenotype than those homozygous for ΔF508, including later diagnosis and lower sweat chloride concentration (The Cystic Fibrosis Genotype-Phenotype Consortium 1993). At the same time, genome-wide association studies in CF patients have uncovered modifier loci in relevant pathways outside of CFTR influencing the severity of lung disease (Corvol et al. 2015). Thus, variation elsewhere in the genome can exert significant effects on disease severity.
In this study, we present the case of a patient who died of adult-onset NPC, including a “genomic autopsy” from whole-genome sequencing (WGS) performed posthumously. We sought to determine if the unusually late onset and long survival of the patient could be accounted for by his previously identified variants in NPC1 alone or if there were other genetic modifiers that contributed to his phenotype. We provide supporting evidence that the variant V950M (rs120074135) causes a milder deficit in NPC1 function and as such is responsible for both the patient's relatively high cholesterol esterification levels and longevity.
RESULTS
Clinical Presentation
The patient was a 54-yr-old man who had apparently enjoyed good health until he presented to a brain injury clinic at the age of 20 complaining of inability to maintain jobs and difficulty controlling finances, which he had attributed to two separate blows to the head from a baseball. He did not experience additional neurological symptoms with either incident and was clinically and radiologically cleared at the time. His mother described a slightly earlier onset of progressive symptoms, around the age of 18, which included increasing incidents of tripping and falling, poor concentration, memory difficulties, and worsening speech articulation. Further investigation revealed a history of neonatal jaundice and hepatomegaly, prompting bone marrow biopsy, which showed evidence of a lipid storage disease consistent with either Gaucher's or Niemann–Pick disease. Neonatal symptoms abated, and the findings were not pursued further. The patient was referred to the medical genetics clinic where physical examination revealed splenomegaly, mild speech slurring, apparent restriction of vertical saccades, and trace gait instability. β-Glucosidase enzyme activity was normal, excluding Gaucher's disease. Skin punch biopsy showed ultrastructural evidence of intracellular lipid storage disorder, and a cholesterol esterification assay was abnormal, indicating NPC. Interestingly, his level of intracellular cholesterol esterification was higher than the average patient with NPC at 357 (normal: 3342 ± 1729; disease: 90 ± 140), meriting his biochemical classification as “variant” NPC.
Therapy with a cholesterol-lowering agent (lovastatin) and a reduced-cholesterol diet was initiated early in his course. He entered an experimental treatment trial with OGT 918 (miglustat) at approximately age 42. His disease was relatively stable with slow progression of findings that included increasing dysarthria, hearing loss, decreased gag reflex, cognitive and behavioral issues, and continued coordination problems with increasing falls (after age 45). His ataxia and spasticity rendered him nonambulatory, and he continued to have increasingly frequent falls out of bed complicated by subdural hemorrhage along with ongoing dysphagia with chronic aspiration. He had an aspiration event requiring hospital admission and was transferred to hospice care, expiring ∼1 mo later. Of interest, his younger brother by 2 yr has also been diagnosed with NPC and has followed a similar course.
Genomic Analyses
At the NPC1 locus, the patient was positive for heterozygous V950M (rs28942105) and N1156S (rs120074135) missense mutations, each of which is known to be associated with NPC (Fig. 1; Carstea et al. 1997; Millat et al. 2001). The V950M mutation has been seen previously in the homozygous form as well as in conjunction with an I1061T mutation (Sévin et al. 2007). The I1061T mutation by itself is often seen in patients with milder disease phenotypes: It is associated with a slowly progressive juvenile form of the disease and was common in a set of adult-onset NPC (Millat et al. 1999; Sévin et al. 2007). On the other hand, the N1156S mutation was previously found to lead to severe infantile disease in conjunction with a Q775P mutation (Fernandez Valero et al. 2005). Sanger DNA sequencing of NPC1 confirmed that the patient's brother, who had a very similar clinical presentation and disease course, carried the same NPC1 coding mutations. With genotype data from the patient's parents unavailable, we cannot say for certain if these occur on the same haplotype; however, a trans orientation is most likely given the recessive nature of NPC.
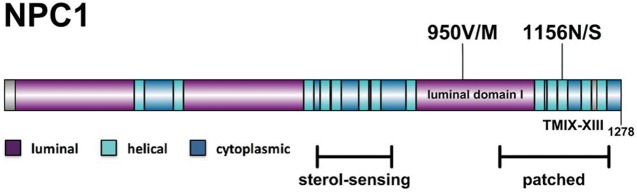
Figure 1.
Location of NPC1 variants. Highlighted are the locations of the patient's two disease-causing variants: 950V/M sits in luminal domain I, the third large luminal domain of NPC, whereas 1156N/S sits in the 11th transmembrane domain, in a region with homology to the patched family of proteins.
Including the V950M and N1156S mutations, the patient possessed five total missense variants and two synonymous variants in the coding region of NPC1, one 5′-UTR variant (Table 1) and 51 single-nucleotide variants (SNVs) and 10 small indels in the gene's intronic regions. The three additional missense mutations, H215R, M642I, and I858V, are considered to be benign: They are highly polymorphic and have been demonstrated to occur in equal abundance in both affected and nonaffected individuals (Millat et al. 1999; Ribeiro et al. 2001; Bauer et al. 2002; Fernandez Valero et al. 2005; Yang et al. 2005).
Table 1.
NPC1 variants
| Gene symbol | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | dbSNP ID | Genotype | CADD score | ExAC MAF |
|---|---|---|---|---|---|---|---|---|
| NPC1 | 18 | g.Chr18:21115443T>C | p.N1156S | Missense | rs28942105 | Heterozygous | 26.9 | 0.002 |
| NPC1 | 18 | g.Chr18:21119382C>T | p.V950M | Missense | rs120074135 | Heterozygous | 12.2 | 0 |
| NPC1 | 18 | g.Chr18:21119777G>A | p.N931N | Synonymous | rs1140458 | Heterozygous | 13.8 | 49.0 |
| NPC1 | 18 | g.Chr18:21120444T>C | p.I858V | Missense | rs1805082 | Heterozygous | 17.2 | 49.5 |
| NPC1 | 18 | g.Chr18:21124945C>G | p.M642I | Missense | n/a | Homozygous | 6.6 | 71.8 |
| NPC1 | 18 | g.Chr18:21140432T>C | p.H215R | Missense | rs1805081 | Heterozygous | 9.7 | 32.9 |
| NPC1 | 18 | g.Chr18:21148863A>G | p.Y129Y | Synonymous | rs12970899 | Heterozygous | 8.8 | 14.4 |
| NPC1 | 18 | g.Chr18:21166545G>C | 5′ UTR | rs8099071 | Heterozygous | 8.3 | n/a |
All coding variants in NPC1 identified in the patient along with one 5′-UTR variant. Coordinates are in GRCh37.
HGVS, Human Genome Variation Society; dbSNP, Database of Single-Nucleotide Polymorphisms; CADD score, Combined Annotation-Dependent Depletion Score; ExAc MAF, Exome Aggregation Consortium minor allele frequency out of 13,006 alleles; n/a, not available.
The patient's two pathogenic mutations are located in different domains of the NPC1 protein (Fig. 1). N1156S is located in the 11th of 13 transmembrane domains, in a region with homology to the patched family of proteins, and 43% sequence similarity to the sterol-sensing domain (Davies and Ioannou 2000; Gong et al. 2016). V950M is located in the third large luminal loop, referred to as luminal domain I. Recent structural studies have shown that both luminal domain I and luminal domain C share a resemblance to the periplasmic porter domains of AcrB, part of the bacterial multidrug efflux pump (Gong et al. 2016). Specifically, these domains are superimposable around a “core” region of tertiary structure conservation, and the mutated residue at V950M falls outside of this core region (Gong et al. 2016). Interestingly, luminal domain I, also referred as the cysteine-rich loop, has been noted before for being the location of a number of mutations associated with late-onset disease or “variant” biochemical phenotype (Millat et al. 2001; Ribeiro et al. 2001).
To prioritize variants found in the patient's genome outside of NPC1, we focused on mutations previously associated with similar neurological diseases, general longevity, and lipid metabolism. Although the etiology of NPC is distinct, the pathological and molecular similarities between NPC and other neurological diseases suggest that genetic modifiers to these more common diseases might also play a role in NPC pathogenesis (Malnar et al. 2014). Variants in APOE are strongly associated with both longevity and late-onset Alzheimer's disease (Broer et al. 2015), and there is a possible link between ApoE status and disease pathology in NPC: Individuals with NPC who were homozygous for ApoE4 have shown diffuse amyloid-β plaques that correlated with extent of tauopathy (Saito et al. 2002), and a study of 15 individuals with NPC demonstrated a correlation between ApoE2/ApoE4 status and the age of neurological onset (Fu et al. 2012). However, both the patient and patient's brother were homozygous for the ApoE3 genotype, which is not associated with risk for Alzheimer's disease, so it is unlikely that the APOE locus played a significant role in the patients’ disease progression.
Neurofibrillary tangles and aggregates of hyperphosphorylated tau protein are a pathological signature of NPC as well as other neurological diseases. Several studies in NPC knockout mice suggest a mechanistic role for tau in the pathogenesis of NPC, as a decrease in phosphorylated tau via inhibition of cyclin-dependent kinases improved NPC phenotypes (Zhang et al. 2004), whereas a complete knockout of the tau protein worsened the phenotypes (Pacheco et al. 2009). Variation in MAPT, the gene encoding the tau protein, has been associated with both a faster rate of decline in Alzheimer's disease (Kauwe et al. 2008) and risk for other neurodegenerative diseases associated with neurofibrillary tangle pathology (Sundar et al. 2006). The patient harbored no rare variants in MAPT and was negative for single-nucleotide polymorphisms (SNPs) associated with decreased levels of tau or phosphorylated tau in cerebrospinal fluid (rs3785883, rs8070723) (Allen et al. 2014). Further, the patient was negative for known pathogenic mutations in genes associated with other lipid storage diseases.
Most loci associated with longevity are common variants with small effect sizes. The patient was found to have 21 of 39 SNPs associated with an increase in life span (Lunetta et al. 2007; Newman et al. 2010; Yashin et al. 2010; Malovini et al. 2011; Walter et al. 2011; Lee et al. 2013). Using the patient's variants and a published model, the patient had a predicted life span of 85 yr (Walter et al. 2011). It is estimated that the genetic component of longevity in the Western World has been limited to survival beyond 85 yr, and it is therefore unlikely that many of these SNPs played a role in this NPC patient's late age of death (Deelen et al. 2014). Therefore, no definitive SNPs or overrepresentation of SNPs conferring either longevity or early mortality were identified in the patient's genome.
Finally, because the pathogenesis of NPC involves impaired cholesterol and lipid transport, we cross-referenced the patient's genome with the National Human Genome Research Institute–European Bioinformatics Institute (NHGRI-EBI) GWAS catalog to identify possible genetic modifiers in lipid metabolism pathways (Welter et al. 2014). Several coding and noncoding SNPs associated with variation in blood cholesterol and triglyceride levels were found, but with mild effect sizes (Supplemental Table 1).
In addition to seeking potential genetic modifiers of the patient's disease course, we also assessed to what extent the patient's genome could have informed his diagnosis. Several tools have been created that attempt to incorporate genome sequence into clinical diagnosis. We tested one such tool, Phenolyzer (Phenotype-Based Gene Analyzer; Yang et al. 2015), which integrates identified genomic variants with symptomology. Because NPC manifests differently according to the age of onset (Patterson et al. 2012), we created lists of common clinical terms associated with varying presentations (Supplemental Table 2). We then randomly downsampled these lists to five phenotypic terms, because the tool limits the number of allowed phenotypic inputs, and performed three permutations. The wANNOVAR-Phenolyzer pipeline (Wang et al. 2010; Chang and Wang 2012; Yang and Wang 2015) performed best with queries sampled from infantile and childhood presentations, listing NPC1 in the top five prioritized genes in five out of six queries and as the top gene in four out of six, most likely because of the specific phenotypes such as vertical supranuclear gaze palsy. Queries derived from the adult-onset presentation, with less specific phenotypic terms, did not perform as well. The requirement for the specific phenotypes, often associated with more infantile presentations, illustrates some of the challenges of trying to apply genetic sequence to less severe or stereotyped forms of Mendelian diseases, where both the effect of the mutations and clinical presentation may be less clear.
Phenotypic Analyses
Gross Neuropathologic Findings
The whole fresh brain weighed 1280 g (1300 g fixed) and the fixed posterior fossa contents weighed 150 g. Evaluation of the dura revealed bilateral, holohemispheric, granular, orange subdural neomembranes (<1-mm-thick) consistent with remote subdural hemorrhage. External examination of the brain showed mild frontal, temporal, and parietal cerebral cortical atrophy with unremarkable cerebellum and brainstem. Coronal sections of cerebral cortex were unremarkable other than mild hippocampal atrophy. Parasagittal sections of cerebellum revealed areas of mild folial atrophy with white matter thinning preferentially involving superior and, to a lesser extent, anterior cerebellar cortex. Axial sections of brainstem demonstrated moderate and mild pallor of the substantia nigra and locus coeruleus, respectively.
Microscopic Findings
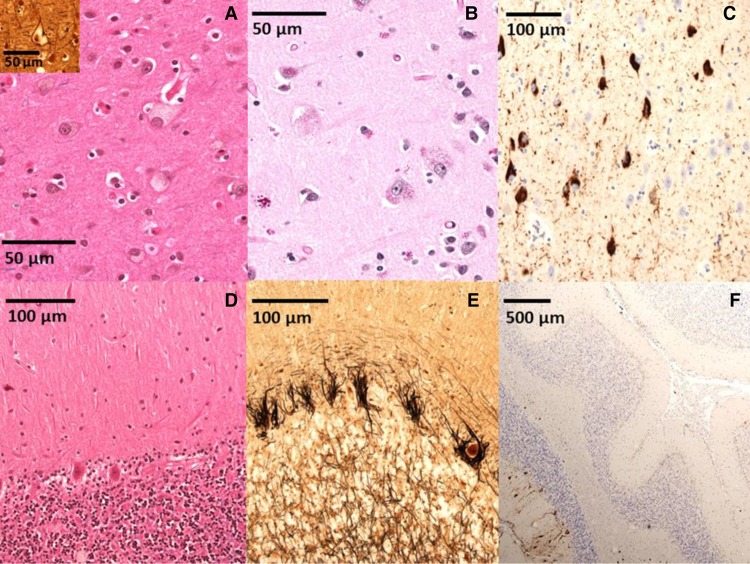
Histologic examination (Fig. 2) of the brain tissue revealed numerous foam cells, including many balloon neurons, with abundant pale cytoplasm containing lightly eosinophilic granular storage material (light staining on periodic acid–Schiff [PAS] stain). Foam cells were found in the majority of the sampled gray matter with the highest density in the substantia nigra (>50% of neuronal population), amygdala, striatum, and entorhinal and cingulate cortices. Variable numbers of foam cells were identified in all additional gray matter samples including cerebral isocortex, hippocampus, subiculum, nucleus basalis, paraventricular thalamus, cerebellar cortex (granular cell, Purkinje, and molecular layers), superior colliculi, oculomotor nerve nuclei, periaqueductal gray matter, locus coeruleus, dorsal pontine gray matter, medulla (including dorsal motor nucleus of the tenth cranial nerve), and rostral spinal cord. Numbers of gray matter lipofuscin or ceroid-laden neurons were increased for age and found in a similar distribution to foam cells (slightly darker staining with PAS). Microscopic analysis of the cerebellum revealed mild white matter atrophy with mild gliosis and extensive spans of Purkinje neuron loss (highlighted by calbindin immunohistochemistry) with Bergmann gliosis and numerous “empty baskets” (highlighted by Bielschowsky silver stain). Sections of cerebral cortex further demonstrated patchy, mild subcortical white matter myelin loss with variable mild gliosis and without significant cortical neuron loss (highlighted by glial fibrillary acid protein [GFAP] immunohistochemistry). These findings are consistent with previously reported structural changes associated with NPC (Ellison et al. 2013). Neurofibrillary tangle deposition, well characterized in NPC, was identified in the hippocampus, subiculum, entorhinal cortex, isocortex, and dentate fascia (indicative of Braak stage VI of VI). However, Alzheimer's disease pathology (i.e., amyloid-β plaques) was not identified (Thal stage 0, CERAD [Consortium to Establish a Registry for Alzheimer's Disease] 0). As such, the Braak score is not applicable. Other significant neuropathological findings include moderate loss of pigmented neurons in the substantia nigra and microscopically confirmed chronic subdural neomembranes. Finally, histologic examination was negative for evidence of Lewy body disease or cerebral microinfarcts.
Figure 2.
Pathological findings. (A) Hematoxylin and eosin (H&E) stain demonstrates scattered balloon neurons distended by lipid material in frontal cortex; Bielschowsky stain (inset) highlights frontal cortex balloon neuron. (B) Periodic acid–Schiff (PAS) stain highlights intracellular glycolipid in frontal cortex. (C) Tau-2 immunohistochemistry highlights neurofibrillary tangles in the subiculum. (D) H&E stain demonstrates Purkinje neuron loss with Bergmann gliosis. (E) Bielschowsky stain demonstrates numerous cerebellar “empty baskets.” (F) Calbindin immunohistochemistry highlights widespread cerebellar Purkinje neuron/arbor loss.
DISCUSSION
We present what is to our knowledge the first full genome sequence of a patient with NPC. Although NPC has been long established as a recessive Mendelian disease, there is wide variability in both onset of disease and age at death (Sévin et al. 2007). The phenotypic variability may be caused by differences in the recessive alleles of NPC or modifiers elsewhere in the genome. In particular, the patient described here exhibited a level of NPC function greater than the average NPC patient and concomitantly a late eventual demise at age 54. WGS of the patient provided an unbiased method to detect potential modifiers and address the question of whether the genetic background might play a role in modulating the phenotype or if instead most of the difference can be accounted for by the variants in NPC1 itself.
Both of the patient’s putative causal variants (V950M and N1156S) have been previously seen in compound heterozygote cases of NPC (Carstea et al. 1997; Fernandez Valero et al. 2005; Millat et al. 2005), although not reported together. That the V950M variant had been previously observed in multiple individuals with the adult-onset phenotype is consistent with its being a hypomorphic allele, whose partial function contributes to both the variant NPC biochemical phenotype and the somewhat later onset and late age at death. Previously, when V950M has been observed in cases of adult-onset NPC, it was either a homozygous mutation or compound heterozygous with I1061T, an allele associated with juvenile-onset NPC. In contrast, the N1156S variant has been observed in severe infantile disease. The observation of the V950M variant paired with N1156S suggests that a single copy of a hypomorphic NPC1 gene provides sufficient function for delayed onset of disease.
One obstacle in interpreting WGS data is identifying genes of interest. In this analysis, we took a simple approach of focusing on genes involved in pathways affected in NPC or linked to similar neurological phenotypes. Looking at the pathways affected in NPC, the patient harbored variants associated with both increased and decreased lipid levels. Although four out of five variants are associated with increased levels, the patient is homozygous for a missense variant in APOB with the strongest effect, a decrease in trigylcerides of 5.99 mg/dL. However, it is hard to say whether or not these could have impacted the patient's clinical outcome. The patient was negative for variants strongly associated with either neurodegenerative disease or longevity. In particular, for APOE—perhaps the best candidate for a gene with a common, high-impact variant—the patient carried the most common ApoE3 variant, which is not associated with increased life span. All together, we found no compelling evidence of strong genetic modifiers outside of the NPC1 gene. This is consistent with the relative mildness of the V950M variant being the main contributor to the patient's phenotype.
The patient's clinical history speaks to the diagnostic challenges that present even with well-characterized genetic diseases. As genomic techniques have become increasingly more common and cost-effective, this type of analysis is coming ever closer to clinical use. An important question with this approach is whether the information gained through WGS could have made an impact on the patient's care had it been performed while he was still alive.
Novel variants in disease genes can be difficult to interpret. In the patient described here, both variants had been previously observed and as such were already presumed to be causal. We may ask, however, how they would have been interpreted were they not seen before in NPC patients. Metrics like conservation, allele frequency, or type of amino acid change can be used to predict the effect of a novel mutation. Newer supervised machine learning approaches have been used to integrate many of these simultaneously. Applied to these variants, commonly used bioinformatics tools like PolyPhen (Polymorphism Phenotyping; Adzhubei et al. 2010) and CADD (Combined Annotation-Dependent Depletion; Kircher et al. 2014) highlight the N1156S mutation as likely damaging: A PolyPhen score of 1 (“possibly damaging”) and a CADD score of 26.9 provide supporting but not overwhelming evidence for the deleteriousness of this variant. However, for the V950M mutation, conservation does not provide strong evidence for disruption of function as a PolyPhen score of 0.007 and a CADD score of 12.2 are low enough such that this variant would likely fail to pass common filtering steps for identifying deleterious missense variants if it had not been previously identified as a disease-causing mutation. In particular, this highlights the challenge in predicting pathogenicity of variants responsible for milder forms of inherited disease, where the impact of the causal variants on gene function may be more subtle. Although it is not possible to draw definitive conclusions from an N = 1 case study, given the role rare, high-impact, variants play in human disease and disease modifiers (Chen et al. 2016), case-only sequencing of patients with atypical phenotypes has the potential to discover novel variants in disease-associated pathways that may play a role in disease pathology. This approach has limited power, especially for detecting low-effect size variants, but is realistic in clinical scenarios when family pedigrees and tissue samples are not available or consent cannot be obtained.
Finally, sequencing and analysis of this patient was done in conjunction with a course for first-year students in a Medical Scientist Training Program. With the ever-growing promise of precision medicine and increasing use of genomic technologies in the clinic, WGS and interpretation allows students to explore both its utility and challenges. We hope this class may serve as a model for the possibilities created by integrating this type of patient analysis into medical education (Kumar et al. 2014). In this course, students took responsibility for every aspect of the study, including the clinical history, gross anatomical dissection, and microscopic analysis of the brain, analysis of sequencing data, and preparation of the manuscript. In addition to furthering our understanding of NPC etiology, we believe that this course and ones like it have the potential for great didactic value for the development of future physicians and scientists.
METHODS
Gross Neuropathological Analysis
Formalin-fixed brain tissue was sectioned into 4-mm-thick slices (brain, coronal; cerebellum, parasagittal; brainstem, axial) for gross examination. Tissue sections were submitted according to 2012 NIA-AA (National Institute on Aging-Alzheimer's Association) criteria with submission of additional tissue sections to demonstrate NPC pathology.
Microscopic Analysis
Microscopic slides were prepared from each of the tissue sections. All sections were stained with hematoxylin and eosin/luxol fast blue (H&E/LFB) with targeted performance of the following immunohistochemical and histochemical stains: amyloid-β (AE10), tau-2, α-synuclein, glial fibrillary acidic protein (GFAP), calbindin, myelin basic protein (MBP), NeuN, CD68, Bielschowsky modified silver, and Congo Red.
Sample Collection
Fresh cerebellar tissue was frozen at the time of autopsy while the remainder of the brain was fixed in formalin for histological sampling.
Genomic Sequencing and Analysis
DNA was extracted from 70 µg frozen cerebellum using QIAGEN DNeasy Blood & Tissue Kit. Shotgun sequencing libraries were prepared using the KAPA library preparation kit (Kapa Biosystems) following manufacturer's instructions. The library sample was sequenced to 50× coverage on an Illumina NextSeq with 150-bp paired-end reads and mapped to GRCh37/hg19 (see Supplemental Table 3 for coverage). SNVs and small indels were called using Genome Analysis Toolkit (GATK) following best practices (McKenna et al. 2010; DePristo et al. 2011) and variants were annotated with SeattleSeq (Ng et al. 2009). We identified 3.3 million SNVs with a transition to transversion ratio of 2.08. Of these, 20,271 were protein-coding mutations, and 1318 of the protein-coding mutations were rare variants (observed in <1% of samples from the Exome Variant Server). We also detected 561,488 small indels with 526 falling in coding regions.
Sanger Sequencing of Patient's Brother
Genomic DNA was isolated from the patient's brother. The V950M and N1156S mutations were amplified and sequenced with the following primers:
V950MF: 5′-GTAAAGGAGAAGGTACCTGAAGCATTGC-3′
V950MR: 5′-ACTAAAGACTTCCTCCCTGTGGAGCAG-3′
N1156SF: 5′-CAAGCGCCAGACTTGGTATCTTACTCC-3′
N1156SR: 5′-TGTGAGCTCTGGTCTGCAGTCATCATG-3′
The ApoE genotype of the patient's brother was amplified and sequenced with the following primers:
ApoE1F: 5′-GCTGATGGACGAGACCATGAAGGAG-3′
ApoE1R: 5′-GCAGGCCATGCTCGGCCAGAGCACC-3′
ApoE2F: 5′-TGCAGGTCATCGGCATCGCGGAGGAG-3′
ApoE2R: 5′-CACCTGCTCCTTCACCTCGTCCAGG-3′
ADDITIONAL INFORMATION
Data Deposition and Access
Patient consent was not explicitly obtained for depositing whole-genome sequencing data in an external archive. Requests for data access can be brought to our institutional review board (IRB). Please direct inquiries to Max Dougherty (mldough@uw.edu), John Lazar (jlazar@uw.edu), and Jason Klein (jcklein@uw.edu). The two NPC1 causal variants (V950M and N1156S) have been deposited to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000297807 and SCV000297806, respectively.
Ethics Statement
The patient was a participant in the University of Washington Alzheimer's Disease Research Center (ADRC) Clinical Core research program. Participation requires prior written consent by the patient with either written or witnessed verbal confirmation of consent by legal next of kin upon the patient's death. Brain autopsy consent specifies that brain tissue removed at autopsy may be retained and used for educational, scientific, and research purposes. A separate brain bank tissue banking consent is also obtained. The University of Washington IRB determined that this research and related tissue banking involving preconsented deceased individuals is not considered human subjects research according to 45 CFR 46.102(f) and, therefore, exempt from IRB review.
Acknowledgments
We deeply appreciate the participation of individuals in the ADRC Clinical Core research program. We also thank Riza Daza for her assistance with DNA sequencing.
Author Contributions
M.D., J.C.K., J.L., J.S., C.D.K., T.B., M.S.H., and D.A.M. conceived and designed the experiments. M.D., J.C.K., J.L., K.D., T.G., E.G., N.H., D.L., M.M., M.N., G.O., S.S., C.D.K., and D.A.M. performed the experiments. M.D., J.C.K., J.L., K.D., T.G., E.G., N.H., D.L., M.M., M.N., G.O., and S.S. analyzed the data. M.D., J.C.K., J.L., K.D., T.G., E.G., N.H., D.L., M.M., M.N., G.O., S.S., J.S., C.D.K., T.B., M.S.H., and D.A.M. wrote the paper.
Funding
This work was supported by the University of Washington School of Medicine and Fred Hutchinson Cancer Research Center Medical Scientist Training Program (National Institutes of Health [NIH] grant T32GM0007266 and institutional educational funds) and the Lambright Fund (to T.B.).
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adebali O, Reznik AO, Ory DS, Zhulin IB. 2016. Establishing the precise evolutionary history of a gene improves prediction of disease-causing missense mutations. Genet Med 10.1038/gim.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Kachadoorian M, Quicksall Z, Zou F, Chai H, Younkin C, Crook JE, Pankratz V, Carrasquillo MM, Krishnan S, et al. 2014. Association of MAPT haplotypes with Alzheimer's disease risk and MAPT brain gene expression levels. Alzheimers Res Ther 6: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Knoblich R, Bauer C, Finckh U, Hufen A, Kropp J, Braun S, Kustermann-Kuhn B, Schmidt DR, Harzer K, et al. 2002. NPC1: complete genomic sequence, mutation analysis, and characterization of haplotypes. Hum Mutat 19: 30–38. [DOI] [PubMed] [Google Scholar]
- Broer L, Buchman AS, Deelen J, Evans DS, Faul JD, Lunetta KL, Sebastiani P, Smith JA, Smith AV, Tanaka T, et al. 2015. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J Gerontol A Biol Sci Med Sci 70: 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, et al. 1997. Niemann–Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277: 228–231. [DOI] [PubMed] [Google Scholar]
- Chang X, Wang K. 2012. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet 49: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Shi L, Hakenberg J, Naughton B, Sklar P, Zhang J, Zhou H, Tian L, Prakash O, Lemire M, et al. 2016. Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases. Nat Biotechnol 34: 531–538. [DOI] [PubMed] [Google Scholar]
- Corvol H, Blackman SM, Boëlle P-Y, Gallins PJ, Pace RG, Stonebraker JR, Accurso FJ, Clement A, Collaco JM, Dang H, et al. 2015. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun 6: 8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA. 2000. Topological analysis of Niemann–Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem 275: 24367–24374. [DOI] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh H-W, Broer L, Ayers KL, Tan Q, Kamatani Y, Bennet AM, Tamm R, Trompet S, et al. 2014. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age. Hum Mol Genet 23: 4420–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D, Love S, Chimelli L, Harding B, Lowe J, Vinters H, Brandner S, Yong WH. 2013. Neuropathology: a reference text of CNS pathology, 3rd ed. Mosby-Elsevier, St. Louis. [Google Scholar]
- Fernandez Valero EM, Ballart A, Iturriaga C, Lluch M, Macias J, Vanier MT, Pineda M, Coll MJ. 2005. Identification of 25 new mutations in 40 unrelated Spanish Niemann–Pick type C patients: genotype-phenotype correlations. Clin Genet 68: 245–254. [DOI] [PubMed] [Google Scholar]
- Fu R, Yanjanin NM, Elrick MJ, Ware C, Lieberman AP, Porter FD. 2012. Apolipoprotein E genotype and neurological disease onset in Niemann–Pick disease, type C1. Am J Med Genet A 158A: 2775–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver WS, Jelinek D, Meaney FJ, Flynn J, Pettit KM, Shepherd G, Heidenreich RA, Vockley CMW, Castro G, Francis GA. 2010. The National Niemann–Pick Type C1 Disease Database: correlation of lipid profiles, mutations, and biochemical phenotypes. J Lipid Res 51: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Qian H, Zhou X, Wu J, Wan T, Cao P, Huang W, Zhao X, Wang X, Wang P, et al. 2016. Structural insights into the Niemann–Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell 165: 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Sidhu R, Mydock-McGrane L, Hsu F-F, Covey DF, Scherrer DE, Earley B, Gale SE, Farhat NY, Porter FD, et al. 2016. Development of a bile acid–based newborn screen for Niemann–Pick disease type C. Sci Transl Med 8: 337ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauwe JSK, Cruchaga C, Mayo K, Fenoglio C, Bertelsen S, Nowotny P, Galimberti D, Scarpini E, Morris JC, Fagan AM, et al. 2008. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-β deposition. Proc Natl Acad Sci 105: 8050–8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dougherty M, Findlay GM, Geisheker M, Klein J, Lazar J, Machkovech H, Resnick J, Resnick R, Salter AI, et al. 2014. Genome sequencing of idiopathic pulmonary fibrosis in conjunction with a medical school human anatomy course. PLoS One 9: e106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Honig LS, Feitosa M, Kammerer CM, Kang MS, Schupf N, Lin SJ, Sanders JL, Bae H, et al. 2013. Genome wide association and linkage analyses identified three loci-4q25, 17q23.2, and 10q11.21-associated with variation in leukocyte telomere length: the Long Life Family Study. Front Genet 4: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunetta KL, D'Agostino RB, Karasik D, Benjamin EJ, Guo C-Y, Govindaraju R, Kiel DP, Kelly-Hayes M, Massaro JM, Pencina MJ, et al. 2007. Genetic correlates of longevity and selected age-related phenotypes: a genome-wide association study in the Framingham Study. BMC Med Genet 8(Suppl. 1): S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnar M, Hecimovic S, Mattsson N, Zetterberg H. 2014. Bidirectional links between Alzheimer's disease and Niemann–Pick type C disease. Neurobiol Dis 72: 37–47. [DOI] [PubMed] [Google Scholar]
- Malovini A, Illario M, Iaccarino G, Villa F, Ferrario A, Roncarati R, Anselmi CV, Novelli V, Cipolletta E, Leggiero E, et al. 2011. Association study on long-living individuals from Southern Italy identifies rs10491334 in the CAMKIV gene that regulates survival proteins. Rejuvenation Res 14: 283–291. [DOI] [PubMed] [Google Scholar]
- Mazzacuva F, Mills P, Mills K, Camuzeaux S, Gissen P, Nicoli ER, Wassif C, Vruchte D, Porter FD, Maekawa M, et al. 2016. Identification of novel bile acids as biomarkers for the early diagnosis of Niemann–Pick C disease. FEBS Lett 590: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Marçais C, Rafi MA, Yamamoto T, Morris JA, Pentchev PG, Ohno K, Wenger DA, Vanier MT. 1999. Niemann–Pick C1 disease: the I1061T substitution is a frequent mutant allele in patients of Western European descent and correlates with a classic juvenile phenotype. Am J Hum Genet 65: 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Marçais C, Tomasetto C, Chikh K, Fensom AH, Harzer K, Wenger DA, Ohno K, Vanier MT. 2001. Niemann–Pick C1 disease: correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am J Hum Genet 68: 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millat G, Baïlo N, Molinero S, Rodriguez C, Chikh K, Vanier MT. 2005. Niemann–Pick C disease: use of denaturing high performance liquid chromatography for the detection of NPC1 and NPC2 genetic variations and impact on management of patients and families. Mol Genet Metab 86: 220–232. [DOI] [PubMed] [Google Scholar]
- Newman AB, Walter S, Lunetta KL, Garcia ME, Slagboom PE, Christensen K, Arnold AM, Aspelund T, Aulchenko YS, Benjamin EJ, et al. 2010. A meta-analysis of four genome-wide association studies of survival to age 90 years or older: the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. J Gerontol A Biol Sci Med Sci 65: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. 2009. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco CD, Elrick MJ, Lieberman AP. 2009. Tau deletion exacerbates the phenotype of Niemann–Pick type C mice and implicates autophagy in pathogenesis. Hum Mol Genet 18: 956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson MC, Hendriksz CJ, Walterfang M, Sedel F, Vanier MT, Wijburg F. 2012. Recommendations for the diagnosis and management of Niemann–Pick disease type C: an update. Mol Genet Metab 106: 330–344. [DOI] [PubMed] [Google Scholar]
- Pentchev PG, Comly ME, Kruth HS, Vanier MT, Wenger DA, Patel S, Brady RO. 1985. A defect in cholesterol esterification in Niemann–Pick disease (type C) patients. Proc Natl Acad Sci 82: 8247–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Scherrer DE, Lanier MH, Langmade SJ, Molugu V, Gale SE, Olzeski D, Sidhu R, Dietzen DJ, Fu R, et al. 2010. Cholesterol oxidation products are sensitive and specific blood-based biomarkers for Niemann–Pick C1 disease. Sci Transl Med 2: 56ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro I, Marcão A, Amaral O, Miranda MS, Vanier MT, Millat G. 2001. Niemann–Pick type C disease: NPC1 mutations associated with severe and mild cellular cholesterol trafficking alterations. Hum Genet 109: 24–32. [DOI] [PubMed] [Google Scholar]
- Saito Y, Suzuki K, Nanba E, Yamamoto T, Ohno K, Murayama S. 2002. Niemann-Pick type C disease: accelerated neurofibrillary tangle formation and amyloid β deposition associated with apolipoprotein E ε4 homozygosity. Ann Neurol 52: 351–355. [DOI] [PubMed] [Google Scholar]
- Sévin M, Lesca G, Baumann N, Millat G, Lyon-Caen O, Vanier MT, Sedel F. 2007. The adult form of Niemann–Pick disease type C. Brain 130: 120–133. [DOI] [PubMed] [Google Scholar]
- Sundar PD, Yu CE, Sieh W, Steinbart E, Garruto RM, Oyanagi K, Craig UK, Bird TD, Wijsman EM, Galasko DR, et al. 2006. Two sites in the MAPT region confer genetic risk for Guam ALS/PDC and dementia. Hum Mol Genet 16: 295–306. [DOI] [PubMed] [Google Scholar]
- The Cystic Fibrosis Genotype-Phenotype Consortium. 1993. Correlation between genotype and phenotype in patients with cystic fibrosis. The Cystic Fibrosis Genotype-Phenotype Consortium. N Engl J Med 329: 1308–1313. [DOI] [PubMed] [Google Scholar]
- Vanier MT, Millat G. 2003. Niemann–Pick disease type C. Clin Genet 64: 269–281. [DOI] [PubMed] [Google Scholar]
- Vanier MT, Gissen P, Bauer P, Coll MJ, Burlina A, Hendriksz CJ, Latour P, Goizet C, Welford RWD, Marquardt T, et al. 2016. Diagnostic tests for Niemann–Pick disease type C (NP-C): a critical review. Mol Genet Metab 118: 244–254. [DOI] [PubMed] [Google Scholar]
- Walter S, Atzmon G, Demerath EW, Garcia ME, Kaplan RC, Kumari M, Lunetta KL, Milaneschi Y, Tanaka T, Tranah GJ, et al. 2011. A genome-wide association study of aging. Neurobiol Aging 32: 2109, e15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassif CA, Cross JL, Iben J, Sanchez-Pulido L, Cougnoux A, Platt FM, Ory DS, Ponting CP, Bailey-Wilson JE, Biesecker LG, et al. 2016. High incidence of unrecognized visceral/neurological late-onset Niemann–Pick disease, type C1, predicted by analysis of massively parallel sequencing data sets. Genet Med 18: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42: D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang K. 2015. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10: 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-C, Su Y-N, Chiou P-C, Fietz MJ, Yu C-L, Hwu W-L, Lee M-J. 2005. Six novel NPC1 mutations in Chinese patients with Niemann–Pick disease type C. J Neurol Neurosurg Psychiatr 76: 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Robinson PN, Wang K. 2015. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods 12: 841–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashin AI, Wu D, Arbeev KG, Ukraintseva SV. 2010. Joint influence of small-effect genetic variants on human longevity. Aging (Albany NY) 2: 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Li J, Chakrabarty P, Bu BT, Vincent I. 2004. Cyclin-dependent kinase inhibitors attenuate protein hyperphosphorylation, cytoskeletal lesion formation, and motor defects in Niemann–Pick type C mice. Am J Pathol 165: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.