Abstract
Listeria (L.) monocytogenes is an opportunistic pathogen causing life-threatening infections in diverse mammalian species including humans and ruminants. As little is known on the link between strains and clinicopathological phenotypes, we studied potential strain-associated virulence and organ tropism in L. monocytogenes isolates from well-defined ruminant cases of clinical infections and the farm environment. The phylogeny of isolates and their virulence-associated genes were analyzed by multilocus sequence typing (MLST) and sequence analysis of virulence-associated genes. Additionally, a panel of representative isolates was subjected to in vitro infection assays. Our data suggest the environmental exposure of ruminants to a broad range of strains and yet the strong association of sequence type (ST) 1 from clonal complex (CC) 1 with rhombencephalitis, suggesting increased neurotropism of ST1 in ruminants, which is possibly related to its hypervirulence. This study emphasizes the importance of considering clonal background of L. monocytogenes isolates in surveillance, epidemiological investigation and disease control.
Listeria (L.) monocytogenes is an opportunistic pathogen that may cause life-threatening infections in many mammalian species upon ingestion1,2. Listeriosis is of major importance in humans and in farmed ruminants3, in which it is associated with gastroenteritis, abortions, bacteremia, mastitis and central nervous system (CNS) infections (neurolisteriosis)4,5,6,7. In humans, L. monocytogenes has the highest hospitalization and mortality rates amongst food-borne pathogens8,9. Surveillance and control prove to be challenging due to the environmental lifestyle of L. monocytogenes, which colonizes diverse environments including soil, water, food processing plants, mammalian intestinal tracts and faeces8,10,11,12. In consequence, L. monocytogenes is frequently found as a contaminant of human food and animal feed.
L. monocytogenes is divided into 13 serotypes13 and four phylogenetic lineages14,15,16,17. Regulations consider all L. monocytogenes strains an equally serious threat for public health18, although an increasing number of studies indicate differences in ecology and virulence between L. monocytogenes strains7,16,19,20,21,22. Most lineage I strains are overrepresented in human clinical infections and in ruminant neurolisteriosis, while a majority of lineage II strains are associated with food contaminations and the environment7,14,23,24. The minor lineages III and IV are rarely isolated and have been linked to animals14. Multilocus sequence typing (MLST), which categorizes isolates into sequence types (STs) and clonal complexes (CCs, hereafter equated to clones) based on sequence data of seven housekeeping genes25,26,27, has shown that L. monocytogenes has a clonal population structure with hypervirulent and hypovirulent clones and that the distribution of certain clones significantly varies between human clinical infection and food7,25,26. However, little is known about strain-associated clinicopathological phenotypes in L. monocytogenes.
To identify clones with higher virulence or organ tropism, we comparatively studied L. monocytogenes isolates from naturally occurring ruminant infections and from their farm environment. The most frequent clinicopathological phenotypes of listeriosis in ruminants are abortions and neurolisteriosis3,8. The latter is one of the most prevalent and fatal CNS infections in ruminants and is typically characterized by encephalitis of the brainstem (rhombencephalitis)28,29,30,31. Other manifestations such as gastroenteritis, bacteremia and mastitis are only sporadically encountered in these species32,33,34,35. Additionally, clinically healthy ruminants may carry L. monocytogenes in their gastrointestinal tract and shed them into the environment36,37,38.
We compared L. monocytogenes isolates from well-defined rhombencephalitis cases and non-encephalitic cases to isolates from healthy ruminants and from the direct ruminant environment in order to identify STs associated with disease or specific tropisms. Additionally, we screened genes that have been implicated in L. monocytogenes virulence39 for polymorphisms and recombination that may explain variations in virulence and organ tropism between strains. Results show that ruminants are exposed to a broad range of genetically diverse strains in the farm environment, yet a particular genotype, ST1, is predominant in rhombencephalitis cases, but not in other disease manifestations. These data suggest increased neurotropism of ST1 strains in ruminants, which might be related to its hypervirulence.
Results
Differential distribution of STs between diseased ruminants and environment
MLST was performed of 187 ruminant clinical isolates from cases with various clinicopathological outcomes (rhombencephalitis, abortions, gastroenteritis, and mastitis), five faecal isolates of clinically healthy ruminants and 56 environmental isolates from ruminant farms (Text. S1, Table S1). Eight to 20 alleles per locus were identified resulting in 45 STs. These STs were distributed over five CCs and 29 singletons within three lineages (lineage I n = 141, lineage II n = 105, lineage III n = 2; Fig. 1). Lineage I showed an increased clustering compared to lineage II, which contained a higher number of distantly related STs. The prevalence varied considerably between STs as 51% (n = 126) of all isolates belonged to only three STs (ST1: 33%, n = 81; ST4: 11%, n = 28; ST412: 7%, n = 17) while the remaining 49% of isolates were distributed over 42 STs (Table S1). The clustering of isolates in these three STs (ST1, ST4, ST412) was particularly striking in clinical isolates (84%) compared to environmental isolates (16%).
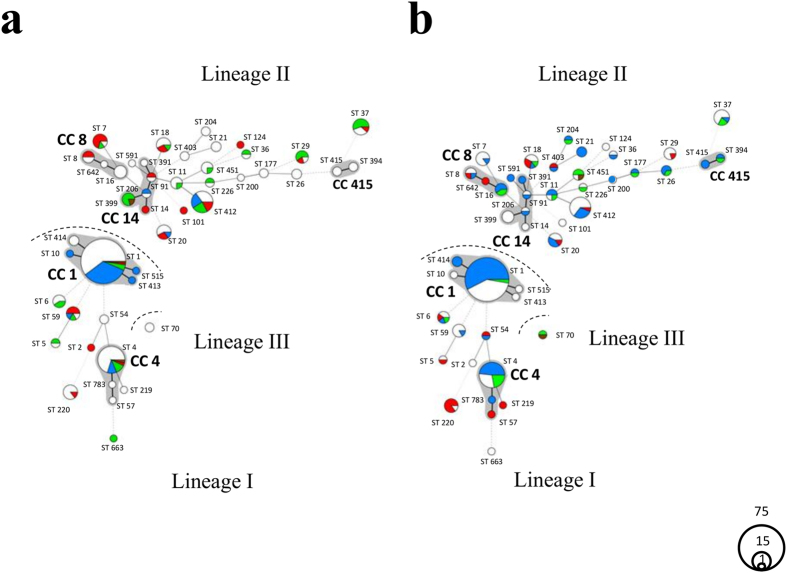
Figure 1. Minimum spanning tree (MST) of 248 L. monocytogenes isolates based on multilocus sequence typing (MLST) analysis.
Circles represent sequence types (STs) and their size corresponds to the number of isolates present in each ST. The lines between different STs represent phylogenetic relationships, bold lines indicate one mismatch in the seven housekeeping genes, plain lines two mismatches, discontinuous lines three mismatches and light discontinuous lines four or more mismatches. Grey zones surrounding multiple STs, represent clonal complexes (CCs), which contain STs with a single mismatch in the seven loci. The three evolutionary lineages are indicated. Isolates of ruminant rhombencephalitis cases are represented in blue (n = 140), non-encephalitic ruminant clinical cases in red (n = 47), ruminant faecal isolates in brown (n = 5) and isolates of the ruminant farm environment in green (n = 56). (a) MST of cattle isolates. Isolates of rhombencephalitis cases are represented in blue (n = 39), non-encephalitic cattle clinical cases in red (n = 28), cattle faecal isolates in brown (n = 3), isolates of their environment in green (n = 33) and small ruminant-associated isolates in white (n = 145). (b) MST of small ruminant isolates. Isolates of rhombencephalitis cases are represented in blue (n = 101), non-encephalitic clinical cases in red (n = 19), faecal isolates in brown (n = 2), isolates of their environment in green (n = 23) and cattle-associated isolates in white (n = 103).
Clinical and environmental isolates partially overlapped in their ST distribution, with 17 STs (38%) containing isolates from both sources. However, the relative prevalence of STs varied strongly between clinical and environmental sources and, notably, between rhombencephalitic and non-encephalitic outcomes of ruminant listeriosis (Figs 1 and 2a). The genetic diversity of isolates was significantly smaller in rhombencephalitis cases (71%; 95% CI ranging from 70.5% to 71.4%) than in non-encephalitic cases (96%; 95% CI ranging from 94.8% to 96.1%) and environmental sources (93%; 95% CI ranging from 90.1% to 95.6%). Rhombencephalitis isolates were significantly overrepresented in lineage I compared to lineage II and, particularly, in ST1 (CC1) of lineage I, which contained 53% (n = 74) of all rhombencephalitis isolates (Figs 1 and 2a, Table 1). Strikingly, 91% of ST1 were of rhombencephalitis origin, while only 36% of the other STs were of rhombencephalitis origin (p-value < 0.001). Relative ST distribution in rhombencephalitis differed between hosts (Fig. 1): while in cattle 84% of ST1, but only 16% of other STs were of rhombencephalitis origin (p-value < 0.001), the variety of other STs contributing to rhombencephalitis was higher in small ruminants. Yet, ST1 was predominant in rhombencephalitis also in these species. Here, 96% of ST1 were of rhombencephalitis origin, while the proportion of rhombencephalitis isolates in other STs was lower (56%, p-value < 0.001). Compared to most other STs, ST1 contained strikingly few isolates from non-encephalitic clinical sources (1.3%), environment (6%) and faeces (1.3%) (Figs 1 and 2a). Two further STs, in which rhombencephalitis isolates clearly prevailed, were ST4 from CC4 (61%) and ST412 from CC412 (53%). However, as they only accounted for 12% (ST4) and 7% (ST412) of isolates, association of rhombencephalitis with these STs was not statistically significant. ST412 was a particular lineage II strain, as in contrast to other lineage II STs it was mainly isolated from rhombencephalitis. ST4, ST412 and rarely ST1 were also isolated from the environment (Fig. 2a). Interestingly, in contrast to ST412, environmental and faecal isolates belonging to ST1 and ST4 were isolated in farms with ongoing35 or recent outbreaks, suggesting recent contamination from clinical cases.
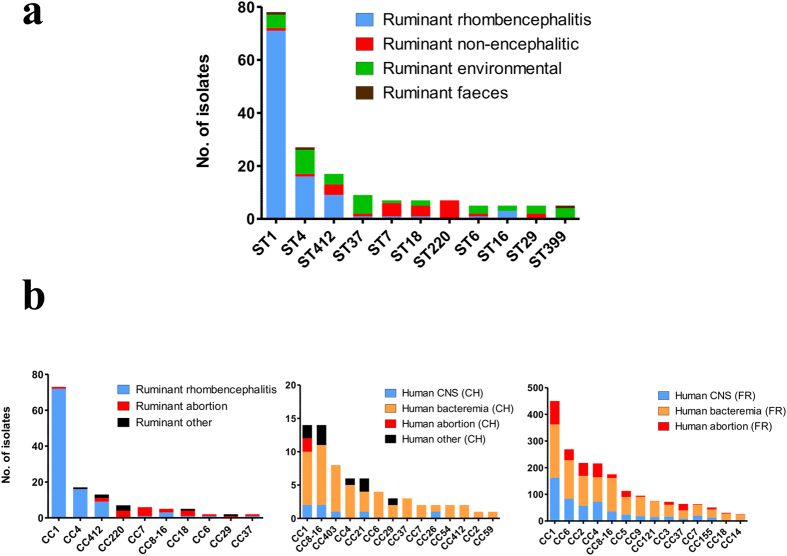
Figure 2. Frequency of L. monocytogenes isolates according to their source of isolation in the most prevalent sequence types (STs) and clonal complexes (CCs).
(a) Non-homogeneous distribution of STs in ruminant-associated isolates. The 11 most prevalent STs are arranged according to their abundance. Blue = ruminant rhombencephalitis (n = 134), red = ruminant non-encephalitic infections (n = 27), green = ruminant-associated environment (n = 40), brown = faeces (n = 4). (b) Divergent distribution of CCs in ruminant (left) and human clinical isolates from Switzerland (CH, middle)40 and France (FR, right)7. Blue = ruminant rhombencephalitis/human central nervous system (CNS) isolates, red = abortions, black = other infection sources, orange = human bacteremia.
Table 1. Prevalence of phylogenetic lineage I and II in rhombencephalitic, non-encephalitic and environmental isolates.
| Source | No. (%) of isolates | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rhombencephalitis |
Non-encephalitic infections |
Environment/faeces |
|||||||
| Total | Cattle | Small ruminants | Total | Cattle | Small ruminants | Total | Cattle | Small ruminants | |
| Total | 140 | 39 | 101 | 47 | 28 | 19 | 61 | 36 | 25 |
| Lineage I | 101 (72)*** | 33 (85)*** | 68 (67)* | 18 (38) | 7 (25) | 11 (58) | 22 (36) | 13 (36) | 9 (36) |
| Lineage II | 39 (28) | 6 (15) | 33 (33) | 29 (62)** | 21 (75) | 8 (42) | 37 (61)*** | 23 (64)* | 14 (56)* |
Fisher’s test: *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001.
In contrast to the rhombencephalitic isolates, isolates from non-encephalitic origin, environment and faeces were significantly associated with lineage II (Table 1). Some STs contained mainly (STs 7, 18) or only (ST220) clinical isolates from non-encephalitic origin. ST220 was significantly associated with non-encephalitic pathologies when compared to all other STs (p-value < 0.001), contrasting with the association of ST1 with rhombencephalitis. STs 399 and 451 were associated only with environmental and faecal isolates, and 78% of ST37 isolates originated from the ruminant environment and faeces. The distribution of these three STs between clinical and environmental/faecal isolates was significantly different (ST37: p-value < 0.001; ST399: p-value < 0.01; ST451: p-value < 0.01) when compared to the remaining STs.
Comparison with multilocus variable number of tandem repeat (MLVA) revealed that (with very few exceptions) lineage II corresponded to MLVA complex C, whereas CC1 and CC4 corresponded to MLVA complexes A and B, respectively23 (Fig. S1). Consistent with the literature, serotypes 1/2b, 3b, 4b, 4d, 4e and 7 corresponded to lineage I, whereas serotypes 1/2a, 1/2c, 3a, and 3c corresponded to lineage II. Within lineage I, most isolates belonged to serotype group 4b, 4d, 4e and within lineage II, most isolates belonged to serotype group 1/2a and 3a (Fig. S1a, Table S1).
Differential distribution of STs in human-associated and ruminant-associated isolates
When compared with Swiss and French human isolates from previous studies7,40, there was some overlap of CCs between human and ruminant clinical isolates, yet the distribution differed (Fig. 2b). Due to the high prevalence of CC1 and CC4 the genetic diversity of ruminant clinical isolates (81%; 95% CI ranging from 80.8% to 81.1%) was significantly lower than the genetic diversity of human clinical isolates (92%; 95% CI ranging from 90.7% to 92.1%). Although CC1 was also amongst the predominant CCs in human clinical isolates (CH: 15%, FR: 20%) (Fig. 2b), its prevalence was significantly higher in ruminant infections (32%, p-value < 0.001). The relative dominance of CC1 in ruminants versus humans was strikingly clear in isolates from neurolisteriosis (Fig. 2b). Additionally, CC220 and CC412 (p-value < 0.001), which were amongst the most common CCs in ruminant clinical infection, were significantly less prevalent in human infection. In contrast, CCs 2, 3, 5, 6, 7, 9, 29, 121 and 155 were significantly associated with human infection when compared to animal infections (p-values < 0.05). Some of these CCs were observed in ruminants, but most were rare or absent amongst the ruminant clinical isolates.
Isolates of rhombencephalitis-associated STs 1, 4 and 412 are hyperinvasive and hyper-replicative in a bovine macrophage cell line compared to isolates from environment-associated STs 18 and 37
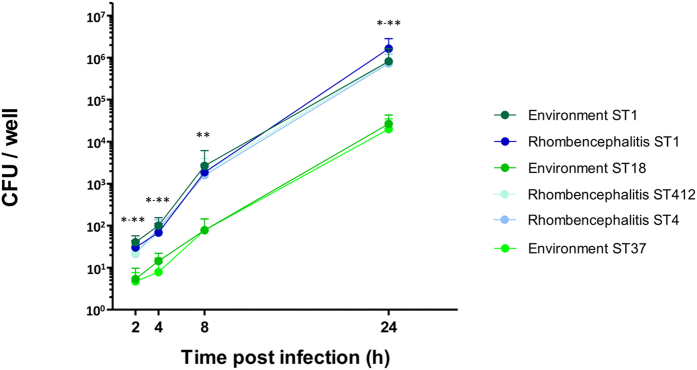
To test whether the association of STs with rhombencephalitis could be attributed to higher invasiveness and increased intracellular replication, we selected six representative cattle isolates based on their ST and source of isolation (rhombencephalitis ST1, ST4 and ST412, environment ST1, ST18 and ST37) and analysed them in a gentamicin protection assay using the bovine macrophage (BoMac) cell line. All six isolates infected BoMac cells. However, invasion and infection kinetics were different between STs (Fig. 3). Significantly more CFUs were recovered from BoMac cells at 2 h when infected with isolates from rhombencephalitis-associated STs 1, 4 and 412, regardless of the source of isolation (rhombencephalitis or environment), indicating that they are more efficient in invasion than isolates from environment-associated STs (ST18, ST37). Intracellular replication between 2 and 24 h post infection was stronger in the ST1, 4 and 412 isolates, which showed a 20 000 to 54 000 (ST1environment/rhombencephalitis) fold increase in CFU numbers over the 22 h incubation period compared to a 4 000 to 5000 fold increase in the ST37 and ST 18 isolates. The difference in recovered CFU numbers between the rhombencephalitis-associated STs and the environment-associated STs increased to up to two orders of magnitude at 24 h post infection. Differences in CFU counts were significant at all time points when ST1, ST4 and ST412 were compared to ST18 and 37 (Fig. 3).
Figure 3. CFUs per well of six isolates in the bovine macrophage (BoMac) cell line.
CFUs were enumerated from cell lysates at indicated time points post infection, three independent experiments were performed in triplicates. Error bars indicate 95% SEM. *p-value < 0.05 (2 h: ST1 rhombencephalitis VS. ST18 and ST37; 4 h: ST1 environment and ST4 Vs. ST18, ST1 rhombencephalitis and ST412 Vs. ST37; 24 h: ST1 Vs. ST18, ST4 Vs. ST37), **p-value < 0.01 (2 h: ST1 environment VS. ST18 and ST37; 4 h: ST1 environment and ST4 Vs. ST37; 8 h: ST1 and ST4 Vs. ST18 and ST37; 24 h: ST1 Vs. ST37).
Virulence-associated gene variations are associated with ST
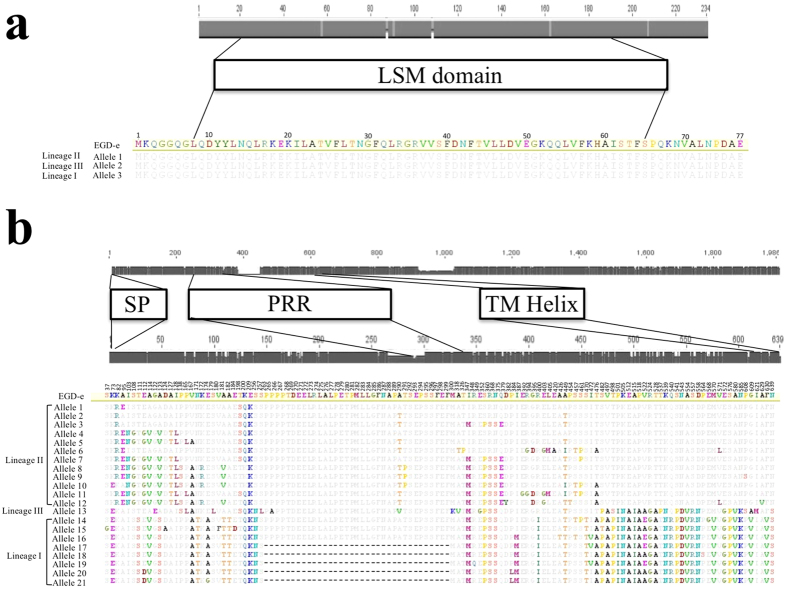
Using a subset of 94 isolates from 25 STs (Table S1) with available next generation sequencing (NGS) data, we analyzed 45 L. monocytogenes virulence-associated genes39 (Table S2). InlF, vip and lapB were absent in few infection and environment-associated STs of lineage II and lineage III (Table S2). Previously described virulence-attenuating point mutations in plcA or plcB were identified in various STs, independently whether the source of isolation was clinical or environmental41 (Table S2). inlA mutations leading to InlA truncations, which are commonly found in food isolates26,42,43,44,45, were not present in any ruminant-associated or environmental isolate. Similarly, mutations of prfA that have been associated with hypovirulence were not identified in any of the isolates46. Phylogenetic analysis with both the concatenated nucleotide and amino-acid sequences clustered isolates according to their lineage or ST, but not according to their source of isolation (Fig. S2). Diversity varied considerably between virulence-associated genes with the number of alleles ranging between 3 (hfq) and 29 (ctaP) resulting in 1 (Hfq, SigB) to 25 (InlJ) protein sequences (Table S2). For example, hfq had only six synonymous polymorphic sites resulting in three alleles correlating with the three phylogenetic lineages (Fig. S3) but encoding for a single protein sequence conserved across lineages (Fig. 4a). In contrast, actA had 26 alleles with 128 non-synonymous polymorphic sites resulting in 21 different protein sequences (Fig. S3, Fig. 4b). Gene sequences of actA, ctaP, lapB, p60 and pycA were ST specific with the exception of STs belonging to CC1, which showed a striking conservation of these genes sharing all the same allele (Table S2, Fig. S3). In some STs more than one allele of these virulence-associated genes was present. In the remaining virulence-associated genes, alleles were shared between some or most STs within the same phylogenetic lineage (Table S2). Importantly, STs involved in rhombencephalitis did not share any common virulence-associated gene allele. Analysis of the 45 virulence genes with ClonalFrame47 estimated the relative frequency of recombination occurrence versus mutation (ρ/θ) from 0.01 (mprF) to 3.91 (inlA), respectively (Table S2). In most virulence genes (n = 37), ρ/θ was below 1 indicating that mutation is more frequent than recombination. Only in four virulence-associated genes (ctpA, gap, gtcA, and inlA), the recombination rate was estimated more than 2-fold higher than mutation rate and of those, gtcA and inlA were subjected to the strongest relative effect of recombination versus point mutation (r/m) (Table S2).
Figure 4. Distribution of polymorphisms in the hfq and actA genes (grey bars).
The scale above the grey bars indicates nucleotide numbers. Below the functional domains, predicted amino acid sequences of the hfq and actA alleles are aligned to the reference strain EGD-e, and scales indicate amino acid number. (a) Polymorphisms within the 234 nucleotides of hfq are indicated as vertical bars in the grey bar. The functional domain LSM (like-Sm) is represented by a white box. While on the nucleotide level every lineage has a specific allele, none of the polymorphisms results in amino-acid changes and the amino-acid sequence is conserved across lineages. (b) Synonymous and non-synonymous nucleotide polymorphisms across the 26 actA alleles are shown as vertical bars in the upper grey bar. The functional domains (signal peptide, SP; proline rich repeats, PRR and transmembrane domain, TM) are shown in white boxes. The scale above the lower grey bar indicates amino-acid numbers and vertical bars correspond to non-synonymous nucleotide polymorphisms. Below, the amino-acid sequences of the actA 21 alleles are aligned to the reference strain EGD-e. Amino acid polymorphisms (or non-synonymous mutations) are represented in colour and gaps by “-”.
Discussion
L. monocytogenes is organized in four phylogenetic lineages with a clonal population structure7,14. Recently, hypervirulent and hypovirulent clones, which are differently distributed between human clinical infections and food, were distinguished by combining epidemiological, clinical and experimental approaches7. Among the various clinicopathological outcomes associated with listeriosis, neurolisteriosis is associated with a high case fatality rate9. Here, we provide evidence for the association of L. monocytogenes ST1 (or clone CC1) with neurotropism in ruminants by analysing L. monocytogenes isolates from clinicopathologically well-defined ruminant infections, from healthy ruminants and from their environment.
Most ruminant-associated isolates belonged to the two phylogenetic lineages I and II, and in line with former studies, lineage IV and III strains were rare15,25,26. We could show that ruminants are potentially exposed to a large diversity of L. monocytogenes strains in their immediate environment, yet not all of these strains are found in clinical infections. We observed that although isolates from ruminant clinical infections belong to both major lineages I and II, lineage I strains are highly prevalent in clinical infections, confirming previous studies in humans and ruminants (Figs 1 and 2)7,24,48. This was particularly true for isolates from neurolisteriosis that strongly clustered in ST1 (CC1) of lineage I. This ST was predominant in and significantly associated with rhombencephalitis suggesting that ST1 has an increased neurotropism in ruminants. Additionally, rhombencephalitis isolates were highly prevalent in ST4 (CC4) and ST412 (CC412), but a significant association was not observed. Bovine macrophage infection assays indicated that STs 1, 4, and 412 are hyperinvasive and hyper-replicative compared to environment-associated STs. Interestingly, ST412 belongs to lineage II indicating that these features are not restricted to strains from lineage I. Results are in line with the hypervirulence of ST1 reported in a previous study7 and may suggest that neurotropism in these STs is determined by their hyperinvasiveness and increased intracellular replication, the mechanisms of which have not yet been clarified. Polymorphisms that have been previously associated with virulence-attenuation were rarely detected in ruminant-associated isolates26,41. Most of them were present in lineage II strains, and they were sporadically detected in ST1 and ST412, which according to their association with clinical infection represent virulent STs. All ruminant associated strains had a full length InlA. Premature stop codons in inlA, which are frequently observed in food isolates40,49,50, were absent in our isolates, which may be linked to the fact that CC9 and 121 that are associated with InlA truncations, were not isolated in our study7,26,40. Hence, given that the ruminant amino acid sequence of the InlA host cell receptor E-cadherin determines permissiveness to L. monocytogenes infection51 and E-cadherin is expressed in ruminant mucosa and nerves52, there may be a role of InlA in the pathogenesis of ruminant listeriosis. Phylogenetic analysis of 45 currently known virulence-associated genes showed that most of these genes clustered according to their ST and clone, but neither to their source of isolation nor to the host species (Fig. S2). Additionally, divergence of most virulence-associated genes was driven by mutation rather than by recombination suggesting co-evolution of virulence associated genes and housekeeping genes and vertical transmission of virulence traits. We did not identify any common allele of a virulence-associated gene shared between STs involved in rhombencephalitis, and horizontal gene transfer between rhombencephalitis-associated STs was not obvious. Altogether these data suggest that these known virulence-associated genes are not determining the differences in neurotropism among L. monocytogenes strains. The association of lineage I and notably ST1 with neurolisteriosis was particularly strong in cattle (Fig. 1), while in small ruminants neurolisteriosis was caused by a variety of STs from lineages I and II suggesting increased susceptibility of small ruminants to develop neurolisteriosis upon infection with diverse L. monocytogenes strains. This interpretation is supported by the high prevalence and mortality and by the frequently fulminant disease and pathology of listeriosis in small ruminants compared to cattle3,6. Hence, our data may suggest specific virulence mechanisms of ST1 that become relevant in the pathogenesis of cattle rhombencephalitis. ST1 was rarely detected in the ruminant environment possibly suggesting that ST1 is not well adapted to the farm environment. However, as there might be a bias due to small sample size of environmental isolates, this finding needs to be confirmed by analysing larger numbers of isolates.
Hypervirulent CC1 and CC4 belong to the predominant clones in human neurolisteriosis, and CCs identified in ruminant infections partially overlapped with those of human infections in Switzerland and France7,40 (Fig. 2b) emphasizing similarities of human and animal neurolisteriosis including common mechanisms of host-pathogen interactions in these host species. Nonetheless, the relative prevalences of CCs are distinct between human and ruminant neurolisteriosis. Although prevalences of ruminant-associated CCs might be biased due to the relatively small sample size compared to human studies, these results indicate possible differences in pathogenesis, ecology, niche adaptation and transmission between CCs. Notably the high prevalence of ST1 and the very low prevalence of CC6 in ruminant neurolisteriosis remain striking (Fig. 2)7,40,53. They may be related to phenotypic differences in ruminant and human neurolisteriosis. In ruminants, the cardinal pathology of neurolisteriosis is rhombencephalitis, a brainstem encephalitis that occurs following invasion via cranial nerves6,31,54. In contrast to cattle, human patients are affected by various forms of neurolisteriosis including meningitis, meningoencephalitis, rhombencephalitis and brain abscesses55,56, of which meningitis and meningoencephalitis clearly prevail57,58. Hence, future studies should address a potential association of STs/CCs with neurolisteriosis subtypes in humans and whether there are commonalities between the CC1 infection mechanisms of rhombencephalitis in cattle and CNS infection in humans. Host differences such as prevalence of comorbidities, feeding habits and immune response might contribute to the divergence in relative prevalences of clones between humans, small ruminants and cattle7,59,60. Both CC220 and CC412 were abundant in the ruminant-associated isolates from various sources including clinical infections, but have been isolated less frequently from human infections7,40 (Fig. 2) suggesting an association of these CCs to ruminant species or a specific niche adaptation. STs 37 and 399 were significantly associated with ruminant environment and ruminant faeces. While ST37 has been identified in food processing plants and environment49,50,61 and was responsible for a Belgian listeriosis outbreak linked to pasteurized cheese62, ST399 has been only rarely reported until now and at low frequencies (0.3%)25. On the other hand, CCs highly prevalent in human infection including CC 2, 3, 5, 6, 7, 29 and 1557,40 (Fig. 2) were strikingly less prevalent in ruminant-associated isolates. Remarkably, CCs 2 and 6 that are associated with human infection and are hypervirulent in a humanized mouse model7, were rarely found in ruminant associated isolates. Similarly, CCs that are highly prevalent in food and food processing plants such as CC9 and CC121 were not represented in our isolates7,50,63,64 (Fig. 2) supporting evidence of specific niche adaptation and increased ability to contaminate food, but not farm environments.
To conclude, we provide evidence for ST1-associated hyperinvasiveness, increased intracellular replication and neurotropism of L. monocytogenes in ruminants. These findings should be considered when studying molecular mechanisms of host-pathogen interactions and epidemiology in rhombencephalitis. The determination of ST-associated virulence and ecology in L. monocytogenes will improve surveillance and control of listeriosis.
Material and Methods
Clinical isolates
A total of 187 clinical isolates from ruminant cases were included in the study. Isolates were collected in Switzerland and UK during surveillance and diagnostic activities between 1996 and 2015 (Table S1). Cases of rhombencephalitis (n = 140, cattle = 39, goats = 31, sheep = 70) were defined as such when L. monocytogenes was isolated from the brainstem of animals with neurological signs and histopathologically confirmed rhombencephalitis6. In abortion cases (n = 33, cattle = 23, goats = 3, sheep = 7), L. monocytogenes was isolated from the placenta and/or the foetus. Gastroenteritis cases (sheep = 8) were defined as cases with diarrhoea and pathologically confirmed neutrophilic gastroenteritis65, in which L. monocytogenes was isolated from the gastrointestinal content. Mastitis cases (n = 6, cattle = 5, goats = 1) were defined by the isolation of L. monocytogenes from an udder quarter.
Environmental isolates
Based on sample size calculations for the detection of differential ST distribution between clinical and environmental isolates (Text. S1) a total of 61 isolates from the direct ruminant environment (n = 56) and faeces of healthy ruminants (n = 5) were included in this study. These isolates originated from cattle (n = 16) and small ruminant (n = 16) farms located in Switzerland (n = 27), Germany (n = 4) and Italy (n = 1), (Table S1).
Multilocus-sequence-typing (MLST)
MLST is based on the sequence determination of loci in the seven house-keeping genes abcZ, bglA, cat, dapE, dat, ldh and lhkA26,27. Each unique nucleotide sequence is assigned to an arbitrarily defined allele number, and the combination of allele numbers at the seven loci defines the allelic profile or ST. These data can be retrieved at the web-based MLST database from Institut Pasteur (http://bigsdb.web.pasteur.fr/listeria/).
To perform MLST, genomic DNA of all isolates was obtained with the guanidium thiocyanate-phenol-chloroform extraction method66. Hundred-twenty-two isolates were subtyped by conventional MLST as previously described27. Briefly, the seven housekeeping genes were amplified by PCR and the obtained PCR products were sequenced using universal sequencing primers26. Allelic profiles and corresponding STs were determined with the BioNumerics software (Version 7.1, Applied Maths, Austin, TX). With the availability of next generation sequencing technologies we moved on to in silico MLST following library preparation and genomic DNA sequencing using either an Illumina HiSeq 2000 or 3000 platform (193 isolates) or an Illumina MiSeq platform (35 isolates) at the Institute of Genetics, Vetsuisse Faculty, University of Bern. The obtained data were assembled using Velvet 1.2.10: algorithms for de novo short read assembly using Bruijn graphs67 and housekeeping gene allele numbers and the respective ST were determined with a specifically created workflow in Geneious R7 (http://www.geneious.com).
Finally, all MLST data were entered into the BioNumerics software in order to construct a minimum spanning tree (MST) and to identify CCs and evolutionary lineages. MSTs were produced using default settings. Bar plots with the most important CCs of ruminant isolates and of human clinical cases of Switzerland (n = 93) and France (n = 2304) from previous studies7,40 were constructed to compare their distribution. We used CCs for these barplots to facilitate comparisons with previous studies. The name of the CC corresponds to the name of the most important ST in this CC.
Multilocus variable number of tandem repeats analysis (MLVA)
Hundred-eighty-three L. monocytogenes isolates from ruminant rhombencephalitis cases (n = 179) and abortion cases (n = 4) had been subtyped by MLVA in a previous study23. Additional isolates from abortion cases (n = 20), mastitis cases (n = 5), gastroenteritis cases (n = 8), rhombencephalitis cases (n = 2) and one environmental isolate were submitted to MLVA according to the protocol described previously23. Briefly, five bacterial colonies grown on tryptic soy agar were lysed (Qiagen, Mericon DNA Bacteria Plus Kit No. 69534) and used for PCR. For each isolate, eight loci were analysed in four multiplex PCR reactions (Qiagen Multiplex PCR Kit No. 206143) combining two primer loci each. Primers were the same as in ref. 23. The size of the PCR products was determined with the Agilent DNA 1000 Kit in an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The number of repeats in each locus was determined according to Sperry et al.68. Missing products were registered as “zero alleles” to facilitate MLVA type and cluster analysis. MLVA data were uploaded into the BioNumerics software (Version 7.1, Applied Maths, Austin, TX). A MST was created from a total of 219 L. monocytogenes isolates to identify complexes that were defined to contain strains differing in a maximum of two MLVA loci. The MST was produced using a maximum number of N-locus variants (N = 1) with a weight of 10000. MLVA complexes were identified in the MLST-based MST and vice versa (Fig. S1).
Serotyping
For two isolates the molecular serovar group type was determined using the PCR based protocols as previously described69. For 226 isolates with available NGS data, the serovar group type was determined in silico using a specifically created workflow in Geneious R7 (http://www.geneious.com) with the primers of ref. 69.
Gentamicin protection assay
The BoMac cell line was cultured in Dulbecco’s modified eagle medium (DMEM; Life Technologies, Zug, Switzerland) supplemented with 10% fetal calf serum (FCS; Bioswisstec, Schaffhausen, Switzerland) and 100 U/ml penicillin/streptomycin (Life Technologies). Cells were seeded (3 × 105 cells/ml) and grown to confluency in 24-well plates in the above mentioned mediums, without antibiotics. Cells were then starved for 1 h in medium without FCS, followed by the infection with a panel of six representative L. monocytogenes cattle isolates at a MOI of 10: ST1, ST4 and ST412 isolated from rhombencephalitis; ST1, ST18 and ST37 isolated from environment. One hour post infection, the inoculum was removed, cells were washed with phosphate buffered saline (PBS) and covered with medium containing 10% FCS and 50 μg/ml gentamicin (Sigma-Aldrich) to inhibit extracellular growth of L. monocytogenes. At indicated time points post infection, cells were washed with PBS and lysed with 0.5% Triton-X100 (Sigma-Aldrich) to harvest CFUs. Serial dilutions were plated on brain heart infusion (BHI) agar and CFUs were quantified. For testing of invasiveness, CFUs were harvested at 2 h and for testing of intracellular replication, CFUs were harvested at 4 h, 8 h and 24 h post infection46. Three independent experiments were performed in triplicates, and results were normalized to the inoculum.
Virulence-associated gene analysis
The nucleotide and amino acid sequences of 45 previously described virulence factors known to be involved in the virulence of L. monocytogenes39 were compared between 94 isolates with NGS data of sufficient quality (Table S1). The 94 isolates belonged to 25 STs and included 35 rhombencephalitis isolates (cattle = 15; small ruminants = 20), seven clinical non-encephalitic isolates (cattle = 5; small ruminants = 2) and 52 environmental isolates (cattle = 33; small ruminants = 19). The data was analyzed for mutations associated with attenuated virulence described in previous studies23,26,41. Using the sequences of the 45 virulence genes from EGD-e (GenBank accession NC_003210), the corresponding genes were extracted from our 94 genome sequences after a MAFFT v7. 222 alignment70. This was then followed by the construction of neighbour-joining trees based on the nucleotide level with synonymous and non-synonymous mutations and on the amino acid level with Geneious R7 (http://www.geneious.com). Additionally, neighbour-joining trees were built based on the concatenated nucleotide and amino acid sequences of all virulence factors following a MAFFT v7. 222 alignment. Recombination events in the 45 virulence genes were computed using the ClonalFrame v1.147 and ML71 as described in ref. 72 in five individual runs. The mean of the five runs was calculated for each virulence gene, and the 95% CI of these means was determined.
Statistical analysis
The R software (R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/) was used to compare the frequency of different STs in different groups of isolates with the Fisher’s exact test. As not all variables were normally distributed using the Shapiro-Wilk test, the non-parametric Kruskal-Wallis test and the Dunn’s multiple comparison post-hoc tests were used to assess significant differences in CFU counts. Differences between groups with a p-value < 0.05 were considered to be statistically significant. Genetic diversity of L. monocytogenes was estimated by the Simpson index of diversity73, which calculates the probability that two isolates belong to different STs and the 95% CI of the analyzed groups was determined74.
Accession numbers
The sequencing data of the 94 isolates used in the virulence associated gene analysis were submitted to the ENA (European Nucleotide Archive) database (Data Set. S1).
Additional Information
How to cite this article: Dreyer, M. et al. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 6, 36419; doi: 10.1038/srep36419 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the farmers for giving us the opportunity to collect samples after obtaining previous information on the study aims. We acknowledge Andreas Thomann and Amandine Ruffieux of the Institute of Veterinary Bacteriology, Bern for useful discussions and help with the laboratory work, as well as the Institute of Genetics, Bern for producing NGS data. We thank Martin Ganter of the University of Veterinary Medicine, Hannover for collecting environmental samples of small ruminant farms in Germany. This work was financially supported by the Swiss National Foundation SNF Sinergia grant CRSII3_147692. The funders had no role in the study design, data collection and interpretation or in the decision to submit this work for publication. Anna Oevermann’s professorship for comparative neuropathology is funded by the Ernst-Frauchiger foundation.
Footnotes
Author Contributions A.O. and J.F. supervised and conceived the project; M.D. collected the environmental and faecal isolates, performed in vitro analyses; M.D. performed MLST typing, serotyping, phylogenetic and virulence gene analyses with support of L.A.-B. and S.R.; J. F., R.S., A.S., A.O. and A.O. provided well characterized clinical isolates; C.G. typed clinical non-encephalitic isolates with MLVA; S.B. and M.L. provided the MLST typing of a portion of strains; S.R. created the Geneious MLST and serotype workflow; M.D. and G.S. produced statistical analyses; M.D. and A.O. wrote the manuscript with the contribution of all co-authors.
References
- Seeliger H. P. R. Listeriosis. Karger, New York, N.Y. (1961). [Google Scholar]
- Farber J. M. & Peterkin P. I. Listeria monocytogenes, a food-borne pathogen. Microbiol.Rev. 55, 476–511 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J. C. & Donachie W. A review of Listeria monocytogenes and listeriosis. Vet.J. 153, 9–29 (1997). [DOI] [PubMed] [Google Scholar]
- Siegman-Igra Y. et al. Listeria monocytogenes infection in Israel and review of cases worldwide. Emerg.Infect.Dis. 8, 305–310 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets D. A. & Bronze M. S. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol.Med.Microbiol. 53, 151–165 (2008). [DOI] [PubMed] [Google Scholar]
- Oevermann A. et al. Neuropathogenesis of naturally occurring encephalitis caused by Listeria monocytogenes in ruminants. Brain Pathol. 20, 378–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury M. M. et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nature genetics 48, 308–313, doi: 10.1038/ng.3501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland J. A. et al. Listeria pathogenesis and molecular virulence determinants. Clin.Microbiol.Rev. 14, 584–640 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan B. & Gerner-Smidt P. The epidemiology of human listeriosis. Microbes.Infect. 9, 1236–1243 (2007). [DOI] [PubMed] [Google Scholar]
- Fox E. et al. Listeria monocytogenes in the Irish dairy farm environment. Journal of food protection 72, 1450–1456 (2009). [DOI] [PubMed] [Google Scholar]
- Fox E., Hunt K., O’Brien M. & Jordan K. Listeria monocytogenes in Irish Farmhouse cheese processing environments. International journal of food microbiology 145, Suppl 1, S39–S45, 10.1016/j.ijfoodmicro.2010.10.012 (2011). [DOI] [PubMed] [Google Scholar]
- Vivant A. L., Garmyn D. & Piveteau P. Listeria monocytogenes, a down-to-earth pathogen. Frontiers in cellular and infection microbiology 3, 87, doi: 10.3389/fcimb.2013.00087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger H. P. R. & Jones D. Listeria. In Bergey’s Manual of Systematic Bacteriology. Edited bySneath P. H. A., Nair N. S., Sharpe N. E. & Holt. J. G.Baltimore: Williams and Wilkins. 2, 1235–1245 (1986).
- Orsi R. H., den Bakker H. C. & Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. International journal of medical microbiology: IJMM 301, 79–96, doi: 10.1016/j.ijmm.2010.05.002 (2011). [DOI] [PubMed] [Google Scholar]
- Rooney A. P. & Ward T. J. Birth-and-death evolution of the internalin multigene family in Listeria. Gene 427, 124–128, doi: 10.1016/j.gene.2008.09.007 (2008). [DOI] [PubMed] [Google Scholar]
- Wiedmann M. et al. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect.Immun. 65, 2707–2716 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffaretti J. C. et al. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proceedings of the National Academy of Sciences of the United States of America 86, 3818–3822 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE, World Organization for Animal Health. Listeria monocytogenes. In: Manual of diagnostic tests and vaccines for terrestrial animals. ed. OIE, 7th ed. OIE, Paris, France. (2008).
- Jeffers G. T. et al. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147, 1095–1104 (2001). [DOI] [PubMed] [Google Scholar]
- Chatterjee S. S. et al. Invasiveness is a variable and heterogeneous phenotype in Listeria monocytogenes serotype strains. Int.J.Med.Microbiol. 296, 277–286 (2006). [DOI] [PubMed] [Google Scholar]
- Pohl M. A., Wiedmann M. & Nightingale K. K. Associations among Listeria monocytogenes genotypes and distinct clinical manifestations of listeriosis in cattle. Am.J.Vet.Res. 67, 616–626 (2006). [DOI] [PubMed] [Google Scholar]
- Tamburro M. et al. Evaluation of transcription levels of inlA, inlB, hly, bsh and prfA genes in Listeria monocytogenes strains using quantitative reverse-transcription PCR and ability of invasion into human CaCo-2 cells. FEMS microbiology letters 362, doi: 10.1093/femsle/fnv018 (2015). [DOI] [PubMed] [Google Scholar]
- Balandyte L., Brodard I., Frey J., Oevermann A. & Abril C. Ruminant rhombencephalitis-associated Listeria monocytogenes alleles linked to a multilocus variable-number tandem-repeat analysis complex. Appl.Environ.Microbiol. 77, 8325–8335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. J. et al. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl.Environ.Microbiol. 70, 5833–5841 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J. K. et al. The ubiquitous nature of Listeria monocytogenes clones: a large-scale Multilocus Sequence Typing study. Environmental microbiology 16, 405–416, doi: 10.1111/1462-2920.12342 (2014). [DOI] [PubMed] [Google Scholar]
- Ragon M. et al. A new perspective on Listeria monocytogenes evolution. PLoS.Pathog. 4, e1000146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo C., Arreaza L., Alcala B., de la Fuente L. & Vazquez J. A. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. Journal of clinical microbiology 41, 757–762 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegraaff R., Borland N. A. & Browning J. W. An outbreak of listerial meningo-encephalitis in sheep. Aust.Vet.J. 57, 94–96 (1981). [DOI] [PubMed] [Google Scholar]
- Wilesmith J. W. & Gitter M. Epidemiology of ovine listeriosis in Great Britain. The Veterinary record 119, 467–470 (1986). [DOI] [PubMed] [Google Scholar]
- Jeffrey M. A neuropathological survey of brains submitted under the Bovine Spongiform Encephalopathy Orders in Scotland. Vet.Rec. 131, 332–337 (1992). [DOI] [PubMed] [Google Scholar]
- Oevermann A. et al. Neuropathological survey of fallen stock: Active surveillance reveals high prevalence of encephalitic listeriosis in small ruminants. Vet.Microbiol. 130, 320–329 (2008). [DOI] [PubMed] [Google Scholar]
- Low J. C. & Renton C. P. Septicaemia, encephalitis and abortions in a housed flock of sheep caused by Listeria monocytogenes type 1/2. Vet.Rec. 116, 147–150 (1985). [DOI] [PubMed] [Google Scholar]
- Schoder D., Winter P., Kareem A., Baumgartner W. & Wagner M. A case of sporadic ovine mastitis caused by Listeria monocytogenes and its effect on contamination of raw milk and raw-milk cheeses produced in the on-farm dairy. J.Dairy Res. 70, 395–401 (2003). [DOI] [PubMed] [Google Scholar]
- Winter P. et al. Clinical and histopathological aspects of naturally occurring mastitis caused by Listeria monocytogenes in cattle and ewes. J.Vet.Med.B Infect.Dis.Vet.Public Health. 51, 176–179 (2004). [DOI] [PubMed] [Google Scholar]
- Dreyer M., Thomann A., Bottcher S., Frey J. & Oevermann A. Outbreak investigation identifies a single Listeria monocytogenes strain in sheep with different clinical manifestations, soil and water. Veterinary microbiology 179, 69–75, doi: 10.1016/j.vetmic.2015.01.025 (2015). [DOI] [PubMed] [Google Scholar]
- Skovgaard N. & Morgen C. A. Detection of Listeria spp. in faeces from animals, in feeds, and in raw foods of animal origin. Int.J.Food Microbiol. 6, 229–242 (1988). [DOI] [PubMed] [Google Scholar]
- Sammarco M. L., Ripabelli G., Fanelli I. & Grasso G. M. Prevalence of Listeria spp. in dairy farm and evaluation of antibiotic-resistance of isolates. Annali di igiene: medicina preventiva e di comunita 17, 175–183 (2005). [PubMed] [Google Scholar]
- Ho A. J., Ivanek R., Grohn Y. T., Nightingale K. K. & Wiedmann M. Listeria monocytogenes fecal shedding in dairy cattle shows high levels of day-to-day variation and includes outbreaks and sporadic cases of shedding of specific L. monocytogenes subtypes. Prev.Vet.Med. 80, 287–305 (2007). [DOI] [PubMed] [Google Scholar]
- Camejo A. et al. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2, 379–394, doi: 10.4161/viru.2.5.17703 (2011). [DOI] [PubMed] [Google Scholar]
- Althaus D. et al. Characterization of Listeria monocytogenes strains isolated during 2011-2013 from human infections in Switzerland. Foodborne pathogens and disease 11, 753–758, doi: 10.1089/fpd.2014.1747 (2014). [DOI] [PubMed] [Google Scholar]
- Roche S. M. et al. Investigation of specific substitutions in virulence genes characterizing phenotypic groups of low-virulence field strains of Listeria monocytogenes. Appl.Environ.Microbiol. 71, 6039–6048 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonquieres R., Bierne H., Mengaud J. & Cossart P. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect.Immun. 66, 3420–3422 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K. K., Windham K., Martin K. E., Yeung M. & Wiedmann M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl.Environ.Microbiol. 71, 8764–8772 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K. K. et al. inlA premature stop codons are common among Listeria monocytogenes isolates from foods and yield virulence-attenuated strains that confer protection against fully virulent strains. Appl.Environ.Microbiol. 74, 6570–6583 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet C. et al. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J.Infect.Dis 189, 2094–2100 (2004). [DOI] [PubMed] [Google Scholar]
- Rupp S. et al. A naturally occurring prfA truncation in a Listeria monocytogenes field strain contributes to reduced replication and cell-to-cell spread. Veterinary microbiology 179, 91–101, doi: 10.1016/j.vetmic.2015.03.002 (2015). [DOI] [PubMed] [Google Scholar]
- Didelot X. & Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics 175, 1251–1266, doi: 10.1534/genetics.106.063305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. et al. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152, 685–693, doi: 10.1099/mic.0.28503-0 (2006). [DOI] [PubMed] [Google Scholar]
- Linke K. et al. Reservoirs of listeria species in three environmental ecosystems. Applied and environmental microbiology 80, 5583–5592, doi: 10.1128/AEM.01018-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckerl I. et al. L. monocytogenes in a cheese processing facility: Learning from contamination scenarios over three years of sampling. International journal of food microbiology 189, 98–105, doi: 10.1016/j.ijfoodmicro.2014.08.001 (2014). [DOI] [PubMed] [Google Scholar]
- Lecuit M. et al. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madarame H. et al. The distribution of E-cadherin expression in listeric rhombencephalitis of ruminants indicates its involvement in Listeria monocytogenes neuroinvasion. Neuropathol.Appl.Neurobiol. (2011). [DOI] [PubMed] [Google Scholar]
- Koopmans M. M. et al. Listeria monocytogenes sequence type 6 and increased rate of unfavorable outcome in meningitis: epidemiologic cohort study. Clin Infect Dis 57, 247–253, doi: 10.1093/cid/cit250 (2013). [DOI] [PubMed] [Google Scholar]
- Otter A. & Blakemore W. F. Observation on the presence of Listeria monocytogenes in axons. Acta Microbiol.Hung. 36, 125–131 (1989). [PubMed] [Google Scholar]
- Mylonakis E., Hohmann E. L. & Calderwood S. B. Central nervous system infection with Listeria monocytogenes. 33 years’ experience at a general hospital and review of 776 episodes from the literature. Medicine (Baltimore). 77, 313–336 (1998). [DOI] [PubMed] [Google Scholar]
- Bartt R. Listeria and atypical presentations of Listeria in the central nervous system. Semin.Neurol. 20, 361–373 (2000). [DOI] [PubMed] [Google Scholar]
- Armstrong R. W. & Fung P. C. Brainstem encephalitis (rhombencephalitis) due to Listeria monocytogenes: case report and review. Clin.Infect.Dis. 16, 689–702 (1993). [DOI] [PubMed] [Google Scholar]
- Pollock S. S., Pollock T. M. & Harrison M. J. Infection of the central nervous system by Listeria monocytogenes: a review of 54 adult and juvenile cases. Q.J.Med. 53, 331–340 (1984). [PubMed] [Google Scholar]
- Di Palma S. et al. Comparative spatiotemporal analysis of the intrathecal immune response in natural listeric rhombencephalitis of cattle and small ruminants. Comp Immunol.Microbiol.Infect.Dis. (2012). [DOI] [PubMed] [Google Scholar]
- Jungi T. W. et al. Comparison of inducible nitric oxide synthase expression in the brains of Listeria monocytogenes-infected cattle, sheep, and goats and in macrophages stimulated in vitro. Infect.Immun. 65, 5279–5288 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoder D. et al. Prevalence of major foodborne pathogens in food confiscated from air passenger luggage. International journal of food microbiology 209, 3–12, doi: 10.1016/j.ijfoodmicro.2014.08.010 (2015). [DOI] [PubMed] [Google Scholar]
- Yde M. et al. Usefulness of the European Epidemic Intelligence Information System in the management of an outbreak of listeriosis, Belgium, 2011. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 17 (2012). [PubMed] [Google Scholar]
- Martin B. et al. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food microbiology 44, 119–127, doi: 10.1016/j.fm.2014.05.014 (2014). [DOI] [PubMed] [Google Scholar]
- Parisi A. et al. Amplified Fragment Length Polymorphism and Multi-Locus Sequence Typing for high-resolution genotyping of Listeria monocytogenes from foods and the environment. Food microbiology 27, 101–108, doi: 10.1016/j.fm.2009.09.001 (2010). [DOI] [PubMed] [Google Scholar]
- Fairley R. A., Pesavento P. A. & Clark R. G. Listeria monocytogenes infection of the alimentary tract (enteric listeriosis) of sheep in New Zealand. J.Comp Pathol. 146, 308–313 (2012). [DOI] [PubMed] [Google Scholar]
- Sambrook J. & Russel D. W. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press (2001). [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome research 18, 821–829, doi: 10.1101/gr.074492.107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry K. E., Kathariou S., Edwards J. S. & Wolf L. A. Multiple-locus variable-number tandem-repeat analysis as a tool for subtyping Listeria monocytogenes strains. J.Clin.Microbiol. 46, 1435–1450 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doumith M., Buchrieser C., Glaser P., Jacquet C. & Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J.Clin.Microbiol. 42, 3819–3822 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. & Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic acids research 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X. & Wilson D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS computational biology 11, e1004041, doi: 10.1371/journal.pcbi.1004041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker H. C., Didelot X., Fortes E. D., Nightingale K. K. & Wiedmann M. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC evolutionary biology 8, 277, doi: 10.1186/1471-2148-8-277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. R. & Gaston M. A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. Journal of clinical microbiology 26, 2465–2466 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H., Hori S. & Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. Journal of clinical microbiology 39, 4190–4192, doi: 10.1128/JCM.39.11.4190-4192.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.