Abstract
Identification of the tissue of origin in cancer of unknown primary (CUP) poses a diagnostic challenge and is critical for directing site-specific therapy. Currently, clinical decision-making in patients with CUP primarily relies on histopathology and clinical features. Comprehensive molecular profiling has the potential to contribute to diagnostic categorization and, most importantly, guide CUP therapy through identification of actionable lesions. We here report the case of an advanced-stage malignancy initially mimicking poorly differentiated soft-tissue sarcoma that did not respond to multiagent chemotherapy. Molecular profiling within a clinical whole-exome and transcriptome sequencing program revealed a heterozygous, highly amplified KRAS G12S mutation, compound-heterozygous TP53 mutation/deletion, high mutational load, and focal high-level amplification of Chromosomes 9p (including PDL1 [CD274] and JAK2) and 10p (including GATA3). Integrated analysis of molecular data and histopathology provided a rationale for immune checkpoint inhibitor (ICI) therapy with pembrolizumab, which resulted in rapid clinical improvement and a lasting partial remission. Histopathological analyses ruled out sarcoma and established the diagnosis of a poorly differentiated adenocarcinoma. Although neither histopathology nor molecular data were able to pinpoint the tissue of origin, our analyses established several differential diagnoses including triple-negative breast cancer (TNBC). We analyzed 157 TNBC samples from The Cancer Genome Atlas, revealing PDL1 copy number gains coinciding with excessive PDL1 mRNA expression in 24% of cases. Collectively, these results illustrate the impact of multidimensional tumor profiling in cases with nondescript histology and immunophenotype, show the predictive potential of PDL1 amplification for immune checkpoint inhibitors (ICIs), and suggest a targeted therapeutic strategy in Chromosome 9p24.1/PDL1-amplified cancers.
Keywords: multifocal breast carcinoma, neoplasm of the gastrointestinal tract
INTRODUCTION
“Omics” technologies, most prominently next-generation sequencing (NGS), have entered clinical medicine in the last decade and are expected to affect the standard of care in oncology. However, the identification of patients who truly benefit from genomics-driven approaches to clinical management and the appropriate timing of therapy are still challenging (Roychowdhury and Chinnaiyan 2014). Moreover, it is currently unclear how to effectively integrate these new tools with conventional technologies such as standard histopathology, and the algorithms for integrated diagnostics and molecularly guided treatment remain to be exactly defined.
Comprehensive cancer profiling can particularly assist diagnosis and treatment planning in patients with malignancies of unresolved histology and in metastatic disease, in which the tissue of origin of the primary tumor cannot be discerned. Cancers of unknown primary (CUP) constitute 3%–5% of all cancer diagnoses and rank fourth in the causes of cancer-related deaths worldwide (Varadhachary and Raber 2014). NGS-based molecular characterization holds the promise to gain insight into the pathobiology of these difficult-to-classify tumors. Compared to targeted sequencing of selected cancer genes, whole-exome sequencing (WES) and RNA sequencing (RNA-seq) allow for superior detection of coding sequence variants, amplifications, deletions, and structural rearrangements of chromosomes as well as their consequences on mRNA transcription. Integration of these data with mutational and transcriptional cancer profiles deposited in public genome databases, such as those generated by The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov) and the International Cancer Genome Consortium (https://icgc.org), can aid in identifying the tissue of origin of cancers of otherwise indefinable histology and nominate candidate therapeutic targets.
Here we describe a patient with refractory, metastatic anaplastic CUP, where integrated analysis of histopathology, immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), WES, and RNA-seq revised the diagnosis and facilitated genotype-informed treatment with an immune checkpoint inhibitor (ICI) that resulted in a durable, very good partial remission.
RESULTS
A 44-yr-old Caucasian female presented to our center with metastatic CUP. Two years before presentation, she had been diagnosed with multiple metastatic lesions in the mesentery of the small intestines and the duodenojejunal flexure. At that time, the tumor lesions had been resected completely. Histopathology analyses were inconclusive, showing a high-grade neoplasm with pleomorphic tumor cells that stained positive for vimentin. MIB-1 was positive in ∼30% of the tumor cells. Following central pathology review, a preliminary diagnosis of CUP with sarcomatoid features was considered with the most likely differential diagnosis of sarcoma, not otherwise specified. The patient was treated with four cycles of doxorubicin and ifosfamide according to the EORTC (European Organisation for Research and Treatment of Cancer) 62012 protocol (Judson et al. 2014) at an outside institution, and subsequent imaging studies revealed no evidence of disease.
Sixteen months after initial diagnosis, the patient developed progressive pain in the right shoulder, and magnetic resonance imaging (MRI) showed a mass of 4 × 2 × 2 cm in the right deltoid muscle and a second metastatic lesion of 1.5 × 2 × 2 cm in the right axilla. Incisional biopsy was performed, and subsequent histopathology assessment with repeat central review suggested metastasis of the previously diagnosed CUP rather than a secondary malignancy. After complete surgical excision of the rapidly progressing lesions, adjuvant radiotherapy was administered to the right shoulder and axilla.
Twenty-one months after initial diagnosis, follow-up MRI studies revealed five new tumor formations (left paravertebral region, right M. infraspinatus, right axilla, right M. latissimus dorsi, left chest wall). Again, surgical resection of multiple lesions was performed. However, the tumor progressed rapidly with new lesions of the right shoulder and thigh; treatment with trabectedin was initiated but had to be stopped because of progressive disease and Grade IV thrombocytopenia.
In an attempt to refine the diagnosis and inform potential therapeutic strategies, the patient was enrolled in NCT (Nationales Centrum für Tumorerkrankungen) MASTER (Molecularly Aided Stratification for Tumor Eradication Research), a cross-entity molecular stratification program for younger adults with advanced-stage cancer and patients with rare tumors (Kordes et al. 2016), and a cryopreserved biopsy of a metastatic shoulder lesion with a tumor cell content of >90% was analyzed by WES and RNA-seq. Peripheral blood mononuclear cells served as a germline control.
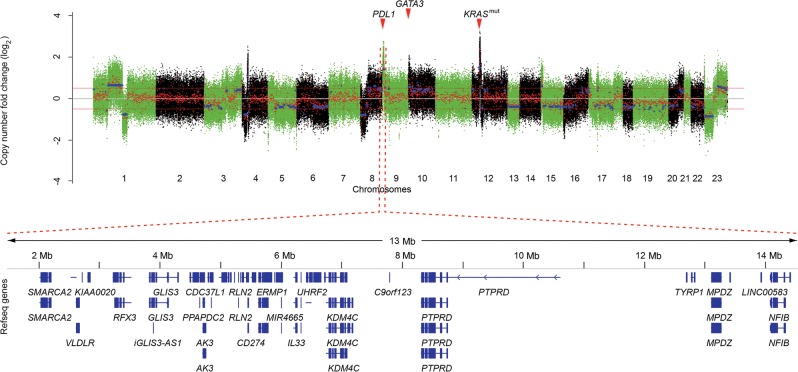
WES analysis revealed a high mutational load of 341 nonsynonymous single-nucleotide variants, 39 insertion/deletions, and multiple chromosomal gains and losses (Fig. 1). Known driver mutations were detected in TP53 (p.E135fs) and KRAS (p.G12S), the latter also being highly expressed on the RNA level because of a focal amplification on Chromosome 12p (Table 1). Mutations of unknown significance occurred in other cancer-related genes (e.g., PIK3CD, CDKN2A, NCOA1, FAT2, EGFR, MSH3, ARID1A, MDC1, SETD1A, SETD3, and TET1) (Supplemental Table S1). A focal high-level amplification on Chromosome 9p encompassed JAK2 and PDL1 (also known as CD274 or B7-H1) together with other genes implicated in carcinogenesis (Fig. 1; Kietz et al. 2009; Moon et al. 2011; Zhang et al. 2012; Hoffman et al. 2014; Jovanovic et al. 2014; Kim et al. 2015).
Figure 1.
PDL1 and GATA3 amplification in a patient with cancer of unknown primary. (Upper panel) Copy number plot showing chromosomal coordinates computed as a set of regions based on whole-exome sequencing data (x-axis) and the log2 ratio of copy number changes (y-axis). Color codes of alternating green and black regions indicate segmentation between chromosomes. (Lower panel) Amplified region on chromosome 9p.23–24.1.
Table 1.
Somatic variants of potential interest
| Chr | GRCh37 position | Ref | Var | Type | Gene | HGVS DNA reference | Predicted effect | HGVS protein reference | Genotype | dbSNP ID |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 27099877 | CA | C | FS-DEL | ARID1A | g.27099877CA>C | p.N1253fs | p.Asn1253fs | het | |
| 1 | 9770641 | G | A | SNV | PIK3CD | g.9770641G>A | p.S43N | p.Ser43Asn | het | |
| 2 | 24991253 | C | A | SNV | NCOA1 | g.24991253C>A | p.T1440N | p.Thr1440Asn | het | |
| 5 | 80160762 | G | A | Splicing | MSH3 | g.80160762G>A | het | |||
| 5 | 150922306 | C | G | SNV | FAT2 | g.150922306C>G | p.R2794S | p.Arg2794Ser | het | |
| 6 | 30671308 | C | A | SNV | MDC1 | g.30671308C>A | p.V1857L | p.Val1857Leu | het | rs149786493 |
| 7 | 55225447 | G | T | Splicing | EGFR | g.55225447G>T | het | |||
| 9 | 1461199–14398642 | AMP | PPAPDC2;PTPRD | |||||||
| 9 | 21971208 | C | A | Splicing | CDKN2A | g.21971208C>A | het | |||
| 10 | 70332663 | C | G | SNV | TET1 | g.70332663C>G | p.Q190E | p.Gln190Glu | het | |
| 10 | 93076–14013840 | AMP | TUBB8; PRPF18 | |||||||
| 12 | 19242142–30289130 | AMP | C12orf39; RASSF8 | |||||||
| 12 | 25398285 | C | T | SNV | KRAS | g.25398285C>T | p.G12S | p.Gly12Ser | het | rs1219135 |
| 14 | 99927578 | A | G | SNV | SETD3 | g.99927578A>G | p.V99A | p.Val99Ala | het | |
| 16 | 30977120 | G | C | SNV | SETD1A | g.30977120G>C | p.G640R | p.Gly640Arg | het | |
| 17 | 7577057 | T | TC | FS-INS | TP53 | g.7577057T>TC | p.E135fs | p.Glu135fs | het |
The complete list of somatic mutations can be found in Supplemental Table S1.
HGVS, Human Genome Variation Society; dbSNP, Database for Short Genomic Variations; AMP, amplification; FS-DEL, frameshift-deletion; FS-INS, frameshift-insertion; het, heterozygous; SNV, single-nucleotide variant.
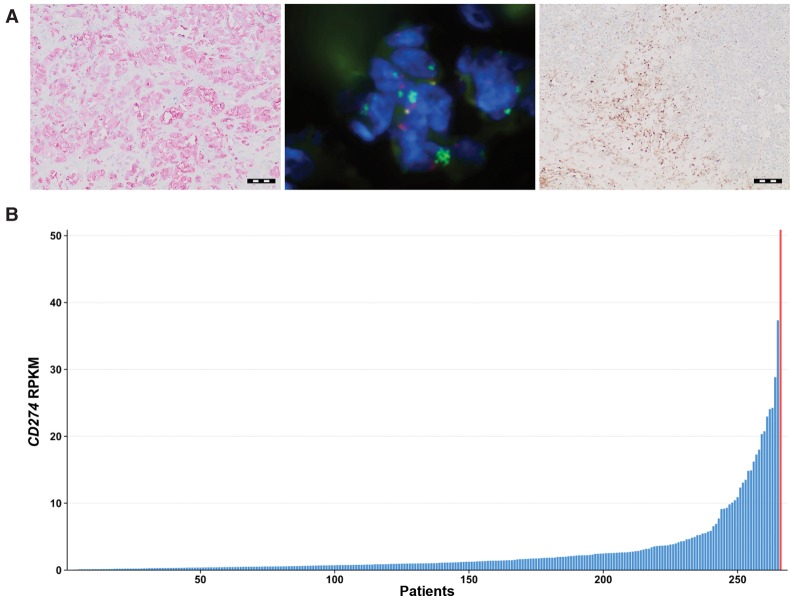
In line with these findings, IHC and FISH analyses showed high-level amplification and subsequent overexpression of PDL1 in the tumor cells. Histologic assessment showed substantial peritumoral infiltration of lymphocytes (Fig. 2A). Correspondingly, PDL1 mRNA levels, as determined by RNA-seq, were highest in this tumor specimen compared to all tumor samples included in NCT MASTER (n = 266; Fig. 2B). A query of publicly available databases for matching cancer mutational profiles comprising TP53 inactivation and amplification of Chromosomes 9p (PDL1, JAK2), 10p (GATA3), and 12p (mutant KRAS) as well as review of the literature suggested lung adenocarcinoma, triple-negative breast cancer (TNBC), or gastric adenocarcinoma as the most probable diagnoses (Cimino-Mathews et al. 2013; Gao et al. 2013; Marquard et al. 2015). Principal component analysis using mRNA expression profiles obtained by RNA-seq data from both TCGA and the NCT MASTER cohorts was inconclusive (data not shown). Based on these findings, we decided to challenge the initial diagnosis of a metastatic sarcoma and reevaluated tissue parameters of the current metastatic lesion. Comprehensive immunohistochemical profiling of 25 different antigens revealed strong expression of cytokeratins (as measured by AE1/3) and CK7. These data are in line with decisive histological features (e.g., focal formation of small tubular structures) and focal positivity with periodic acid–Schiff (PAS) staining leading to the diagnosis of an adenocarcinoma. We also noticed focal positivity of vimentin, which is consistent with poor differentiation and epithelial–mesenchymal transition from a histological and biological point of view, respectively. Tumor cells showed strong GATA3 expression (Supplemental Fig. 1) consistent with the genetic amplification and were negative for estrogen receptor (ER), progesterone receptor (PR), and ERBB2 (also known as HER2). In the context of poor differentiation, negativity for TTF1 and napsin does not rule out the differential diagnosis of lung adenocarcinoma but is not supportive either. Although neither IHC profiling nor molecular analyses allowed us to unequivocally pinpoint the tissue of origin, reevaluation of all parameters suggested metastatic TNBC among the top differential diagnoses, although a definitive categorization based on conventional diagnostic critera of this CUP case still remained tentative, given the clinical presentation without a primary tumor detectable in either breast, lung, or stomach.
Figure 2.
PDL1 protein expression, PDL1 amplification, peritumoral lymphocyte infiltration, and PDL1 mRNA expression in a patient with anaplastic cancer of unknown primary. (A) (Left) Photomicrograph showing PDL1 protein expression in a metastasis (scale bar, 100 µm); (middle) representative fluorescence in situ hybridization signal pattern showing amplification of the PDL1 locus (green signals) relative to the centromere of Chromosome 9 (red signals); (right) CD4 lymphocyte staining (scale bar, 200 µm). (B) Ranking of 266 patient samples analyzed in the NCT MASTER study according to PDL1 (CD274) mRNA levels, as determined by RNA-seq. The red bar indicates the described index patient. RPKM, reads per kilobase of exon model per million mapped reads.
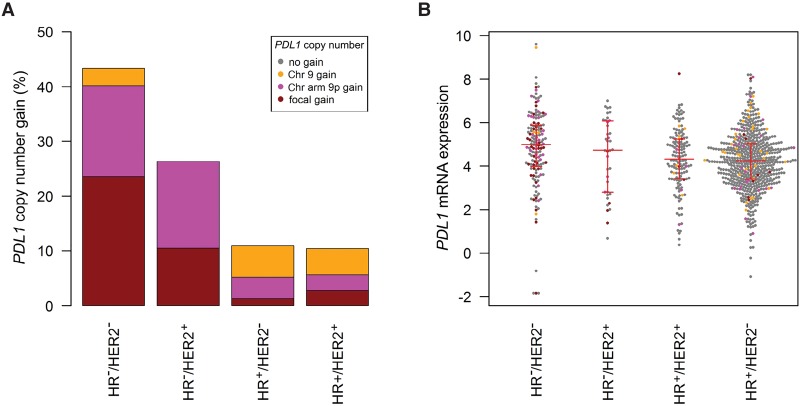
In similarity to our case, immune cell infiltrates and Chromosome 9p amplification have recently been described in a small cohort of TNBC (Barrett et al. 2015). We thus aimed to systematically address the question whether genomic PDL1 gains are recurrent in TNBC and associated with elevated PDL1 expression as observed in our case. To this end, we analyzed PDL1 copy number variation in a TCGA cohort of 937 breast carcinomas (Fig. 3A). Copy number gains were more frequent in TNBC (43.3%) compared to hormone receptor (HR)+/HER2−, HR+/HER2+, and HR−/HER2+ breast cancer (10.9%, 10.3%, and 26.3%; p = 0.00045). Focal amplifications occurred in 23.6% of TNBC samples compared to 1.3%, 2.8%, and 10.5% in HR+/HER2−, HR+/HER2+, and HR−/HER2+ breast cancer specimens. PDL1 mRNA expression analysis was performed in 934 tumors (Fig. 3B) and revealed significantly higher PDL1 levels in tumors with PDL1 copy number gains compared to tumors that were diploid for the PDL1 locus (fold change, 1.25; p = 0.019). Furthermore, triple-negative tumors had significantly increased PDL1 expression compared to HR+/HER2− and HR+/HER2+ tumors (fold change, 1.49; p = 0.00030 and fold change, 1.42; p = 0.0068).
Figure 3.
PDL1 DNA copy number and PDL1 mRNA expression in the molecular subtypes of breast cancer (TCGA data sets). (A) PDL1 copy number gains are more frequent in triple-negative breast cancer (TNBC) (43.3%) compared to HR−/HER2+, HR+/HER2−, and HR+/HER2+ breast cancer (26.3%, 10.9%, and 10.3%, respectively). (B) PDL1 mRNA expression is higher in TNBC compared to HR+/HER2− and HR+/HER2+ breast cancer (fold change, 1.49; p = 0.00030 and fold change, 1.42; p = 0.0068). In the beeswarm plot, each colored dot represents a tumor. The horizontal red lines indicate the first quartile, the median, and the third quartile.
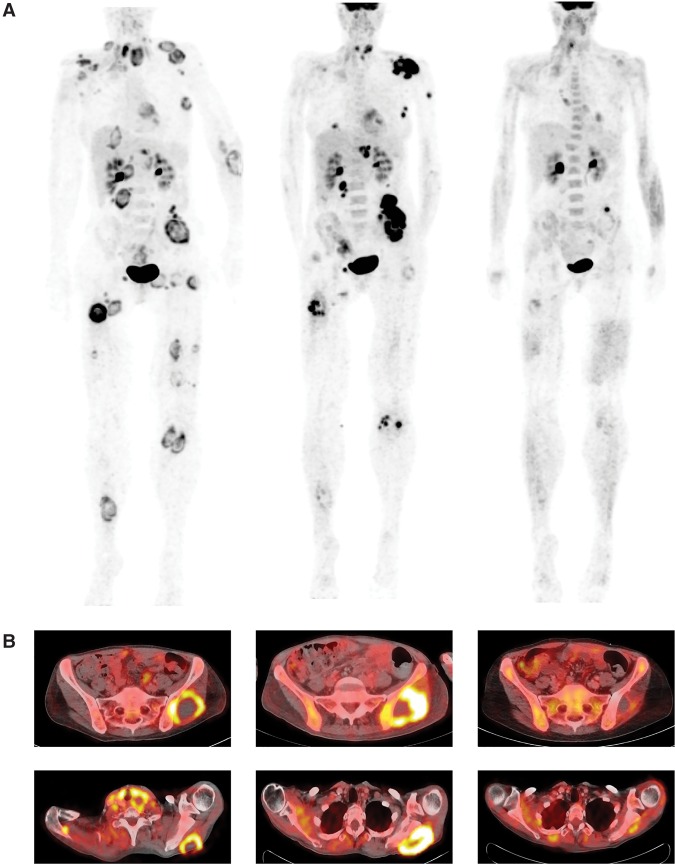
Both amplification and overexpression of PDL1 and the hypermutated tumor genome provided a strong rationale for ICI treatment with the anti-PD1 monoclonal antibody pembrolizumab. The patient received pembrolizumab at a dose of 2 mg/kg body weight for repeated cycles of 3 wk starting from month 24 after initial diagnosis. At the start of treatment, baseline positron emission tomography/computed tomography (PET/CT) imaging showed widely disseminated tumor manifestations (Fig. 4A). Treatment was well tolerated and could be administered without serious side effects. After 2 mo of therapy, PET imaging showed evidence of tumor regression in most lesions while a left gluteal and a left axillar lesion increased in size and metabolic activity, likely representing an immune-mediated phenomenon. Clinically, performance status was improving rapidly, and tumor-related symptoms such as pain and motoric impairment of the left arm decreased. Six months after initiation of pembrolizumab, PET imaging results suggested a near-complete remission, formally qualifying as a very good partial remission according to Response Evaluation Criteria in Solid Tumors (Fig. 4). The patient has remained in continuous near-complete remission and free of disease-specific symptoms while being on immunotherapy at the time this report was written (14 mo after commencement of pembrolizumab).
Figure 4.
Response assessment after 2 and 6 mo of immunotherapy with pembrolizumab in a patient with metastatic cancer of unknown primary. (A) Maximum intensity plots computed from positron emission tomography/computed tomography (PET/CT) imaging showing multifocal tumor manifestations that almost vanish after 6 mo of PD1 inhibitor treatment. (B) Axial PET/CT images showing target lesions in the left gluteal (upper panels) and scapular (lower panels) regions. Time line from left to right: baseline, 2 mo, 6 mo.
DISCUSSION
“Multi-omic” approaches and integrative analyses allow tissue-of-origin molecular profiling and can uncover new diagnostic and therapeutic opportunities in patients with high-risk and refractory cancers (Roychowdhury and Chinnaiyan 2014). Matching the genetic data obtained in individual patients to the rapidly expanding public repositories of genome, exome, and transcriptome data from large cohorts of well-annotated cancers can inform histopathologic assessment, including characteristic tissue-of-origin IHC and FISH markers, and facilitate the identification of the primary cancer site in difficult cases. In our patient, molecular analysis and histopathology suggested several possible tissues of origin, but a definitive diagnosis could not be reached. This caveat may in part be explained by the limitations associated with comparison of molecular profiles of metastastic tissue with data sets obtained in treatment-naïve tumors, such as those available from TCGA.
Besides its diagnostic implications, WES can identify driver genomic alterations that are missed by interrogating single genes or a panel of cancer-related genes but may represent targets for individualized treatment. Importantly, tissue context may modulate the efficacy of therapies directed against specific molecular lesions, as shown, for example, by the variable responses to vemurafenib seen among different tumor entities with BRAF V600E mutations (Samuel et al. 2014; Corcoran et al. 2015; Hyman et al. 2015; Kopetz et al. 2015). Hence, responses can be difficult to predict, particularly to drugs not developed for a given indication (Le Tourneau et al. 2015). Despite this purported predicament, targeted therapies have become the mainstay of treatment in a range of malignancies (e.g., gastrointestinal stromal tumor, chronic myeloid leukemia, or non-small-cell lung cancer), underscoring the utility of personalized genomic medicine and the incorporation of “omics” data in the clinical decision algorithm on an individual-case basis in patients otherwise refractory to conventional treatment modalities (Tothill et al. 2013; Knoechel et al. 2015; Schwaederle et al. 2015; Kordes et al. 2016). Our diagnostic approach ruled out sarcoma and rendered the diagnosis of poorly differentiated adenocarcinoma. We were, however, unable to trace the definite tissue of origin even when employing both molecular and conventional pathology. This result may in part be explained by the fact that our case is a patient with metastatic CUP who underwent several lines of treatment before molecular profiling, a setting that hampers direct comparison with molecular profiles of treatment-naïve tumors, such as those deposited in the TCGA database. Furthermore, novel data-sharing initiatives, such as AACR (American Association for Cancer Research) Project GENIE (Genomics, Evidence, Neoplasia, Information Exchange; http://www.aacr.org/Research/Research/Pages/aacr-project-genie.aspx), as well as innovative interventional clinical trials, such as NCI-MATCH (National Cancer Institute-Molecular Analysis for Therapy Choice; http://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match), are expected to broaden the basis for greater individualization of therapy, particularly in the case of rare cancers and rare mutations in common cancers.
In our patient, the finding of PDL1 amplification and overexpression in conjunction with the high mutational load, and the failure of previous conventional chemotherapeutic approaches, prompted us to administer off-label immunotherapy. Based on our patient's favorable clinical course, our exploration of potential differential diagnoses, and an analysis of the TCGA data set on TNBC, we speculate that ICIs might be a new treatment modality in this aggressive and clinically challenging tumor type (Buisseret et al. 2015; Le et al. 2015). PD1-inhibitory agents are currently being investigated in TNBC in phase II/III trials that are prospectively collecting data on tumoral PDL1 expression, allowing correlative evaluation of PDL1 copy number changes as a predictive marker (Sharma and Allison 2015; Topalian et al. 2015). The mechanistic rationale underlying PD1 blockade is to reverse tumor immune evasion mediated by PDL1-PD1 engagement of the tumor with antitumor CD8+ T cells, which consequently become anergic. The choice of immunotherapy in our patient was supported by the high density of tumor-infiltrating lymphocytes, PDL1 amplification resulting in strong intratumoral PDL1 expression both on the RNA and protein level, and a high mutational load, which was reported as being predictive for response to ICI in a range of cancers (Garon et al. 2015; Le et al. 2015; Robert et al. 2015). Although future studies are needed to further establish robust and reliable determinants of response to ICI (Carbognin et al. 2015; Patel and Kurzrock 2015), PDL1 amplification might serve as a rapidly available and inexpensive surrogate marker across histologic entities, as supported by the striking therapeutic activity of PD1 blockade in TNBC (this study) and refractory classical Hodgkin lymphoma (Ansell et al. 2015; Roemer et al. 2016) characterized by Chromosome 9p24.1 gain. In a recent pan-cancer analysis, PDL1 gains were shown to occur in many cancer types and to result in PDL1 mRNA overexpression in some tumors (Budczies et al. 2016). Moreover, a PDL1 core amplification region of 7.8 Mb on Chromosome 9p that extended to the telomere was identified, consistent with the finding in this report. JAK2 is coamplified in this region (Balko et al. 2016), which is of potential interest, because JAK2 has been reported previously as a positive regulator of PDL1 and PDL2 genes (Green et al. 2010). Inactivating JAK2 mutations have recently been implicated in acquired resistance to PD1 inhibitor treatment and immune evasion of cancer cells, possibly related to down-regulation of PDL1 and antigen presentation as a consequence of abolished interferon signaling due to loss-of-function alterations in JAK2 (Zaretsky et al. 2016).
In summary, our observations show that clinical NGS can uncover new therapeutic targets and may eventually help to change the management of a considerable number of patients with CUP. The steadily decreasing turnaround time of WES and even whole-genome analysis, declining sequencing costs, and continuous workflow standardizations create a strong imperative to implement this methodology for the diagnostic workup of CUP patients. Furthermore, our experience illustrates how integration of different methods can optimize diagnostics and identify targets for intervention especially in cases in which it is difficult to arrive at a consensus diagnosis by conventional means.
METHODS
WES, RNA-seq, and Bioinformatic Analysis
Exome capture was performed using SureSelect Human All Exon V5+UTRs in-solution capture reagents (Agilent). One and one-half micrograms of genomic DNA was sheared sonically to 150–200 bp (paired-end) insert size with a Covaris S2 device. Two hundred and fifty nanograms of Illumina adapter-containing library was hybridized with exome baits at 65°C for 16 h. Paired-end sequencing (101 bp) was performed with a HiSeq 2500 instrument (Illumina) in a rapid mode. RNA-seq libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina). Briefly, mRNA was purified from 1 µg total RNA using oligo(dT) beads, poly(A)+ RNA was fragmented to 150 bp and converted into cDNA, and cDNA fragments were end-repaired, adenylated on the 3′ end, adapter-ligated, and amplified with 12 cycles of polymerase chain reaction. The final libraries were validated using a Qubit 2.0 Fluorometer (Life Technologies) and a Bioanalyzer 2100 system (Agilent). Paired-end sequencing (2 × 101 bp) was performed with a HiSeq 2500 instrument (Illumina) in a rapid mode. Sequencing data were analyzed using a previously reported bioinformatics workflow (Kordes et al. 2016; see Table 2 for coverage metrics).
Table 2.
Whole-exome sequencing (WES) and RNA sequencing (RNA-seq) coverage metrics
| WES | RNA-seq | |||||
|---|---|---|---|---|---|---|
| Sample | Total reads | Percentage of reads aligned | Percentage of duplicate reads | Average on-target read coverage | Total reads | Percentage of reads aligned |
| Buffy coat | 94,580,016 | 99.49 | 5.6 | 125.04 | n.a. | n.a. |
| Tumor | 113,842,258 | 99.58 | 7.4 | 149.78 | 243,198,700 | 94.14 |
n.a., not applicable.
Triple-Negative Breast Cancer Data Set
Breast cancer data sets (study brca_tcga_pub2015) including data on DNA copy number status (GISTIC) and mRNA expression (RNA Seq V2 RSEM) of PDL1 were retrieved from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal) and analyzed as described previously (Budczies et al. 2016). Immunohistochemistry data for ER, PR, and HER2 as well as FISH data for HER2 were retrieved from the TCGA data portal (https://tcga-data.nci.nih.gov). The study cohort comprised 937 breast carcinomas with available copy number data. Cases were classified into molecular subtypes: 597 (63.7%) HR+/HER2− tumors, 145 (15.5%) HR+/HER2+ tumors, 38 HR−/HER2+ tumors (4.1%), and 157 (16.8%) HR−/HER2− tumors. RNA-seq data were available for 934 of these tumors.
Immunohistochemistry and Fluorescence In Situ Hybridization
Immunohistochemical staining was performed on formalin-fixed and paraffin-embedded whole-tissue slides using a BenchMark XT device (Ventana Medical Systems) according to quality-controlled standard procedures. After antigen retrieval, the following primary antibodies were used: AE1/3: 1:100 dilution, DAKO (#M3515); GATA3: ready to use, Ventana (#760–4897); CK7: 1:50 dilution, DAKO (#M7018); PDL1: 1:100 dilution, Spring (#07309457001); CD4 mouse monoclonal antibody: 1:20 dilution, Novocastra/Leica (clone 1F6); napsin: 1:400 dilution, Novacastra/Leica (clone IP64, #NCL-L-Napsin A); TTF1: 1:100 dilution, Novocastra/Leica (clone SPT24, #NCL-L-TTF1). Either 3,3′-diaminoenzidine peroxide substrate or 3-amino-9-ethylcarbazole served as chromogens.
FISH using a commercially available dual color probe for the PDL1 (CD274) gene (Zytovision; # Z-2179–200) was performed on whole slides according to the manufacturer's instructions. Four-micrometer-thick sections were cut, mounted on SuperFrost slides, and deparaffinized. The PDL1 probe was labeled with a green fluorochrome and the classical satellite III region of Chromosome 9 (CEN9, D9Z3) was labeled with an orange fluorochrome.
ADDITIONAL INFORMATION
Data Deposition and Access
WES and RNA-seq data derived from the patient specimen have been deposited at the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/), which is hosted by the European Bioinformatics Institute (EBI), under accession number EGAS00001001846. Point substitution variants were submitted to the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic, identifier COSP41747).
Ethics Statement
Patient tissue samples were collected after informed consent was obtained and provided by the NCT Heidelberg Tissue Bank under protocol NCT MASTER, S-206/2011 in accordance with its regulations and after approval by the Ethics Committee of Heidelberg University. Specifically, patients provided written informed consent including permission of tissue banking of tumor and germline specimens and analysis by next-generation sequencing technology (i.e., whole-exome sequencing and RNA sequencing and ancillary molecular and histopathological methodologies for validation of results).
Acknowledgments
The authors thank the DKFZ-HIPO and NCT Precision Oncology Program (POP) Sample Processing Laboratory, the DKFZ Genomics and Proteomics Core Facility, and the DKFZ-HIPO Data Management Group for technical support and expertise. We also thank Katja Beck, Karolin Willmund, Roland Eils, and Peter Lichter for infrastructure and program development within DKFZ-HIPO and NCT POP.
Author Contributions
S.G., M.B., A.S., W.W., and S.F. collected and interpreted patient data and were involved in clinical management. B.H., D.B., J.B., M.F., D.H., and S.U. contributed to data analysis and interpretation. S.G., M.B., B.H., J.B., D.B., C.H., P.H., M.F., S.U., D.H., C.G., D.R., N.P., K.P., S.W., P.S., D.J., C.v.K., B.B., H.G., W.W., A.S., and S.F. wrote the manuscript.
Funding
This work was supported by grants H021 and H063 from DKFZ-HIPO.
Competing Interest Statement
The authors have declared no competing interest.
Referees
Anonymous
James Hicks
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al. 2015. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med 372: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balko JM, Schwarz LJ, Luo N, Estrada MV, Giltnane JM, Davila-Gonzalez D, Wang K, Sanchez V, Dean PT, Combs SE, et al. 2016. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 8: 334ra353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, Dueck AC, McCullough AE, Reddy SK, Ramanathan RK, et al. 2015. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 6: 26483–26493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J, Denkert MBC, Klauschen F, Gröschel S, Darb-Esfahani S, Pfarr N, Leichsenring J, Onozato ML, Lennerz JK, Dietel M, et al. 2016. Pan-cancer analysis of copy number changes in programmed death-ligand 1 (PD-L1, CD274)—associations with gene expression, mutational load and survival. Genes Chromosomes Cancer 55: 626–639. [DOI] [PubMed] [Google Scholar]
- Buisseret L, Specht J, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Gause CK, Karantza V, Nanda R. 2015. 14P * KEYNOTE-012: a phase Ib study of pembrolizumab (MK-3475) in patients (pts) with metastatic triple-negative breast cancer (mTNBC). Ann Oncol 26suppl 3: ii6–iii6. [Google Scholar]
- Carbognin L, Pilotto S, Milella M, Vaccaro V, Brunelli M, Caliò A, Cuppone F, Sperduti I, Giannarelli D, Chilosi M, et al. 2015. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS One 10: e0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino-Mathews A, Subhawong AP, Illei PB, Sharma R, Halushka MK, Vang R, Fetting JH, Park BH, Argani P. 2013. GATA3 expression in breast carcinoma: utility in triple-negative, sarcomatoid, and metastatic carcinomas. Hum Pathol 44: 1341–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Atreya CE, Falchook GS, Kwak EL, Ryan DP, Bendell JC, Hamid O, Messersmith WA, Daud A, Kurzrock R, et al. 2015. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J Clin Oncol 33: 4023–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, et al. 2015. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372: 2018–2028. [DOI] [PubMed] [Google Scholar]
- Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O'Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, et al. 2010. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116: 3268–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, Bagdasarian L, Huber J, Lindeman A, Chen D, et al. 2014. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci 111: 3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay J-Y, Wolf J, Raje NS, Diamond EL, Hollebecque A, et al. 2015. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med 373: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic IP, Pejnovic NN, Radosavljevic GD, Pantic JM, Milovanovic MZ, Arsenijevic NN, Lukic ML. 2014. Interleukin-33/ST2 axis promotes breast cancer growth and metastases by facilitating intratumoral accumulation of immunosuppressive and innate lymphoid cells. Int J Cancer 134: 1669–1682. [DOI] [PubMed] [Google Scholar]
- Judson I, Verweij J, Gelderblom H, Hartmann JT, Schöffski P, Blay JY, Kerst JM, Sufliarsky J, Whelan J, Hohenberger P, et al. 2014. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol 15: 415–423. [DOI] [PubMed] [Google Scholar]
- Kietz S, Feng S, Agoulnik A, Hombach-Klonisch S. 2009. Estrogen and TCDD influence RLN2 gene activity in estrogen receptor–positive human breast cancer cells. Ann NY Acad Sci 1160: 367–373. [DOI] [PubMed] [Google Scholar]
- Kim MS, Kim E, Heo JS, Bae DJ, Lee JUW, Lee TH, Lee HJ, Chang HS, Park JS, Jang AS, et al. 2015. Circulating IL-33 level is associated with the progression of lung cancer. Lung Cancer 90: 346–351. [DOI] [PubMed] [Google Scholar]
- Knoechel B, Bhatt A, Pan L, Pedamallu CS, Severson E, Gutierrez A, Dorfman DM, Kuo FC, Kluk M, Kung AL, et al. 2015. Complete hematologic response of early T-cell progenitor acute lymphoblastic leukemia to the γ-secretase inhibitor BMS-906024: genetic and epigenetic findings in an outlier case. Cold Spring Harb Mol Case Stud 1: a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetz S, Desai J, Chan E, Hecht JR, O'Dwyer PJ, Maru D, Morris V, Janku F, Dasari A, Chung W, et al. 2015. Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol 33: 4032–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes M, Röring M, Heining C, Braun S, Hutter B, Richter D, Geörg C, Scholl C, Gröschel S, Roth W, et al. 2016. Cooperation of BRAF(F595L) and mutant HRAS in histiocytic sarcoma provides new insights into oncogenic BRAF signaling. Leukemia 30: 937–946. [DOI] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372: 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tourneau C, Delord J-P, Gonçalves A, Gavoille C, Dubot C, Isambert N, Campone M, Trédan O, Massiani M-A, Mauborgne C, et al. 2015. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16: 1324–1334. [DOI] [PubMed] [Google Scholar]
- Marquard AM, Birkbak NJ, Thomas CE, Favero F, Krzystanek M, Lefebvre C, Ferté C, Jamal-Hanjani M, Wilson GA, Shafi S, et al. 2015. TumorTracer: a method to identify the tissue of origin from the somatic mutations of a tumor specimen. BMC Med Genomics 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HG, Hwang KT, Kim JA, Kim HS, Lee MJ, Jung EM, Ko E, Han W, Noh DY. 2011. NFIB is a potential target for estrogen receptor-negative breast cancers. Mol Oncol 5: 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SP, Kurzrock R. 2015. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther 14: 847–856. [DOI] [PubMed] [Google Scholar]
- Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al. 2015. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 372: 2521–2532. [DOI] [PubMed] [Google Scholar]
- Roemer MGM, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, Connelly CF, Sun HH, Daadi SE, Freeman GJ, et al. 2016. PD-L1 and PD-L2 genetic alterations define classical Hodgkin lymphoma and predict outcome. J Clin Oncol 34: 2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychowdhury S, Chinnaiyan AM. 2014. Translating genomics for precision cancer medicine. Annu Rev Genomics Hum Genet 15: 395–415. [DOI] [PubMed] [Google Scholar]
- Samuel J, Macip S, Dyer MJS. 2014. Efficacy of vemurafenib in hairy-cell leukemia. N Engl J Med 370: 286–288. [DOI] [PubMed] [Google Scholar]
- Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. 2015. Impact of precision medicine in diverse cancers: a meta-analysis of phase II clinical trials. J Clin Oncol 33: 3817–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Allison JP. 2015. The future of immune checkpoint therapy. Science 348: 56–61. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Drake CG, Pardoll DM. 2015. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothill RW, Li J, Mileshkin L, Doig K, Siganakis T, Cowin P, Fellowes A, Semple T, Fox S, Byron K, et al. 2013. Massively-parallel sequencing assists the diagnosis and guided treatment of cancers of unknown primary. J Pathol 231: 413–423. [DOI] [PubMed] [Google Scholar]
- Varadhachary GR, Raber MN. 2014. Cancer of unknown primary site. N Engl J Med 371: 757–765. [DOI] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. 2016. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Ng SC, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. 2012. Glycine decarboxylase activity drives non–small cell lung cancer tumor–initiating cells and tumorigenesis. Cell 148: 259–272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.