Abstract
Hereditary spastic paraplegias (HSPs) are a group of heterogeneous neurodegenerative disorders, which are often presented with overlapping phenotypes such as progressive paraparesis and spasticity. To assist the diagnosis of HSP subtypes, next-generation sequencing is often used to provide supporting evidence. In this study, we report the case of two probands from the same family with HSP symptoms, including bilateral lower limb weakness, unsteady gait, cognitive decline, dysarthria, and slurring of speech since the age of 14. Subsequent whole-genome sequencing revealed that the patients are compound heterozygous for variants in the SPG11 gene, including the paternally inherited c.6856C>T (p.Arg2286*) variant and the novel maternally inherited c.2316+5G>A splice-donor region variant. Variants in SPG11 are the common cause of autosomal recessive spastic paraplegia type 11. According to the ClinVar database, there are already 101 reported pathogenic variants in SPG11 that are associated with HSPs. To our knowledge, this is the first report of SPG11 variants in our local population. The novel splice variant identified in this study enriches the catalog of SPG11 variants, potentially leading to better genetic diagnosis of HSPs.
Keywords: gait imbalance, slowed slurred speech, spastic dysarthria, spastic gait, spastic paraparesis, spastic paraplegia
CASE PRESENTATION
Spastic paraplegia type 11 (SPG11) is a type of autosomal recessive neurological disease characterized by the progression of lower limb muscle stiffness and spasticity and often accompanied with thinning of the corpus callosum, intellectual disability, neuropathy, and a variety of neurological symptoms. Onset time is childhood to early adult (Shibasaki et al. 2000; Casali et al. 2004; Winner et al. 2004; Olmez et al. 2006; Hehr et al. 2007). A majority of SPG11 cases are caused by homozygous recessive or compound heterozygous variants in the SPG11 gene (Stevanin et al. 2007, 2008), which codes for the Spatacsin protein with a role in axonal maintenance, cargo trafficking (Pérez-Brangulí et al. 2014), and autophagy (Chang et al. 2014). Because of overlapping phenotypes in different hereditary spastic paraplegia (HSP) subtypes (Pensato et al. 2014), diagnosis of SPG11 is often supplemented with evidence from molecular genetics testing. In particular, next-generation sequencing is gaining traction as a tool for assisting the diagnosis and treatment of neurological diseases (Tsoi et al. 2014; Petrovski et al. 2015; Yang et al. 2015; Ye et al. 2015).
We report the case study of a family with two probands that showed HSP symptoms (Fig. 1A). Both parents were asymptomatic without history of consanguineous marriages. The age of disease onset for the probands was 14, when both of them showed bilateral lower limb weakness and unsteady gait (Table 1). Subsequently, they developed slurring of speech, dysarthria, and cognitive decline. The elder female proband (II:2) presented with slower disease progression, and she is still able to walk with aids. On the other hand, the younger male proband (II:1) suffered from rapid deterioration over a few years after onset and became bedridden. Extensive workup had been done including serum copper, cortisol, cholestanol, campesterol, stigmasterol, and β-sitosterol levels; however, all of these indicators were normal. Further skin biopsy and urine testing for porphyrin also showed negative results. The magnetic resonance imaging (MRI) brain imaging for II:1 showed mild cerebral and cerebellar atrophy at the age of 25, which is compatible with his symptoms of cognitive decline and unsteady gait (Supplemental Fig. 1).
Figure 1.
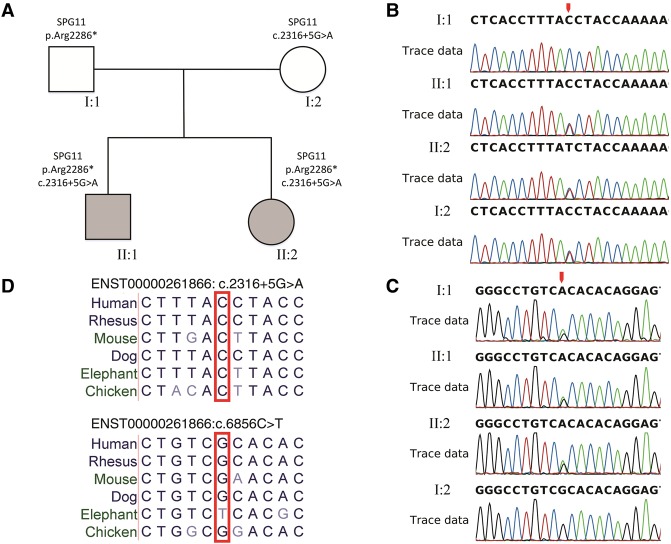
Compound heterozygous variants in SPG11 in probands. (A) Pedigree of family under study. The two probands inherited compound heterozygous variants (ENST00000261866:c.6856C>T;p.Arg2286* and ENST00000261866:c.2316+5G>A) from unaffected parents. Sanger sequencing validation of (B) the ENST00000261866:c.2316+5G>A variant and (C) the ENST00000261866:c.6856C>T;p.Arg2286* variant. Mutated positions are marked with red arrows. (D) Multiz alignments of vertebrates showed a high degree of conservation at the mutated location.
Table 1.
Clinical parameters of the two probands investigated in this study
| II:2 | II:1 | |
|---|---|---|
| Age | 31 | 29 |
| Gender | Female | Male |
| Age of onset | 14 years old | 14 years old |
| Presenting symptoms | Unsteady gait | Unsteady gait |
| Cognitive decline | Yes | Yes |
| Psychosis | No | Yes |
| Spasticity | Yes | Yes |
METHODS
To elucidate the diagnosis of the HSP subtype and potentially inform clinical decisions, whole-genome sequencing was performed using the Illumina HiSeq X Ten platform for all four family members, including the two probands and two unaffected parents. On average, ∼214.8 Gb (∼66.4× depth of coverage) of DNA sequence was generated for each individual (Supplemental Table 1). The short read sequences were aligned to the human genome (version GRCh38) using Burrows–Wheeler alignment (BWA) (version 0.7.12) (Li and Durbin 2009), followed by polymerase chain reaction (PCR) duplicate marking, local realignment around indels, and base quality score recalibration using Picard (version 1.141) tools and the Genome Analysis Toolkit (GATK, version 3.4) (McKenna et al. 2010). Sequence variants and small indels were called according to the best practices for using the GATK HaplotypeCaller (McKenna et al. 2010), resulting in ∼4,790,000 sequence variants and small indels. Because HSP is a rare disease affecting 1–10 in 100,000 individuals depending on the geographical location (Fink 2006), common variants with a minor allele frequency (MAF) >5% were filtered, leaving ∼450,000 variants on average per individual (Supplemental Table 1). Finally, variant annotation was performed using SnpEff (Cingolani et al. 2012) and dbNSFP (Database for Nonsynonymous SNPs’ Functional Predictions; Liu et al. 2013). Published guidelines from the American College of Medical Genetics and Genomics (ACMG) were used for interpretation of the variants (Richards et al. 2015). Copy-number variants and structural variants were called using LUMPY (version 0.2.11) (Layer et al. 2014), yet none of these larger scale variants match the inheritance pattern. Sequence variants focused in this study were validated using conventional Sanger sequencing analysis (Table 2).
Table 2.
List of variants in SPG11
| Genomic location (GRCh38) | dbSNP/ClinVar | HGVS | 1000G MAF | Variant interpretation | CADD | MutationTaster | I:2 | I:1 | II:1 | II:2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Chr15: 44565997 | rs312262785/41353 | ENST00000261866:c.6856C>T; p.Arg2286* | 0.0002 | Pathogenic (PVS1, PM1, PM2, PP3, PP4) | Pathogenic (47) | Disease causing (1) | N/A | 0/1 | 0/1 | 0/1 |
| Chr15: 44622723 | rs879255274/252959 | ENST00000261866:c.2316+5G>A ENST00000559193:c.2321G>A; p.Gly774Asp |
N/A | Likely pathogenic (PM2, PM3, PP1, PP3, PP4) | Pathogenic (19.36) | Disease causing (1) | 0/1 | N/A | 0/1 | 0/1 |
| Chr15: 44651599 | rs3759873/130364 | ENST00000261866:c.1348A>G; p.Ile450Val | 0.0389 | Benign (BA1, BS1, BS4, BP4) | Neutral (0.002) | Polymorphism (0.994) | N/A | 0/1 | N/A | N/A |
Genotypes for each family member are shown in the right-most columns, in which 0/1 represents heterozygous. Parenthetical codes in the Variant interpretation column denote the pathogenic criteria in the ACMG (American College of Genetics and Genomics) guidelines 2015 (Richards et al. 2015). Predicted functional impact on the transcript and protein was calculated by SnpEff, CADD, and MutationTaster. Numbers in the CADD column denote the degree of pathogenicity in Phred scale. Numbers in the MutationTaster column denote the confidence of pathogenicity classification, in which 1 is the most confident and 0 is the least confident.
dbSNP, Database for Short Genetic Variations; HGVS, Human Genome Variation Society; 1000G, 1000 Genomes; MAF minor allele frequency; CADD, Combined Annotation-Dependent Depletion; N/A, not applicable.
VARIANT INTERPRETATION
After annotation of variants in accordance to the ACMG standards (Richards et al. 2015), one pathogenic nonsense variant was discovered in SPG11 (ENST00000261866:c.6856C>T; p.Arg2286*). The variant is extremely rare, where the global MAF of the variant is 0.0002 (1/5008) in the 1000 Genomes Project Phase III or 1.664 × 10−5 (2/120202) in the Exome Aggregation Consortium (ExAC). The p.Arg2286* variant is known to be associated with autosomal recessive SPG11 (Denora et al. 2009) and recorded as a pathogenic variant in the ClinVar database (Variation ID: 41353). Subsequent Sanger sequencing validation confirmed that all probands are heterozygous carriers of the allele (Fig. 1A,B). However, the heterozygous variant alone cannot fully explain the observed autosomal recessive inheritance pattern, because the unaffected father also carries the p.Arg2286* variant (Fig. 1A,B). This suggests the possibility of compound heterozygosity, in which more than one variant contributes to the autosomal recessive pattern.
To explore the possibility of compound heterozygosity, we expanded the search of SPG11 sequence variants and indels to the ACMG “Likely Pathogenic” and “Unknown Significance” tiers. One heterozygous variant in the splice-donor region of SPG11 exon 12 (ENST00000261866:c.2316+5G>A) was found to be shared by two probands and the unaffected mother, which was subsequently validated using Sanger sequencing (Fig. 1C). This novel variant was not described in the 1000 Genomes Project Phase III, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP), nor the ExAC database. Based on multiple sequence alignment of SPG11 sequences from six vertebrates, c.6856C>T and c.2316+5G>A variants were located in a conserved location (Fig. 1D). The variant's impact on splice pattern was further assessed using Human Splicing Finder (HSF) (Desmet et al. 2009) and Alternative Splice Site Predictor (ASSP) (Wang and Marín 2006). HSF showed that the mutant splice site would be 92% weaker than wild type based on the MaxEnt model, whereas ASSP suggested that the variant would lead to the loss of the splice-donor site. Because of alternative splicing, this novel variant can also be found in the coding region of the alternative transcript (ENST00000559193:c.2321G>A; p.Gly774Asp). However, ENST00000559193 is not expressed in brain, whereas ENST00000261866 is the most abundant transcript among those with Transcription Support Level 1 in the Genotype-Tissue Expression (GTEx) project (Lonsdale et al. 2013). This suggests that the variant mainly exerts its impact through ENST00000261866:c.2316+5G>A. The novel maternally inherited c.2316+5G>A variant, in combination with the paternally inherited c.6856C>T (p.Arg2286*) variant, supports the compound heterozygous diagnosis of SPG11.
SUMMARY
The SPG11 gene encodes for the Spatacsin protein, yet the detailed molecular function of Spatacsin is not well understood. It was suggested that axonal defects were observed in patients with nonsense and splice variants in SPG11, which is further supported by a functional study using a mouse model (Pérez-Brangulí et al. 2014). Loss of Spatacsin also causes accumulation of autolysosomes and deprivation of free lysosomes, thereby disrupting the autophagic lysosome reformation pathway, ultimately leading to neurodegeneration (Chang et al. 2014). In this study, we have identified compound heterozygous variants in SPG11 that were predicted to cause truncation of the corresponding protein. The p.Arg2286* nonsense variant was previously reported to be associated with SPG11 (Denora et al. 2009); while in the vicinity of the novel c.2316+5G>A variant identified in this study, a splice variant (c.2316+1G>A) was previously linked to SPG11 (Stevanin et al. 2008). In a recent large-scale investigation of SPG11 cases in London, 79.5% of SPG11 variants were found to be nonsense, frameshift, or splice-site variants that could cause large-scale amino acid sequence changes (Kara et al. 2016). Because both variants identified in this study were predicted to cause the truncation of Spatacsin, findings in this study further supported the major role of loss-of-function variants in SPG11 pathogenesis (Pensato et al. 2014; Pérez-Brangulí et al. 2014). To our knowledge, our study is the first report of SPG11 variants in the Hong Kong population. The novel splice variant identified in this study expands the repertoire of SPG11 variants, facilitating the molecular genetic testing of HSP.
ADDITIONAL INFORMATION
Data Deposition and Access
Raw sequencing data were deposited to the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega) under accession number EGAS00001001849. The variant was deposited in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) under accession number SCV000292372.
Ethics Statement
Informed and signed consent forms were obtained for all sequenced individuals of this study. The project is approved by The Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee (CRE-2012.361).
Author Contributions
Patients were recruited and phenotyped by A.Y.-Y.C. and W.C.A. Data analysis, interpretation, and validation were performed by A.C.-S.Y. and Y.S. The manuscript was prepared by A.C.-S.Y., A.Y.-Y.C., Y.S., H.-Y.E.C., and T.F.C. All authors contributed to the reviewing of the final version.
Funding
This work was partially supported by the Chow Tai Fook Charity Foundation (6903898), Hong Kong Spinocerebellar Ataxia Association (6903291), and Gerald Choa Neuroscience Centre (7105306) grants to H.-Y.E.C. A.C.-S.Y. and T.F.C. are supported by the Research Grants Council (RGC) General Research Fund (GRF14102014) and Collaborative Research Fund (CRF: C4042-14G).
Competing Interest Statement
T.F.C. and H.-Y.E.C. are scientific advisors of Codex Genetics Limited (Codex), and A.C.-S.Y. is the genetic scientist at the same company. However, neither financial relationships nor activities with Codex appear to have influenced the submitted work.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Casali C, Valente EM, Bertini E, Montagna G, Criscuolo C, De Michele G, Villanova M, Damiano M, Pierallini A, Brancati F, et al. 2004. Clinical and genetic studies in hereditary spastic paraplegia with thin corpus callosum. Neurology 62: 262–268. [DOI] [PubMed] [Google Scholar]
- Chang J, Lee S, Blackstone C. 2014. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest 124: 5249–5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denora PS, Schlesinger D, Casali C, Kok F, Tessa A, Boukhris A, Azzedine H, Dotti MT, Bruno C, Truchetto J, et al. 2009. Screening of ARHSP-TCC patients expands the spectrum of SPG11 mutations and includes a large scale gene deletion. Hum Mutat 30: E500–E519. [DOI] [PubMed] [Google Scholar]
- Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. 2009. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JK. 2006. Hereditary spastic paraplegia. Curr Neurol Neurosci Rep 6: 65–76. [DOI] [PubMed] [Google Scholar]
- Hehr U, Bauer P, Winner B, Schule R, Olmez A, Koehler W, Uyanik G, Engel A, Lenz D, Seibel A, et al. 2007. Long-term course and mutational spectrum of spatacsin-linked spastic paraplegia. Ann Neurol 62: 656–665. [DOI] [PubMed] [Google Scholar]
- Kara E, Tucci A, Manzoni C, Lynch DS, Elpidorou M, Bettencourt C, Chelban V, Manole A, Hamed SA, Haridy NA, et al. 2016. Genetic and phenotypic characterization of complex hereditary spastic paraplegia. Brain 139: 1904–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol 15: R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. 2013. dbNSFP v2.0: A database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34: E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmez A, Uyanik G, Ozgül RK, Gross C, Cirak S, Elibol B, Anlar B, Winner B, Hehr U, Topaloglu H, et al. 2006. Further clinical and genetic characterization of SPG11: hereditary spastic paraplegia with thin corpus callosum. Neuropediatrics 37: 59–66. [DOI] [PubMed] [Google Scholar]
- Pensato V, Castellotti B, Gellera C, Pareyson D, Ciano C, Nanetti L, Salsano E, Piscosquito G, Sarto E, Eoli M, et al. 2014. Overlapping phenotypes in complex spastic paraplegias SPG11, SPG15, SPG35 and SPG48. Brain 137: 1907–1920. [DOI] [PubMed] [Google Scholar]
- Pérez-Brangulí F, Mishra HK, Prots I, Havlicek S, Kohl Z, Saul D, Rummel C, Dorca-Arevalo J, Regensburger M, Graef D, et al. 2014. Dysfunction of spatacsin leads to axonal pathology in SPG11-linked hereditary spastic paraplegia. Hum Mol Genet 23: 4859–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Shashi V, Petrou S, Schoch K, McSweeney KM, Dhindsa RS, Krueger B, Crimian R, Case LE, Khalid R, et al. 2015. Exome sequencing results in successful riboflavin treatment of a rapidly progressive neurological condition. Mol Case Stud 1: a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki Y, Tanaka H, Iwabuchi K, Kawasaki S, Kondo H, Uekawa K, Ueda M, Kamiya T, Katayama Y, Nakamura A, et al. 2000. Linkage of autosomal recessive hereditary spastic paraplegia with mental impairment and thin corpus callosum to chromosome 15A13-15. Ann Neurol 48: 108–112. [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, Martin E, Ouvrard-Hernandez A-M, Tessa A, Bouslam N, et al. 2007. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 39: 366–372. [DOI] [PubMed] [Google Scholar]
- Stevanin G, Azzedine H, Denora P, Boukhris A, Tazir M, Lossos A, Rosa AL, Lerer I, Hamri A, Alegria P, et al. 2008. Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain 131: 772–784. [DOI] [PubMed] [Google Scholar]
- Tsoi H, Yu ACS, Chen ZS, Ng NKN, Chan AYY, Yuen LYP, Abrigo JM, Tsang SY, Tsui SKW, Tong TMF, et al. 2014. A novel missense mutation in CCDC88C activates the JNK pathway and causes a dominant form of spinocerebellar ataxia. J Med Genet 51: 590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Marín A. 2006. Characterization and prediction of alternative splice sites. Gene 366: 219–227. [DOI] [PubMed] [Google Scholar]
- Winner B, Uyanik G, Gross C, Lange M, Schulte-Mattler W, Schuierer G, Marienhagen J, Hehr U, Winkler J. 2004. Clinical progression and genetic analysis in hereditary spastic paraplegia with thin corpus callosum in spastic gait gene 11 (SPG11). Arch Neurol 61: 117–121. [DOI] [PubMed] [Google Scholar]
- Yang H, Douglas G, Monaghan KG, Retterer K, Cho MT, Escobar LF, Tucker ME, Stoler J, Rodan LH, Stein D, et al. 2015. De novo truncating variants in the AHDC1 gene encoding the AT-hook DNA-binding motif-containing protein 1 are associated with intellectual disability and developmental delay. Mol Case Stud 1: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Cho MT, Retterer K, Alexander N, Ben-Omran T, Al-Mureikhi M, Cristian I, Wheeler PG, Crain C, Zand D, et al. 2015. De novo POGZ mutations are associated with neurodevelopmental disorders and microcephaly. Mol Case Stud 1: a000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.